PlTem1, a Key Cell Cycle Regulator, Serves as an Important Bridge Between Cell Division and Autophagy in Peronophythora litchii
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Pathogenicity Assays
2.3. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.4. Bioinformatic Analyses
2.5. Targeted Deletion and Complementation of PlTEM1
2.6. Staining and Microscopic Observation
2.7. Yeast-Two-Hybrid (Y2H) Assay
2.8. Statistical Analyses
3. Results
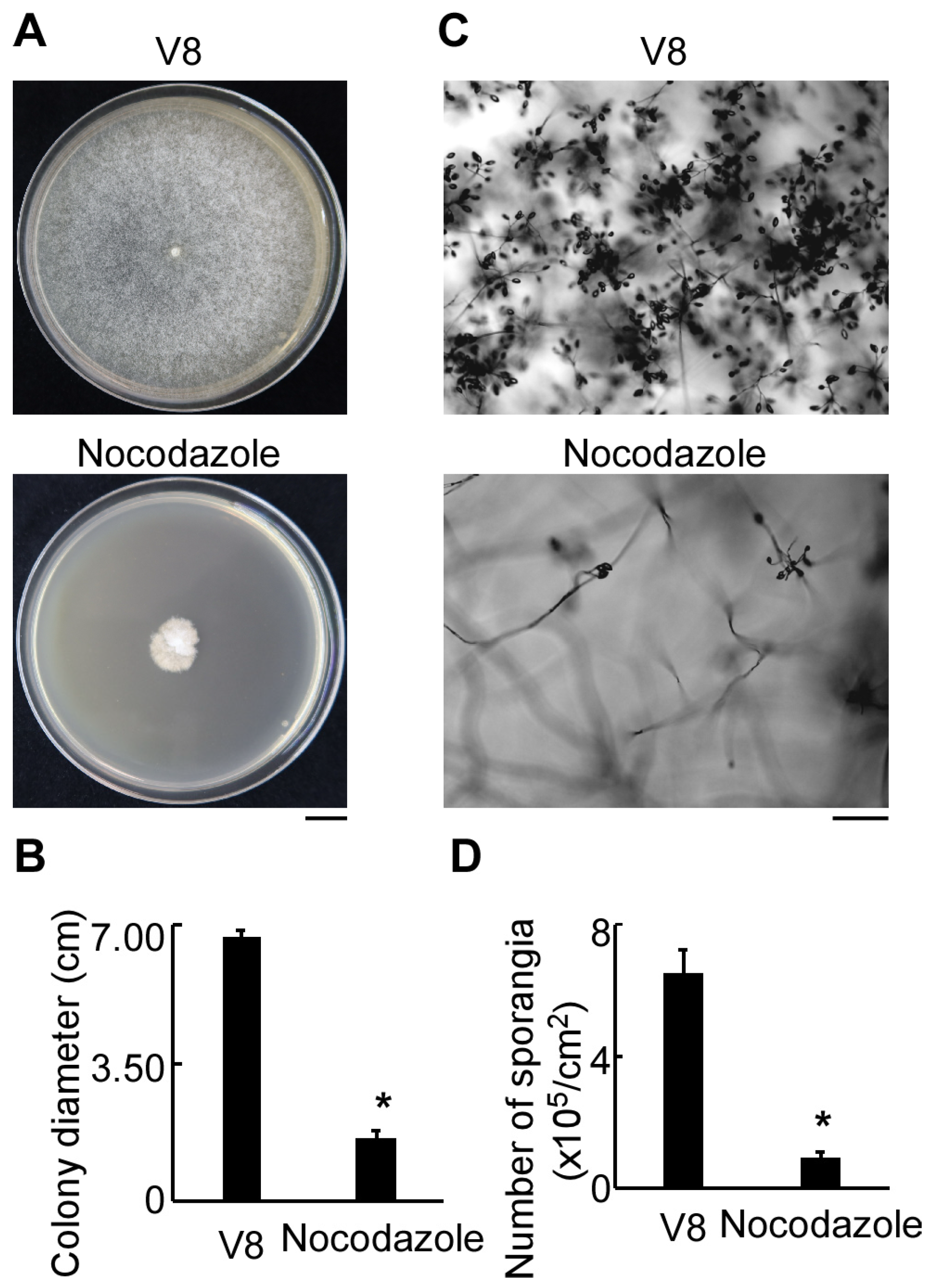
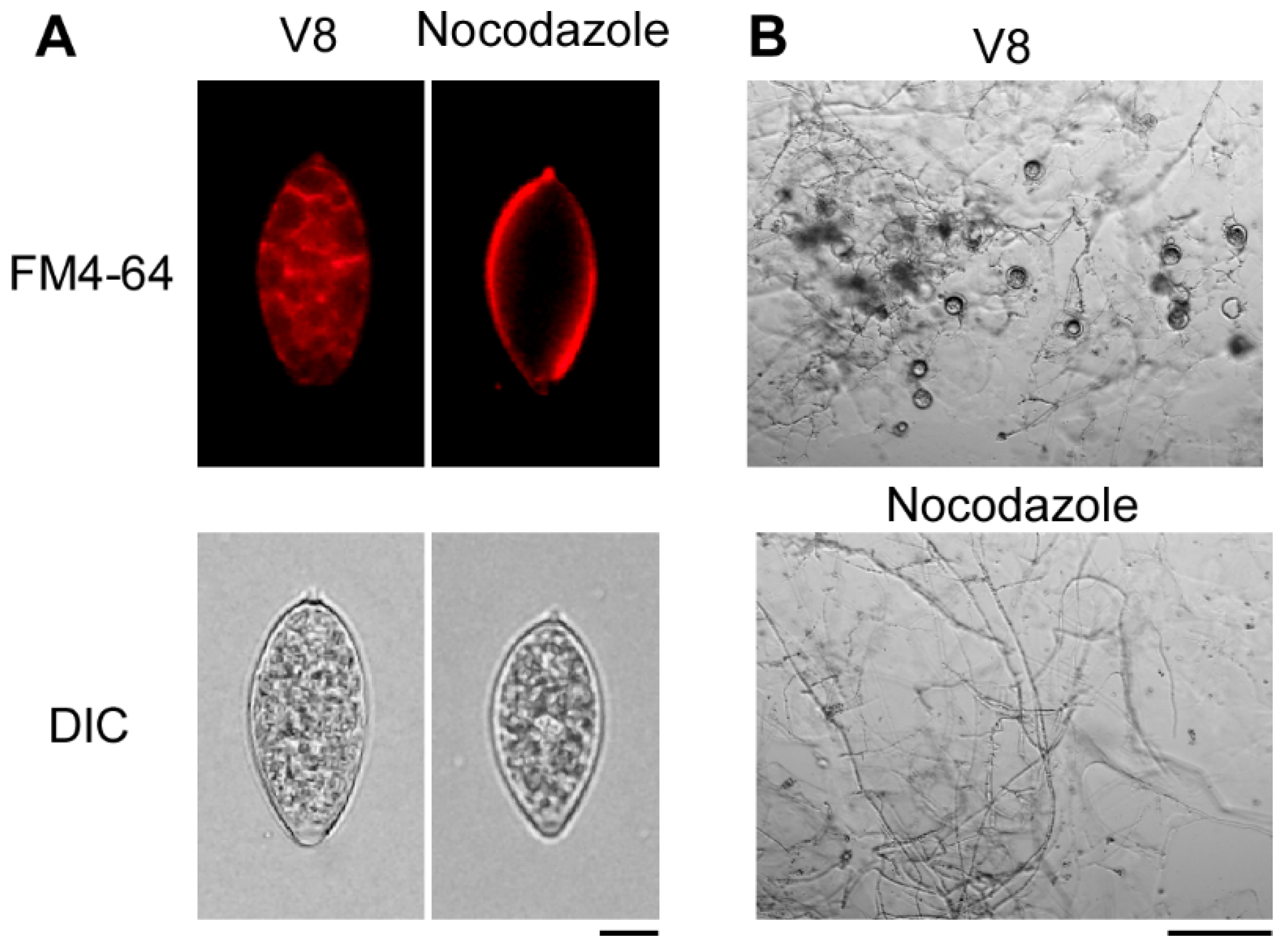
3.1. Cell Cycle Inhibition Impaired Vegetative Growth, Sporangia Production, Zoospore Release, and Oospore Formation in P. litchii
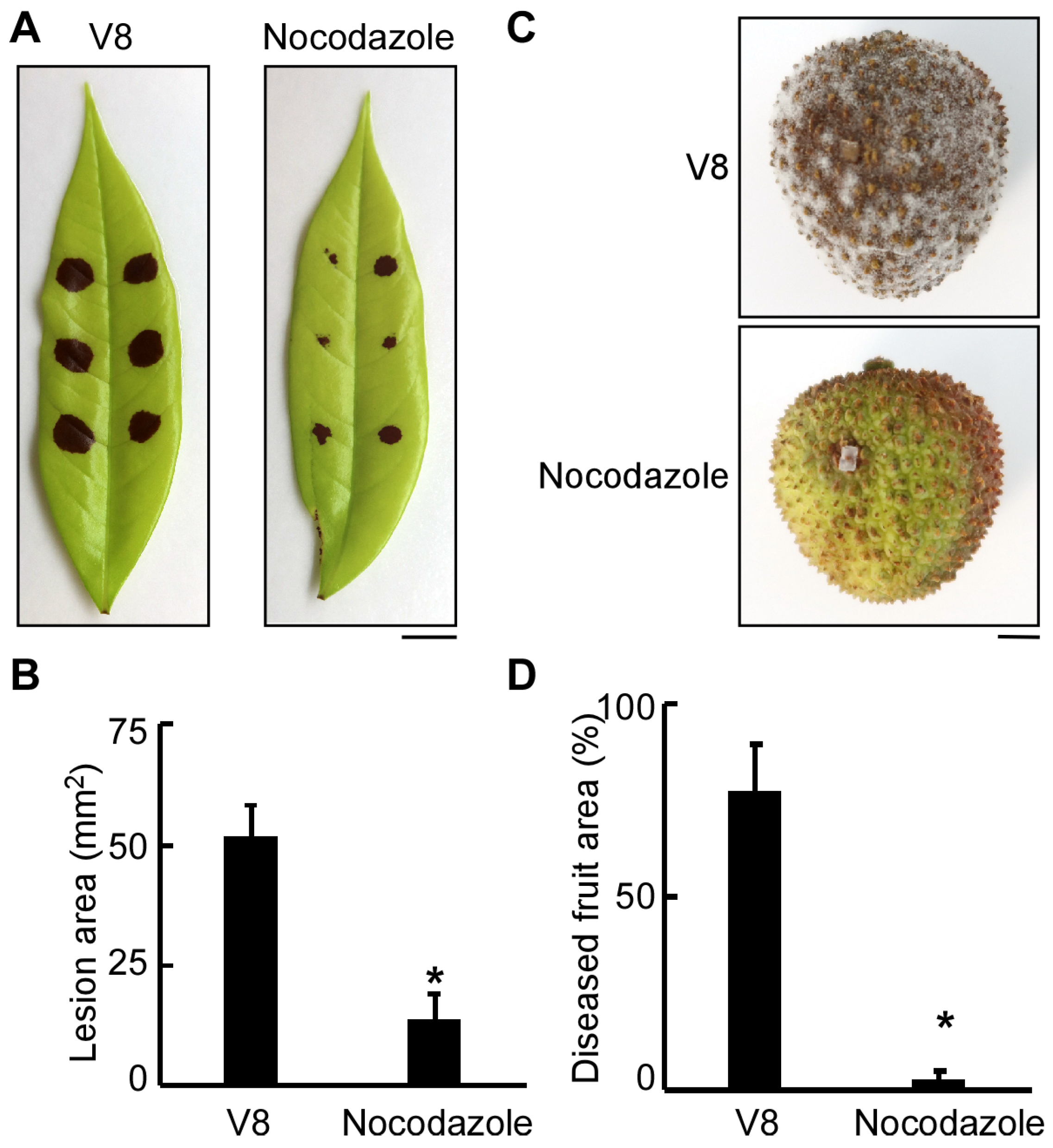
3.2. Normal Progression of Cell Division Is Essential for the Pathogenicity of P. litchii
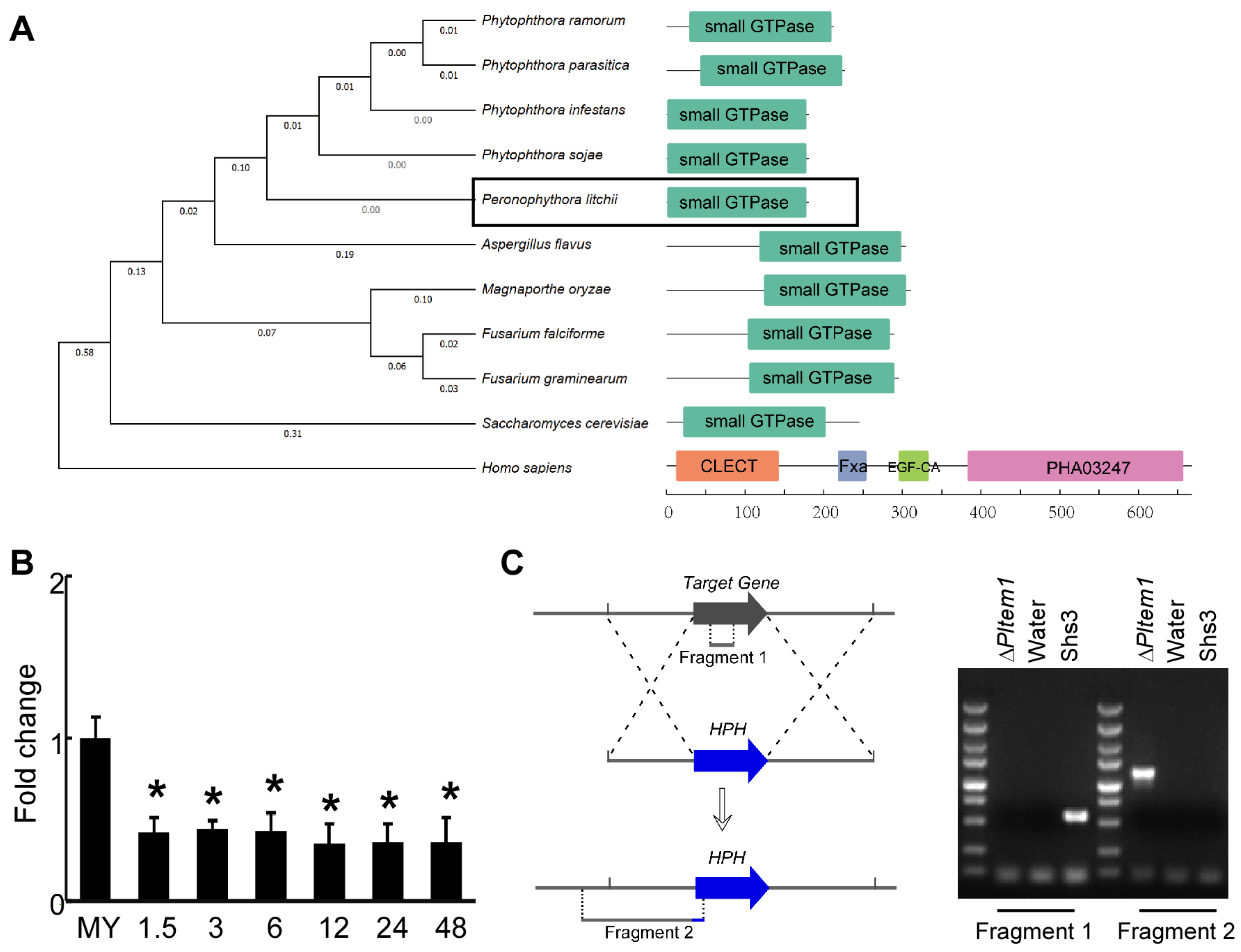
3.3. Characterization of Small GTPase PlTem1 in P. litchii
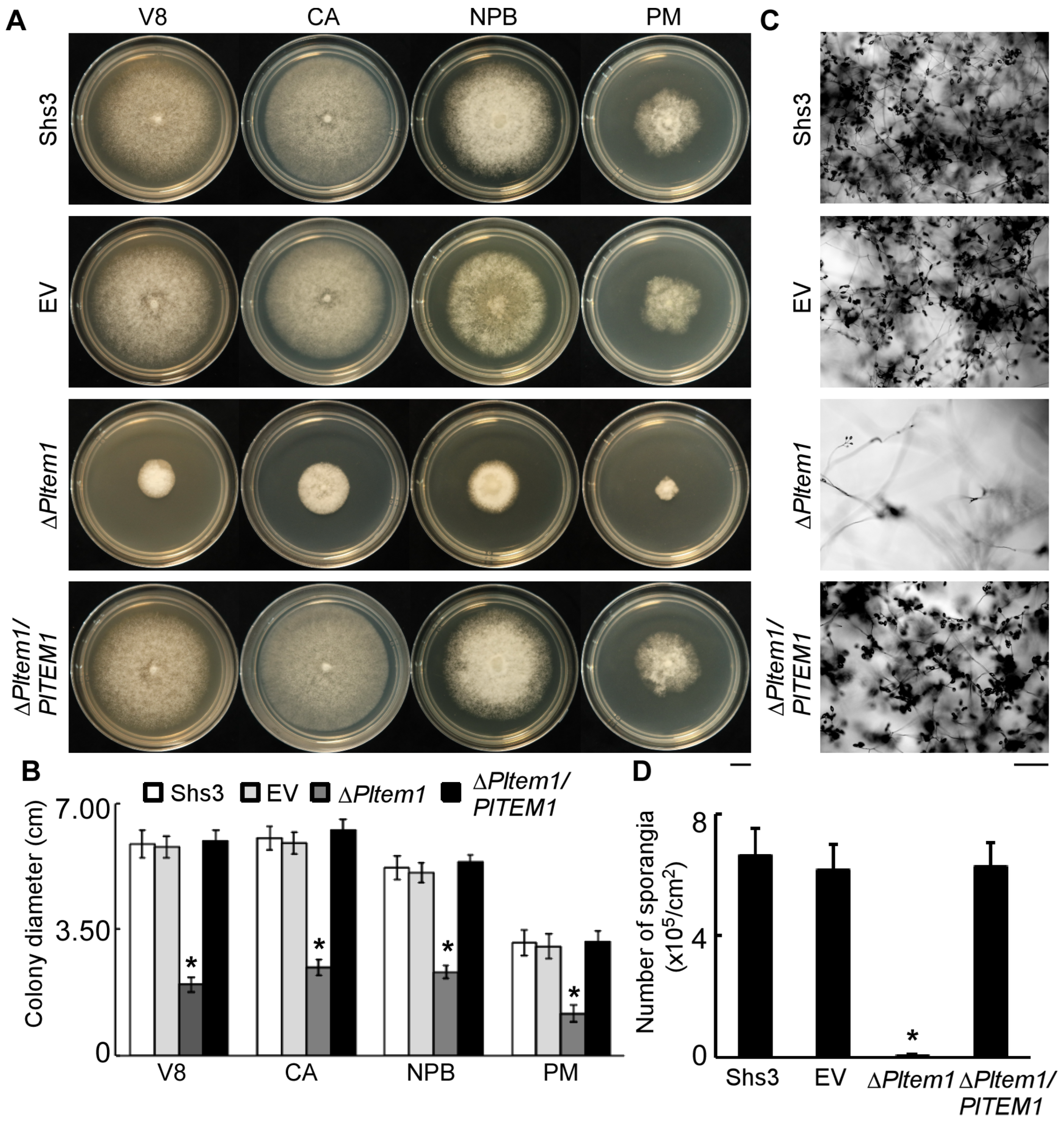
3.4. Tem1 Plays an Important Role in the Vegetative Hyphae and Sporangia of P. litchii
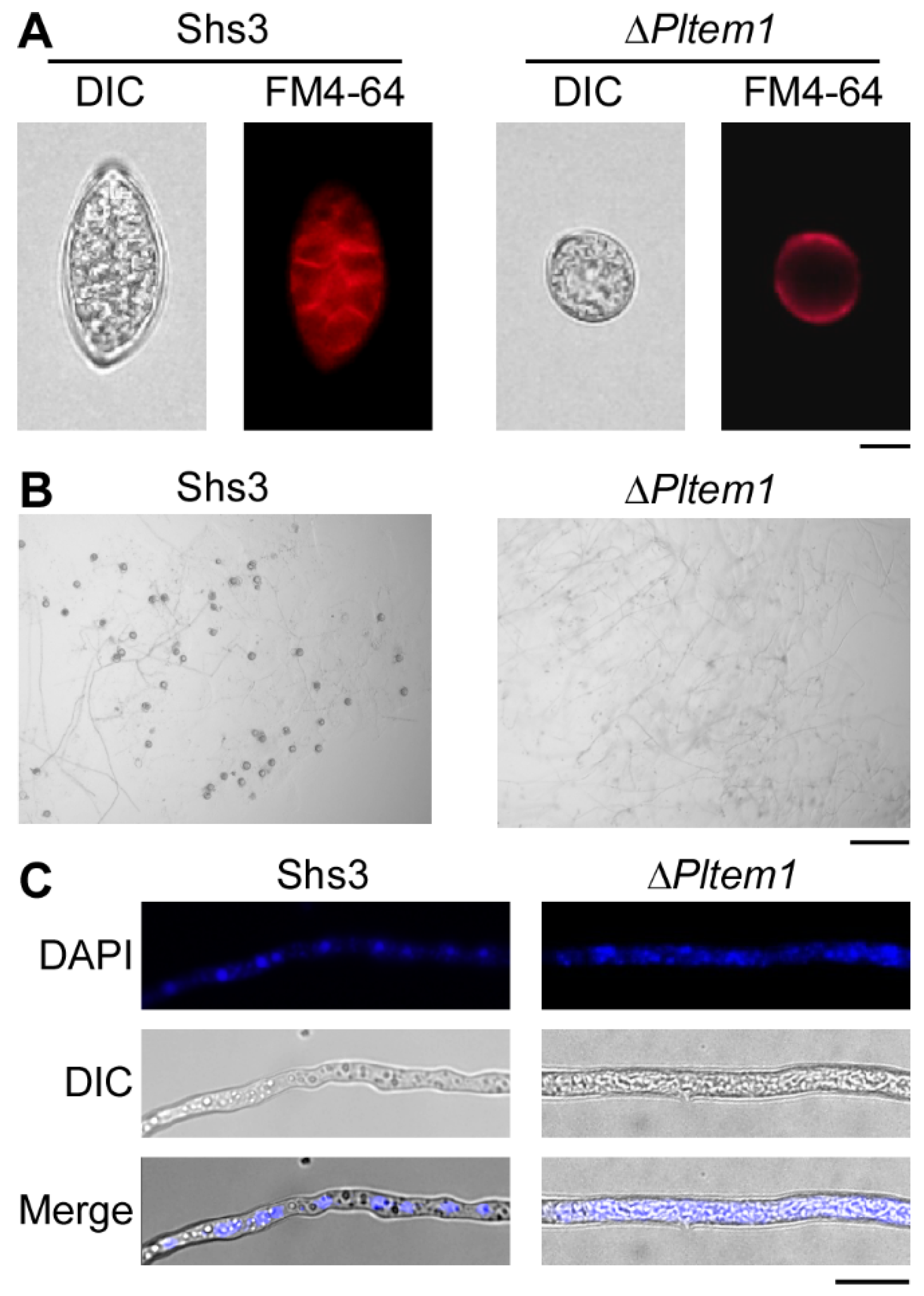
3.5. The Deletion of PlTEM1 Inhibits P. litchii to Produce Zoospores and Oospores, and Its Nuclei Cannot Divide with Abnormal Morphology
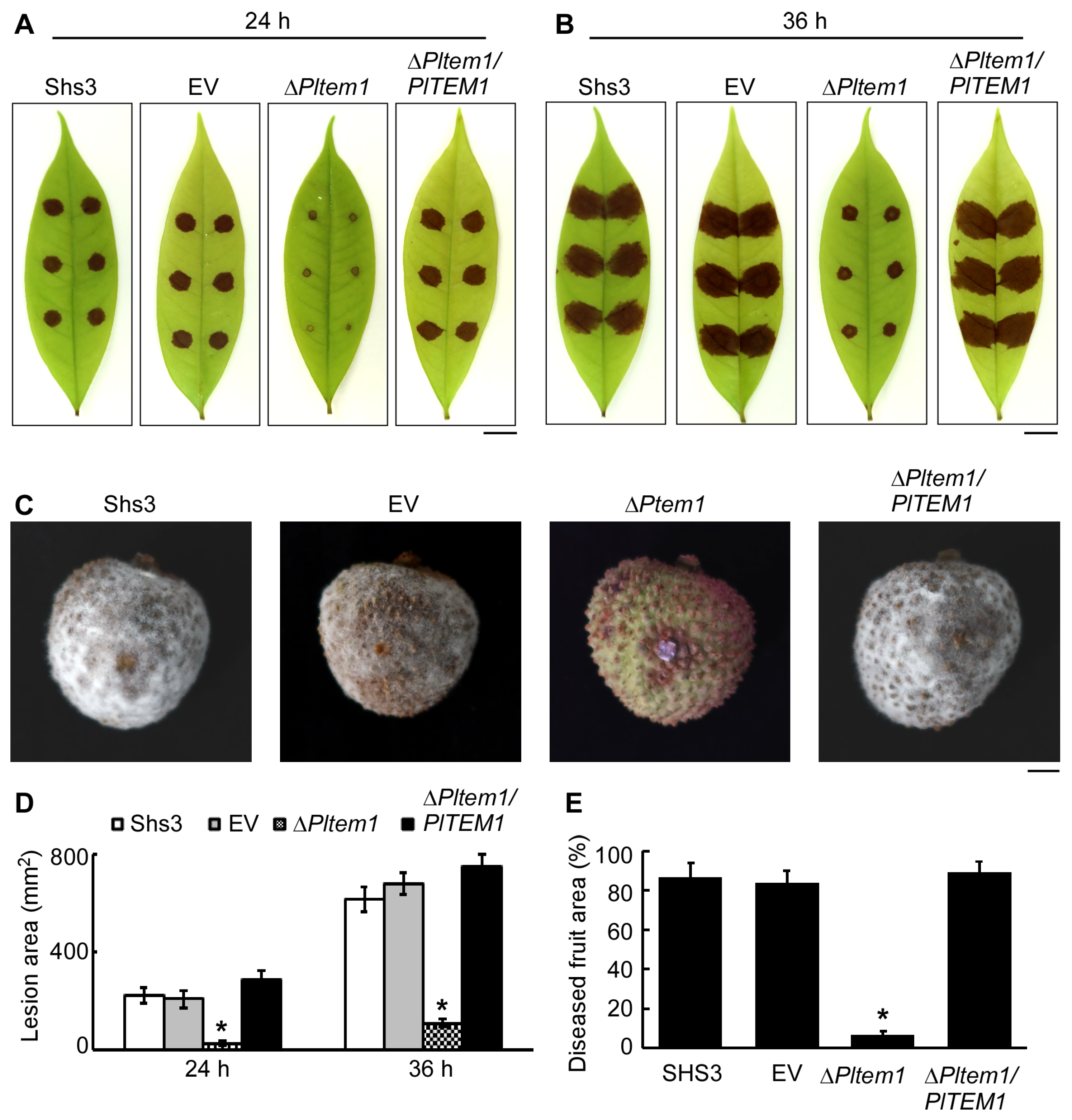
3.6. PlTem1 Is Essential for P. litchii Pathogenicity
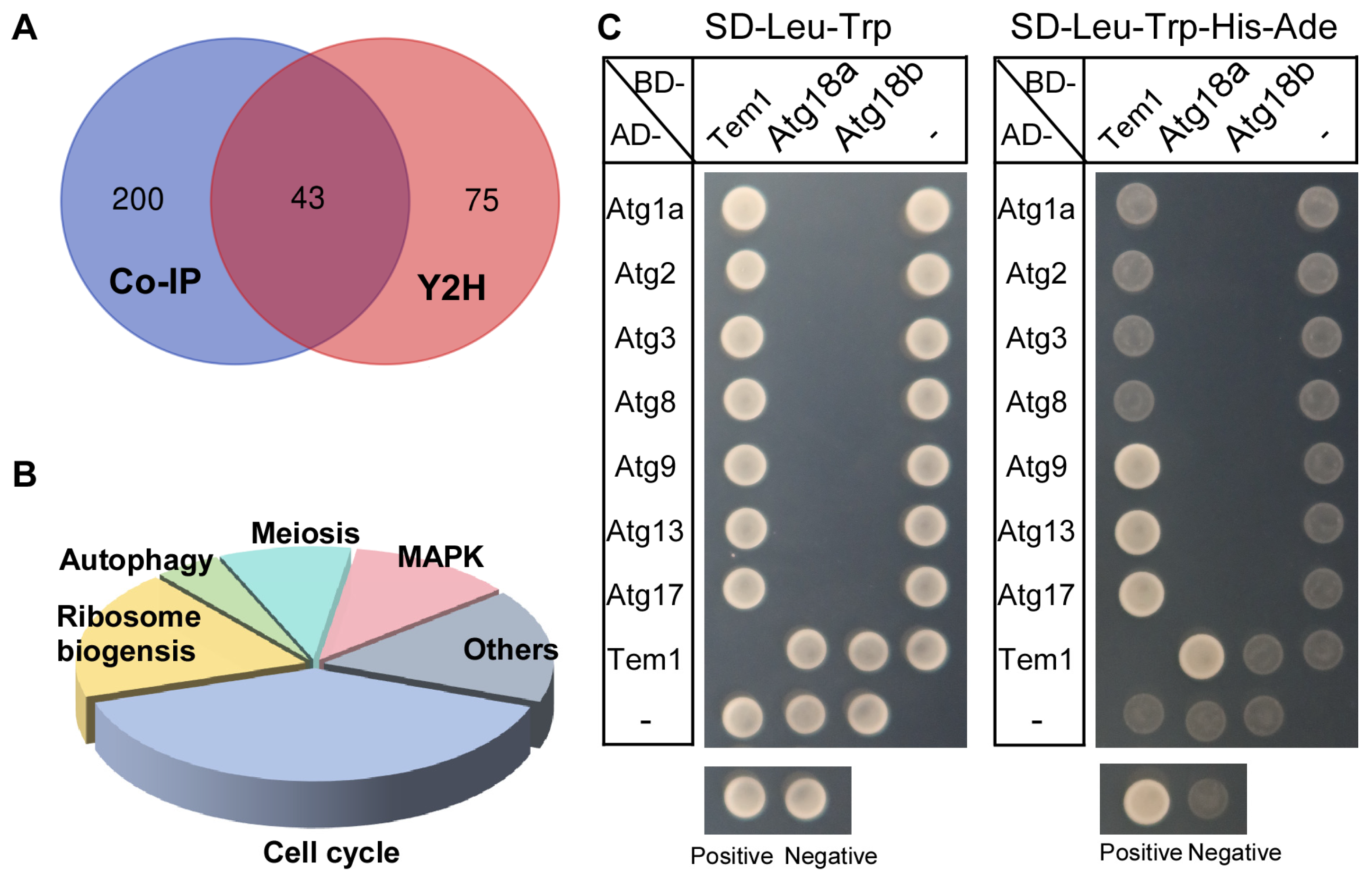
3.7. PlTem1 Interacts with Multiple Autophagy-Related Proteins
4. Discussion
4.1. The Cell Cycle Is Necessary for the Growth and Pathogenicity of Fungi and Oomycetes
4.2. Tem1 Plays a Conserved Role in Regulating the Virulence of Multiple Pathogens
4.3. PlTem1 Has the Potential to Be an Important Bridge Between Autophagy and Cell Division in P. litchii
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jiang, Y.; Wang, Y.; Song, L.; Liu, H.; Lichter, A.; Kerdchoechuen, O.; Joyce, D.C.; Shi, J. Postharvest characteristics and handling of litchi fruit. Aust. J. Exp. Agric. 2006, 46, 1541–1556. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, K.; Wang, K.; Zhu, J.; Hu, Z. Nutrient components, health benefits, and safety of litchi (Litchi chinensis Sonn.): A review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2139–2163. [Google Scholar] [CrossRef] [PubMed]
- Chi, P.K.; Pang, X.P.; Liu, R. On downy blight of Litchi chinensis (Sonn. I). Pathog. Its Infect. Process. Acta Phytopathol. Sin. 1984, 2, 113–119. [Google Scholar]
- Wang, H.; Sun, H.; Ma, J.; Stammler, G.; Zhou, M. Fungicide effectiveness during the various developmental stages of Peronophythora litchii in vitro. J. Phytopathol. 2009, 157, 407–412. [Google Scholar] [CrossRef]
- Jing, G.; Huang, H.; Yang, B.; Li, J.; Zheng, X.; Jiang, Y. Effect of pyrogallol on the physiology and biochemistry of litchi fruit during storage. Chem. Cent. J. 2013, 7, 19–29. [Google Scholar] [CrossRef]
- Zheng, L.; Situ, J.; Zhu, Q.; Xi, P.; Zheng, Y.; Liu, H.; Zhou, X.; Jiang, Z. Identification of volatile organic compounds for the biocontrol of postharvest litchi fruit pathogen Peronophythora litchii. Postharvest Biol. Technol. 2019, 155, 37–46. [Google Scholar] [CrossRef]
- Fang, Y.; Tyler, B.M. Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 2016, 17, 127–139. [Google Scholar] [CrossRef]
- Ye, W.; Wang, Y.; Shen, D.; Li, D.; Pu, T.; Jiang, Z.; Zhang, Z.; Zheng, X.; Tyler, B.; Wang, Y. Sequencing of the litchi downy blight pathogen reveals it is a Phytophthora species with downy mildew-like characteristics. Mol. Plant-Microbe Interact. 2016, 29, 573–583. [Google Scholar] [CrossRef]
- Fang, Y.; Cui, L.; Gu, B.; Arredondo, F.; Tyler, B.M. Efficient genome editing in the oomycete Phytophthora sojae using CRISPR/cas9. Curr. Protoc. Microbiol. 2017, 44, 21A.1.1–21A.1.26. [Google Scholar] [CrossRef]
- Situ, J.; Jiang, L.; Shao, Y.; Kong, G.; Xi, P.; Jiang, Z. Establishment of CRISPR/Cas9 genome editing system in Peronophythora litchii. J. Fungal Res. 2020, 18, 181–188. [Google Scholar]
- Guo, H.; Bao, J.; Lin, L.; Wang, Z.; Shi, M.; Huang, Y.; Wang, R.; Li, B.; Liu, P.; Chen, Q. Genome sequence data of Peronophythora litchii, an oomycete pathogen causing litchi downy blight. Mol. Plant-Microbe Interact. 2021, 34, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001, 2, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Stegmeier, F.; Amon, A. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004, 38, 203–232. [Google Scholar] [CrossRef] [PubMed]
- Hotz, M.; Barral, Y. The mitotic exit network: New turns on old pathways. Trends Cell Biol. 2014, 24, 145–152. [Google Scholar] [CrossRef]
- Scarfone, I.; Piatti, S. Coupling spindle position with mitotic exit in budding yeast: The multifaceted role of the small GTPase Tem1. Small GTPases 2015, 6, 196–201. [Google Scholar] [CrossRef]
- Rock, J.M.; Amon, A. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev. 2011, 25, 1943–1954. [Google Scholar] [CrossRef]
- Musacchio, A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef]
- Stukenberg, P.T.; Burke, D.J. Connecting the microtubule attachment status of each kinetochore to cell cycle arrest through the spindle assembly checkpoint. Chromosoma 2015, 124, 463–480. [Google Scholar] [CrossRef]
- Fukada, F.; Kodama, S.; Nishiuchi, T.; Kajikawa, N.; Kubo, Y. Plant pathogenic fungi Colletotrichum and Magnaporthe share a common G1 phase monitoring strategy for proper appressorium development. New Phytol. 2019, 222, 1909–1923. [Google Scholar] [CrossRef]
- Feng, W.; Yin, Z.; Wu, H.; Liu, P.; Liu, X.; Liu, M.; Yu, R.; Gao, C.; Zhang, H.; Zheng, X.; et al. Balancing of the mitotic exit network and cell wall integrity signaling governs the development and pathogenicity in Magnaporthe oryzae. PLoS Pathogens 2021, 17, e1009080. [Google Scholar] [CrossRef]
- Feng, W.; Wang, J.; Liu, X.; Wu, H.; Liu, M.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. Distinctive phosphorylation pattern during mitotic exit network (MEN) regulation is important for the development and pathogenicity of Magnaporthe oryzae. Stress Biol. 2022, 2, 41. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, X.; Zhang, W.; Feng, W.; Liu, M.; Yang, L.; Yang, Z.; Zhang, H.; Zhang, Z.; Wang, P. Hydrophobic cue-induced appressorium formation depends on MoSep1-mediated MoRgs7 phosphorylation and internalization in Magnaporthe oryzae. PLoS Genet. 2023, 19, e1010748. [Google Scholar] [CrossRef] [PubMed]
- Miao, P.; Mao, X.; Chen, S.; Abubakar, Y.S.; Li, Y.; Zheng, W.; Zhou, J.; Wang, Z.; Zheng, H. The mitotic exit mediated by small GTPase Tem1 is essential for the pathogenicity of Fusarium graminearum. PLoS Pathog. 2023, 19, e1011255. [Google Scholar] [CrossRef]
- Just, W.W.; Peränen, J. Small GTPases in peroxisome dynamics. Biochim. Biophys. Acta 2016, 1863, 1006–1013. [Google Scholar] [CrossRef]
- Dautt-Castro, M.; Rosendo-Vargas, M.; Casas-Flores, S. The small GTPases in fungal signaling conservation and function. Cells 2021, 10, 1039. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D.L. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Qiu, M.; Li, Y.; Ye, W.W.; Zheng, X.B.; Wang, Y.C. A CRISPR/Cas9-mediated in situ complementation method for Phytophthora sojae mutants. Mol. Plant Pathol. 2021, 22, 373–381. [Google Scholar] [CrossRef]
- Lin, L.; Ye, W.; Wu, J.; Xuan, M.; Li, Y.; Gao, J.; Wang, Y.; Wang, Y.; Dong, S.; Wang, Y. The MADS-box transcription factor PsMAD1 is involved in zoosporogenesis and pathogenesis of Phytophthora sojae. Front. Microbiol. 2018, 9, 2259. [Google Scholar] [CrossRef]
- Huang, J.; Xi, P.; Deng, Y.; Huang, W.; Wang, J.; Zhao, Q.; Yang, W.; Li, W.; Situ, J.; Jiang, L.; et al. The mitogen-activated protein kinase PlMAPK2 is involved in zoosporogenesis and pathogenicity of Peronophythora litchii. Int. J. Mol. Sci. 2021, 22, 3524. [Google Scholar] [CrossRef]
- Yin, Z.; Feng, W.; Chen, C.; Xu, J.; Li, Y.; Yang, L.; Wang, J.; Liu, X.; Wang, W.; Gao, C.; et al. Shedding light on autophagy coordinating with cell wall integrity signaling to govern pathogenicity of Magnaporthe oryzae. Autophagy 2020, 16, 900–916. [Google Scholar] [CrossRef]
- Situ, J.; Song, Y.; Feng, D.; Wan, L.; Li, W.; Ning, Y.; Huang, W.; Li, M.; Xi, P.; Deng, Y.; et al. Oomycete pathogen pectin acetylesterase targets host lipid transfer protein to reduce salicylic acid signaling. Plant Physiol. 2024, 194, 1779–1793. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, P.; Deng, Y.; Situ, J.; He, Z.; Zhou, W.; Li, M.; Xi, P.; Liang, X.; Kong, G.; et al. A plant cell death-inducing protein from litchi interacts with Peronophythora litchii pectate lyase and enhances plant resistance. Nat. Commun. 2024, 15, 22. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, G.; Huang, W.; Li, W.; Feng, D.; Liu, L.; Xi, P.; Jiang, Z.; Kong, G. Autophagy-related gene PlATG6a is involved in mycelial growth, asexual reproduction and tolerance to salt and oxidative stresses in Peronophythora litchii. Int. J. Mol. Sci. 2022, 23, 1839. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Li, W.; Yang, C.; Zhang, X.; Luo, M.; Chen, T.; Wang, X.; Wang, R.; Chen, Q. PlAtg8-mediated autophagy regulates vegetative growth, sporangial cleavage, and pathogenesis in Peronophythora litchii. Microbiol. Microbiol. Microbiol. Spectr. 2024, 12, e0353123. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Situ, J.; Deng, Y.Z.; Wan, L.; Xu, D.; Chen, Y.; Xi, P.; Jiang, Z. PlMAPK10, a mitogen-activated protein kinase (MAPK) in Peronophythora litchii, is required for mycelial growth, sporulation, laccase activity, and plant infection. Front. Microbiol. 2018, 9, 426. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, W.; Wang, H.; Hong, D.; Liao, G.; Yu, G.; Yang, L.; Yang, C.; Chen, Q. PlTem1, a Key Cell Cycle Regulator, Serves as an Important Bridge Between Cell Division and Autophagy in Peronophythora litchii. Agronomy 2025, 15, 1619. https://doi.org/10.3390/agronomy15071619
Feng W, Wang H, Hong D, Liao G, Yu G, Yang L, Yang C, Chen Q. PlTem1, a Key Cell Cycle Regulator, Serves as an Important Bridge Between Cell Division and Autophagy in Peronophythora litchii. Agronomy. 2025; 15(7):1619. https://doi.org/10.3390/agronomy15071619
Chicago/Turabian StyleFeng, Wanzhen, Han Wang, Danlu Hong, Guoliang Liao, Ge Yu, Lina Yang, Chengdong Yang, and Qinghe Chen. 2025. "PlTem1, a Key Cell Cycle Regulator, Serves as an Important Bridge Between Cell Division and Autophagy in Peronophythora litchii" Agronomy 15, no. 7: 1619. https://doi.org/10.3390/agronomy15071619
APA StyleFeng, W., Wang, H., Hong, D., Liao, G., Yu, G., Yang, L., Yang, C., & Chen, Q. (2025). PlTem1, a Key Cell Cycle Regulator, Serves as an Important Bridge Between Cell Division and Autophagy in Peronophythora litchii. Agronomy, 15(7), 1619. https://doi.org/10.3390/agronomy15071619





