Genome-Wide Identification and Abiotic Stress Response Analysis of C2H2 Zinc Finger Protein Genes in Foxtail Millet (Setaria italica)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification of C2H2 Genes in Foxtail Millet
2.3. Phylogenetic Tree, Chromosomal Location, and Conserved Motifs of SiC2H2
2.4. Whole-Genome Duplication and Synteny Analysis
2.5. MiRNA and Cis-Acting Element Prediction
2.6. Protein–Protein Interaction Network and GO Enrichment
2.7. Expression Pattern Analysis of SiC2H2
2.8. RNA Extraction, Reverse Transcription, and qRT-PCR Analysis
2.9. Data Processing and Statistical Analysis
3. Results
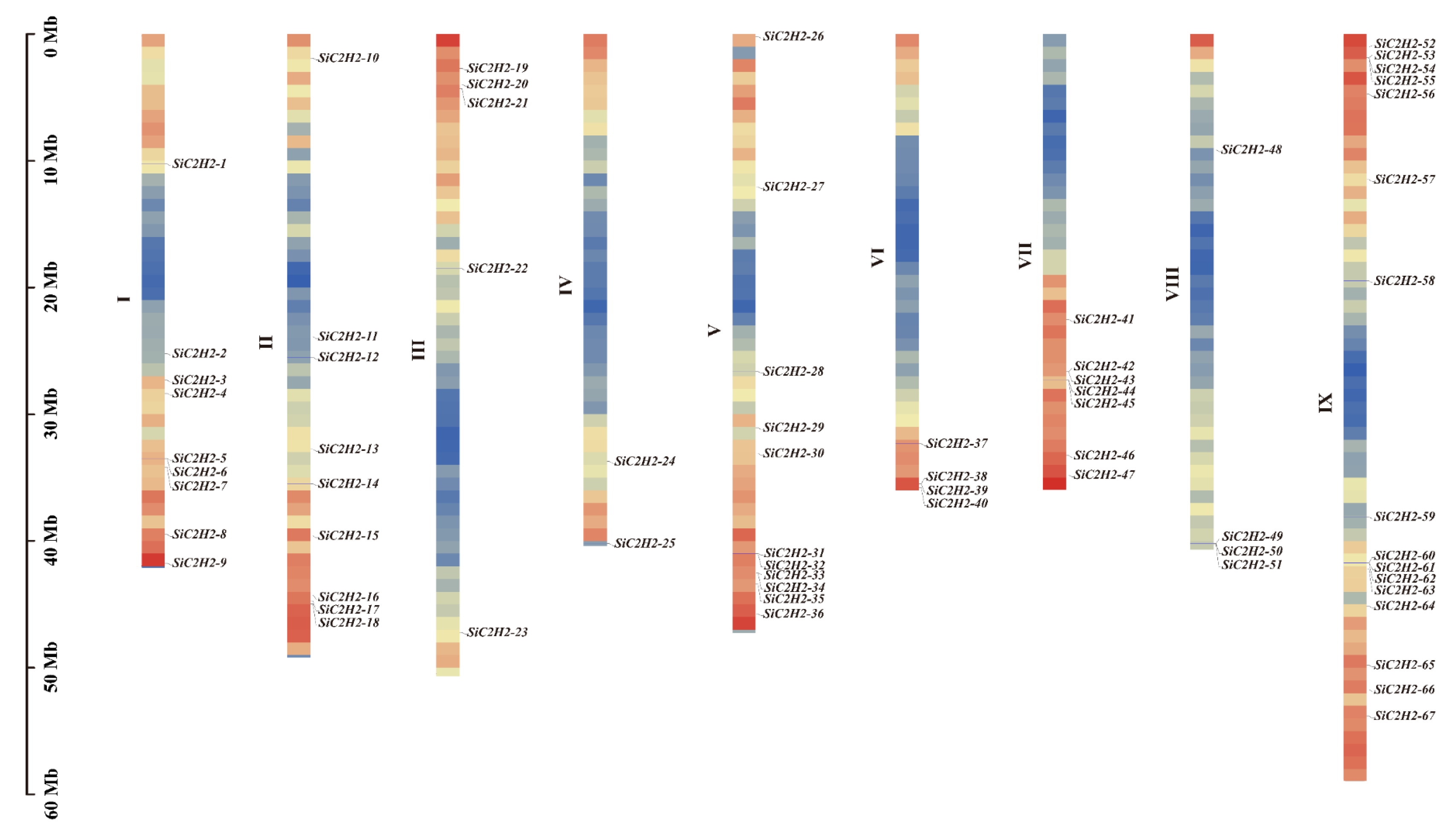
3.1. Genomic Identification, Chromosomal Localization, and Physicochemical Characterization of C2H2-ZFP Gene Family Members in Foxtail Millet
3.2. Phylogenetic and Evolutionary Analysis of C2H2-ZFPs Gene Family Members
3.3. SiC2H2 Gene Structure and Conserved Domains
3.4. Gene Duplication Events of the SiC2H2 Gene
3.5. Analysis of Cis-Acting Elements in Promoter of Foxtail Millet C2H2-ZFP Gene Family and miRNA Prediction
3.6. SiC2H2 Protein–Protein Interaction Network and GO Enrichment Analysis
3.7. SiC2H2 Gene Expression Pattern Analysis Based on RNA-Seq Data
3.8. Expression Levels of SiC2H2 Gene in Foxtail Millet Under Abiotic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Khan, A.R.; Gan, Y. C2H2 zinc finger proteins response to abiotic stress in plants. Int. J. Mol. Sci. 2022, 23, e2730. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Wang, H.L.; Zhang, T.; Zeng, X.Q.; Chen, H.; Wang, Z.W.; Zhang, J.; Zheng, H.; Tang, J.; Ling, Y.H.; et al. NONSTOP GLUMES1 encodes a C2H2 zinc finger protein that regulates spikelet development in rice. Plant Cell 2020, 32, 392–413. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cao, X.; Li, J.; Niu, Q.; Mo, Y.; Xiao, L. Genome-wide characterization of C2H2 zinc-finger gene family provides insight into the mechanisms and evolution of the dehydration-rehydration responses in Physcomitrium and Arabidopsis. Front. Plant Sci. 2022, 13, e953459. [Google Scholar] [CrossRef] [PubMed]
- Ji, E.; Hu, S.; Lu, Q.; Zhang, M.; Jiang, M. Hydrogen peroxide positively regulates ABA signaling via oxidative modification of the C2H2-type zinc finger protein ZFP36 in rice. Plant Physiol. Biochem. 2024, 213, e108844. [Google Scholar] [CrossRef]
- Li, Y.; Sun, A.; Wu, Q.; Zou, X.; Chen, F.; Cai, R.; Xie, H.; Zhang, M.; Guo, X. Comprehensive genomic survey, structural classification and expression analysis of C2H2-type zinc finger factor in wheat (Triticum aestivum L.). BMC Plant Biol. 2021, 21, e380. [Google Scholar] [CrossRef]
- Shahriari, A.G.; Soltani, Z.; Tahmasebi, A.; Poczai, P. Integrative system biology analysis of transcriptomic responses to drought stress in soybean (Glycine max L.). Genes 2022, 13, e1732. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Cai, Q.; Li, X.; Sun, Y.; Yu, T.; Yang, J.; Zhang, J. Analysis of the C2H2 gene family in maize (Zea mays L.) under cold stress: Identification and expression. Life 2023, 13, e122. [Google Scholar] [CrossRef]
- Du, T.; Zhou, Y.; Qin, Z.; Li, A.; Wang, Q.; Li, Z.; Hou, F.; Zhang, L. Genome-wide identification of the C2H2 zinc finger gene family and expression analysis under salt stress in sweetpotato. Front. Plant Sci. 2023, 14, e1301848. [Google Scholar] [CrossRef]
- Cui, H.; Chen, J.; Liu, M.; Zhang, H.; Zhang, S.; Liu, D.; Chen, S. Genome-wide analysis of C2H2 zinc finger gene family and its response to cold and drought stress in Sorghum [Sorghum bicolor (L.) Moench]. Int. J. Mol. Sci. 2022, 23, e5571. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.; Xu, X.; Zhang, H.; Li, C. Genome-wide analysis of C2H2 zinc-finger family transcription factors and their responses to abiotic stresses in Poplar (Populus trichocarpa). PLoS ONE 2015, 10, e0134753. [Google Scholar] [CrossRef]
- Yin, M.; Wang, Y.; Zhang, L.; Li, J.; Quan, W.; Yang, L.; Wang, Q.; Chan, Z. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Bot. 2017, 68, 2991–3005. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; He, H.; Yang, L.; Zhang, H.; Li, J.; Zhu, Y.; Xu, J.; Jiao, G.; Xiang, C.; Wang, C.; et al. AtZAT10/STZ1 improves drought tolerance and increases fiber yield in cotton. Front. Plant Sci. 2024, 15, e1464828. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.M.; Nguyen, X.C.; Kim, K.E.; Han, H.J.; Yoo, J.; Lee, K.; Kim, M.C.; Yun, D.J.; Chung, W.S. Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem. Biophys. Res. Commun. 2013, 430, e1054-9. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Luo, C. Overexpression of zinc finger transcription factor zat6 enhances salt tolerance. Open Life Sci. 2018, 13, 431–445. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, H.; Sun, C.; Ma, Q.; Bu, H.; Chong, K.; Xu, Y. A C2H2 zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice. J. Plant Physiol. 2018, 229, 100–110. [Google Scholar] [CrossRef]
- Sun, S.J.; Guo, S.Q.; Yang, X.; Bao, Y.M.; Tang, H.J.; Sun, H.; Huang, J.; Zhang, H.S. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 2010, 61, 2807–2818. [Google Scholar] [CrossRef]
- de Freitas, G.M.; Thomas, J.; Liyanage, R.; Lay, J.O.; Basu, S.; Ramegowda, V.; do Amaral, M.N.; Benitez, L.C.; Bolacel Braga, E.J.; Pereira, A. Cold tolerance response mechanisms revealed through comparative analysis of gene and protein expression in multiple rice genotypes. PLoS ONE 2019, 14, e0218019. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, R.; Guo, B.; Huang, K.; Wang, L.; Han, Y.; Li, H.; Hou, S. Ectopic expression of GmZAT4, a putative C2H2-type zinc finger protein, enhances PEG and NaCl stress tolerances in Arabidopsis thaliana. 3 Biotech 2019, 9, e166. [Google Scholar] [CrossRef]
- Cheng, Z.; Sun, Y.; Yang, S.; Zhi, H.; Yin, T.; Ma, X.; Zhang, H.; Diao, X.; Guo, Y.; Li, X.; et al. Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica). Plant Biotechnol. J. 2021, 19, 1089–1091. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Lu, P.; Wu, B.; Liu, M.; Gao, J.; Wang, C.; Bai, K.; Guo, G. Six underutilized grain crops for food and nutrition in China. Plants 2022, 11, 2451. [Google Scholar] [CrossRef]
- Kalsi, R.; Bhasin, J.K. Nutritional exploration of foxtail millet (Setaria italica) in addressing food security and its utilization trends in food system. eFood 2023, 4, e111. [Google Scholar] [CrossRef]
- Lamlom, S.F.; Abdelghany, A.M.; Farouk, A.S.; Alwakel, E.S.; Makled, K.M.; Bukhari, N.A.; Hatamleh, A.A.; Ren, H.; El-Sorady, G.A.; Shehab, A.A. Biochemical and yield response of spring wheat to drought stress through gibberellic and abscisic acids. BMC Plant Biol. 2025, 25, e5. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jiao, L.; Lian, X. Changes in compound extreme events and their impacts on cropland productivity in China, 1985–2019. Earth’s Future 2025, 13, e2024EF005038. [Google Scholar] [CrossRef]
- Wu, J.; Chen, L.; Yang, Z.; Lu, J.; Yang, J.; Li, N.; Shi, H. Transcriptomics uncovers pathways mediating low-nitrogen stress tolerance in two foxtail millet varieties. Agriculture 2025, 15, 628. [Google Scholar] [CrossRef]
- Yuan, J.; Amend, A.; Borkowski, J.; DeMarco, R.; Bailey, W.; Liu, Y.; Xie, G.; Blevins, R. MULTICLUSTAL: A systematic method for surveying Clustal W alignment parameters. Bioinformatics 1999, 15, 862–863. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Kayum, M.A.; Park, J.I.; Nath, U.K.; Saha, G.; Biswas, M.K.; Kim, H.T.; Nou, I.S. Genome-wide characterization and expression profiling of PDI family gene reveals function as abiotic and biotic stress tolerance in Chinese cabbage (Brassica rapassp. pekinensis). BMC Genom. 2017, 18, 885–920. [Google Scholar] [CrossRef] [PubMed]
- Khaja, R.; MacDonald, J.R.; Zhang, J.; Scherer, S.W. Methods for identifying and mapping recent segmental and gene duplications in eukaryotic genomes. Methods Mol. Biol. 2006, 338, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Xie, S.J.; Liu, Y.W.; Qi, X.; Yu, J.J. Genome-wide characterization of microRNA in foxtail millet (Setaria italica). BMC Plant Biol. 2013, 13, e212. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; Van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Lescot, M.; De’ hais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Tang, S.; Li, L.; Wang, Y.Q.; Chen, Q.N.; Zhang, W.Y.; Jia, G.Q.; Zhi, H.; Zhao, B.H.; Diao, X.M. Genotype-specific physiological and transcriptomic responses to drought stress in Setaria italica (an emerging model for Panicoideae grasses). Sci. Rep. 2017, 17, e10009. [Google Scholar] [CrossRef]
- Han, F.; Sun, M.J.; He, W.; Guo, S.Q.; Feng, J.Y.; Wang, H.; Yang, Q.G.; Pan, H.; Lou, Y.H.; Zhuge, Y.P. Transcriptome analysis reveals molecular mechanisms under salt stress in leaves of foxtail millet (Setaria italica L.). Plants 2022, 7, e1864. [Google Scholar] [CrossRef]
- Pfafff, M.W. A new mathematical model for relative quantification in real- time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Chen, L.; Wang, R.; Hu, X.; Wang, D.; Wang, Y.; Xue, R.; Wu, M.; Li, H. Overexpression of wheat C2H2 zinc finger protein transcription factor TaZAT8-5B enhances drought tolerance and root growth in Arabidopsis thaliana. Planta 2024, 260, e126. [Google Scholar] [CrossRef]
- Zhang, D.; Ding, X.; Wang, Z.; Li, W.; Li, L.; Liu, L.; Zhou, H.; Yu, J.; Zheng, C.; Wu, H.; et al. A C2H2 zinc finger protein, OsZOS2-19, modulates aba sensitivity and cold response in rice. Plant Cell Physiol. 2025, 66, 753–765. [Google Scholar] [CrossRef]
- Yuan, S.; Li, X.; Li, R.; Wang, L.; Zhang, C.; Chen, L.; Hao, Q.; Zhang, X.; Chen, H.; Shan, Z.; et al. Genome-wide identification and classification of soybean C2H2 zinc finger proteins and their expression analysis in legume-rhizobium symbiosis. Front. Microbiol. 2018, 9, e126. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Li, Z.; He, X.; Zhang, Z.; Wang, D.; Cui, L.; Xie, M.; Zhao, Z.; Sun, Q.; Wang, D.; et al. Comparative transcriptome analysis reveals genes involved in trichome development and metabolism in tobacco. BMC Plant Biol. 2024, 24, e541. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Li, Y.; Qiao, Z.; Wang, C.; Zhao, Y.; Guo, J.; Chen, M.; Wang, B. Advances in the regulation of epidermal cell development by C2H2 zinc finger proteins in plants. Front. Plant Sci. 2021, 12, e754512. [Google Scholar] [CrossRef] [PubMed]
- Faraji, S.; Rasouli, S.H.; Kazemitabar, S.K. Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp. Durum): Insights into the roles in biological processes especially stress response. Biometals 2018, 31, 1019–1042. [Google Scholar] [CrossRef]
- Dai, X.; Wang, Z.; Bao, Y.; Jia, C.; Bai, F.; Hasi, A.; Che, G. Identification and functional characterization of the C2H2 ZFP transcription factor CmSUP7 in regulating melon plant growth and fruit development. Plant Physiol. Biochem. 2025, 220, e109513. [Google Scholar] [CrossRef]
- Qiu, T.; Liu, Z.; Liu, B. The effects of hybridization and genome doubling in plant evolution via allopolyploidy. Mol. Biol. Rep. 2020, 47, 5549–5558. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef]
- Seoighe, C.; Gehring, C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004, 20, 461–464. [Google Scholar] [CrossRef]
- Zan, Y.; Chen, S.; Ren, M.; Liu, G.; Liu, Y.; Han, Y.; Dong, Y.; Zhang, Y.; Si, H.; Liu, Z.; et al. The genome and GeneBank genomics of allotetraploid Nicotiana tabacum provide insights into genome evolution and complex trait regulation. Nat. Genet. 2025, 57, 986–996. [Google Scholar] [CrossRef]
- Schulthess, A.W.; Kale, S.M.; Liu, F.; Zhao, Y.; Philipp, N.; Rembe, M.; Jiang, Y.; Beukert, U.; Serfling, A.; Himmelbach, A.; et al. Genomics-informed prebreeding unlocks the diversity in genebanks for wheat improvement. Nat. Genet. 2022, 54, 1544–1552. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, L.; Pan, Z.; Zhao, M.; Zhu, L.; Han, Y.; Li, L.; Wang, Y.; Wang, K.; Liu, S.; et al. Genome-wide analysis of the C2H2 zinc finger protein gene family and its response to salt stress in ginseng, Panax ginseng Meyer. Sci. Rep. 2022, 12, e10165. [Google Scholar] [CrossRef]
- Liu, Z.; Coulter, J.A.; Li, Y.; Zhang, X.; Meng, J.; Zhang, J.; Liu, Y. Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 153, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shi, Y.; Zhang, W.; Zhu, K.; Feng, L.; Wang, J. C2H2 zinc finger protein family analysis of Rosa rugosa identified a salt-tolerance regulator, RrC2H2-8. Plants 2024, 13, 3580. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, S.D.; Novak, N.G. Comparative analysis of the genetic variability within the Q-type C2H2 zinc-finger transcription factors in the economically important cabbage, canola and Chinese cabbage genomes. Hereditas 2018, 155, e29. [Google Scholar] [CrossRef]
- Zhou, X.; Gao, T.; Zhang, Y.; Han, M.; Shen, Y.; Su, Y.; Feng, X.; Wu, Q.; Sun, G.; Wang, Y. Genome-wide identification, characterization and expression of C2H2 zinc finger gene family in Opisthopappus species under salt stress. BMC Genom. 2024, 25, e385. [Google Scholar] [CrossRef]
- Wu, D.; Shen, E.; Jiang, B.; Feng, Y.; Tang, W.; Lao, S.; Jia, L.; Lin, H.; Xie, L.; Weng, X.; et al. Genomic insights into the evolution of Echinochloa species as weed and orphan crop. Nat. Commun. 2022, 13, 689. [Google Scholar] [CrossRef]
- Liao, X.; Wang, L.; Zhu, S.; Zheng, F.; Yang, C. Identification, genomic organization, and expression profiles of single C2H2 zinc finger transcription factors in tomato (Solanum lycopersicum). J. Appl. Genet. 2021, 62, 1–15. [Google Scholar] [CrossRef]
- Xie, M.; Sun, J.; Gong, D.; Kong, Y. The roles of Arabidopsis C1-2i subclass of C2H2-type zinc-finger transcription factors. Genes 2019, 10, 653. [Google Scholar] [CrossRef]
- Agarwal, P.; Arora, R.; Ray, S.; Singh, A.K.; Singh, V.P.; Takatsuji, H.; Kapoor, S.; Tyagi, A.K. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2007, 65, 467–485. [Google Scholar] [CrossRef]







| Gene Name | Length (aa) | MW (KDa) | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|
| SiC2H2-1 | 865 | 97.56 | 7.68 | 40.59 | 78.66 | −0.298 |
| SiC2H2-2 | 488 | 53.59 | 7.64 | 59.66 | 63.01 | −0.633 |
| SiC2H2-3 | 350 | 39.44 | 7.96 | 55.17 | 53.6 | −0.957 |
| SiC2H2-4 | 392 | 41.62 | 6.25 | 57.43 | 64.44 | −0.442 |
| SiC2H2-5 | 569 | 60.54 | 5.96 | 53.71 | 68.49 | −0.417 |
| SiC2H2-6 | 340 | 35.13 | 6.06 | 64.13 | 54.56 | −0.44 |
| SiC2H2-7 | 585 | 60.21 | 8.89 | 52.87 | 59.2 | −0.4 |
| SiC2H2-8 | 271 | 28.91 | 8.22 | 70.78 | 53.47 | −0.577 |
| SiC2H2-9 | 315 | 32.77 | 8.65 | 61.41 | 70.51 | −0.3 |
| SiC2H2-10 | 300 | 30.51 | 9.27 | 61.53 | 68.17 | −0.32 |
| SiC2H2-11 | 140 | 16.05 | 10.97 | 69.53 | 53.79 | −0.689 |
| SiC2H2-12 | 348 | 37.66 | 6.67 | 57.78 | 52.21 | −0.565 |
| SiC2H2-13 | 471 | 50.40 | 9.21 | 71.32 | 63.99 | −0.537 |
| SiC2H2-14 | 433 | 46.66 | 6.06 | 58.62 | 53.37 | −0.749 |
| SiC2H2-15 | 511 | 52.80 | 9.17 | 49.48 | 55.03 | −0.471 |
| SiC2H2-16 | 711 | 78.53 | 6.93 | 73.56 | 52.74 | −0.86 |
| SiC2H2-17 | 187 | 19.25 | 8.93 | 73.92 | 65.99 | −0.243 |
| SiC2H2-18 | 419 | 43.70 | 5.89 | 69.33 | 60.02 | −0.563 |
| SiC2H2-19 | 378 | 42.92 | 8.84 | 55.77 | 52.06 | −0.873 |
| SiC2H2-20 | 188 | 20.12 | 6.78 | 41.83 | 72.77 | −0.232 |
| SiC2H2-21 | 378 | 42.40 | 7.65 | 51.26 | 48.78 | −0.981 |
| SiC2H2-22 | 390 | 42.81 | 7.23 | 53.31 | 57.36 | −0.67 |
| SiC2H2-23 | 257 | 26.63 | 7.64 | 92.22 | 64.12 | −0.375 |
| SiC2H2-24 | 393 | 42.20 | 7.27 | 52.62 | 46.36 | −0.78 |
| SiC2H2-25 | 182 | 18.89 | 9.05 | 69.11 | 66.21 | −0.295 |
| SiC2H2-26 | 402 | 42.79 | 9.36 | 54.86 | 71.02 | −0.388 |
| SiC2H2-27 | 465 | 49.36 | 9.04 | 48.76 | 57.7 | −0.529 |
| SiC2H2-28 | 436 | 46.36 | 8.94 | 54.15 | 55.53 | −0.559 |
| SiC2H2-29 | 497 | 51.14 | 5.54 | 60.26 | 64.08 | −0.429 |
| SiC2H2-30 | 474 | 50.15 | 8.79 | 56.86 | 57.45 | −0.556 |
| SiC2H2-31 | 214 | 23.14 | 9.04 | 69.47 | 75.42 | −0.481 |
| SiC2H2-32 | 184 | 19.37 | 6.16 | 68.78 | 58.1 | −0.622 |
| SiC2H2-33 | 517 | 56.73 | 5.62 | 46.29 | 59.81 | −0.616 |
| SiC2H2-34 | 517 | 56.73 | 5.62 | 46.29 | 59.81 | −0.616 |
| SiC2H2-35 | 454 | 47.36 | 6.6 | 72.58 | 58.33 | −0.595 |
| SiC2H2-36 | 469 | 49.75 | 8.86 | 53.8 | 60.43 | −0.464 |
| SiC2H2-37 | 419 | 45.23 | 5.66 | 57.26 | 57.21 | −0.492 |
| SiC2H2-38 | 529 | 54.70 | 9.44 | 48.71 | 53.69 | −0.53 |
| SiC2H2-39 | 526 | 54.44 | 9.33 | 49.84 | 53.99 | −0.522 |
| SiC2H2-40 | 392 | 41.91 | 8.7 | 54.69 | 63.16 | −0.42 |
| SiC2H2-41 | 503 | 51.99 | 5.83 | 57.32 | 63.12 | −0.477 |
| SiC2H2-42 | 562 | 59.07 | 6.18 | 54.31 | 63.79 | −0.423 |
| SiC2H2-43 | 319 | 33.31 | 7.09 | 66.17 | 55.92 | −0.571 |
| SiC2H2-44 | 607 | 62.56 | 8.77 | 45.56 | 58.07 | −0.442 |
| SiC2H2-45 | 607 | 62.56 | 8.77 | 45.56 | 58.07 | −0.442 |
| SiC2H2-46 | 477 | 52.21 | 5.99 | 45.5 | 59.92 | −0.534 |
| SiC2H2-47 | 304 | 31.44 | 9.62 | 75.81 | 65.03 | −0.471 |
| SiC2H2-48 | 134 | 14.71 | 7.09 | 42.67 | 67.16 | −0.754 |
| SiC2H2-49 | 89 | 9.64 | 10.27 | 66.31 | 72.36 | −0.319 |
| SiC2H2-50 | 165 | 17.54 | 10.36 | 37.02 | 72.85 | −0.351 |
| SiC2H2-51 | 142 | 14.78 | 10.16 | 36.05 | 71.69 | −0.149 |
| SiC2H2-52 | 331 | 36.41 | 6.31 | 50.75 | 64.92 | −0.463 |
| SiC2H2-53 | 239 | 24.87 | 10.04 | 54.83 | 71.21 | −0.313 |
| SiC2H2-54 | 186 | 19.83 | 8.43 | 59.24 | 63.71 | −0.565 |
| SiC2H2-55 | 142 | 14.67 | 7.82 | 70.17 | 69.08 | −0.389 |
| SiC2H2-56 | 408 | 42.89 | 9.18 | 67.93 | 64.73 | −0.346 |
| SiC2H2-57 | 230 | 24.47 | 9.14 | 78.52 | 61.78 | −0.489 |
| SiC2H2-58 | 399 | 42.93 | 9.48 | 52.1 | 71.33 | −0.396 |
| SiC2H2-59 | 340 | 36.41 | 8.98 | 81.1 | 47.74 | −0.791 |
| SiC2H2-60 | 224 | 23.12 | 8.56 | 66.55 | 68.62 | −0.197 |
| SiC2H2-61 | 220 | 23.05 | 8.7 | 63.99 | 72.09 | −0.217 |
| SiC2H2-62 | 208 | 21.39 | 9.3 | 69.03 | 65.48 | −0.249 |
| SiC2H2-63 | 405 | 43.95 | 5.72 | 65.79 | 60.35 | −0.757 |
| SiC2H2-64 | 144 | 15.17 | 6.96 | 53.24 | 72.08 | −0.419 |
| SiC2H2-65 | 188 | 19.89 | 7.09 | 46.14 | 70.69 | −0.401 |
| SiC2H2-66 | 454 | 48.14 | 10.95 | 75.99 | 67.44 | −0.502 |
| SiC2H2-67 | 537 | 55.82 | 9 | 71.58 | 60.32 | −0.383 |
| MiRNA | Target | Expectation | MiRNA Length | Target_Start | Target_End |
|---|---|---|---|---|---|
| SitmiR156c | SiC2H2-58 | 4.0 | 22 | 94 | 115 |
| SitmiR156c | SiC2H2-5 | 5.0 | 22 | 458 | 479 |
| SitmiR156c | SiC2H2-13 | 5.0 | 22 | 995 | 1016 |
| SitmiR166a-1/2/3/4/5 | SiC2H2-29 | 4.0 | 21 | 1313 | 1333 |
| SitmiR166c/d | SiC2H2-29 | 4.0 | 21 | 1313 | 1333 |
| SitmiR2118a-1/2/3 | SiC2H2-2 | 4.5 | 22 | 1298 | 1319 |
| SitmiR2118d | SiC2H2-44 | 4.5 | 22 | 1249 | 1270 |
| SitmiR2118d | SiC2H2-45 | 4.5 | 22 | 1249 | 1270 |
| SitmiR164a/b | SiC2H2-50 | 5.0 | 21 | 164 | 184 |
| SitmiR166a-1/2/3 | SiC2H2-18 | 5.0 | 21 | 11 | 31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Q.; Zhang, Y.; Xing, X.; Li, S.; Sun, R.; Zhang, W.; Zhang, J.; Jiang, L.; Zang, Z.; Gao, M.; et al. Genome-Wide Identification and Abiotic Stress Response Analysis of C2H2 Zinc Finger Protein Genes in Foxtail Millet (Setaria italica). Agronomy 2025, 15, 1618. https://doi.org/10.3390/agronomy15071618
Zhao Q, Zhang Y, Xing X, Li S, Sun R, Zhang W, Zhang J, Jiang L, Zang Z, Gao M, et al. Genome-Wide Identification and Abiotic Stress Response Analysis of C2H2 Zinc Finger Protein Genes in Foxtail Millet (Setaria italica). Agronomy. 2025; 15(7):1618. https://doi.org/10.3390/agronomy15071618
Chicago/Turabian StyleZhao, Qian, Yingxin Zhang, Xiangyu Xing, Shuyao Li, Ruidong Sun, Weilong Zhang, Jun Zhang, Liangyu Jiang, Zhenyuan Zang, Ming Gao, and et al. 2025. "Genome-Wide Identification and Abiotic Stress Response Analysis of C2H2 Zinc Finger Protein Genes in Foxtail Millet (Setaria italica)" Agronomy 15, no. 7: 1618. https://doi.org/10.3390/agronomy15071618
APA StyleZhao, Q., Zhang, Y., Xing, X., Li, S., Sun, R., Zhang, W., Zhang, J., Jiang, L., Zang, Z., Gao, M., & Zhang, J. (2025). Genome-Wide Identification and Abiotic Stress Response Analysis of C2H2 Zinc Finger Protein Genes in Foxtail Millet (Setaria italica). Agronomy, 15(7), 1618. https://doi.org/10.3390/agronomy15071618







