Intelligent Detection and Control of Crop Pests and Diseases: Current Status and Future Prospects
Abstract
1. Introduction
2. Classic Machine Learning
2.1. Decision Tree
2.2. Support Vector Machine
2.3. Random Forest
2.4. K-Means Clustering
2.5. RL
3. Deep Learning
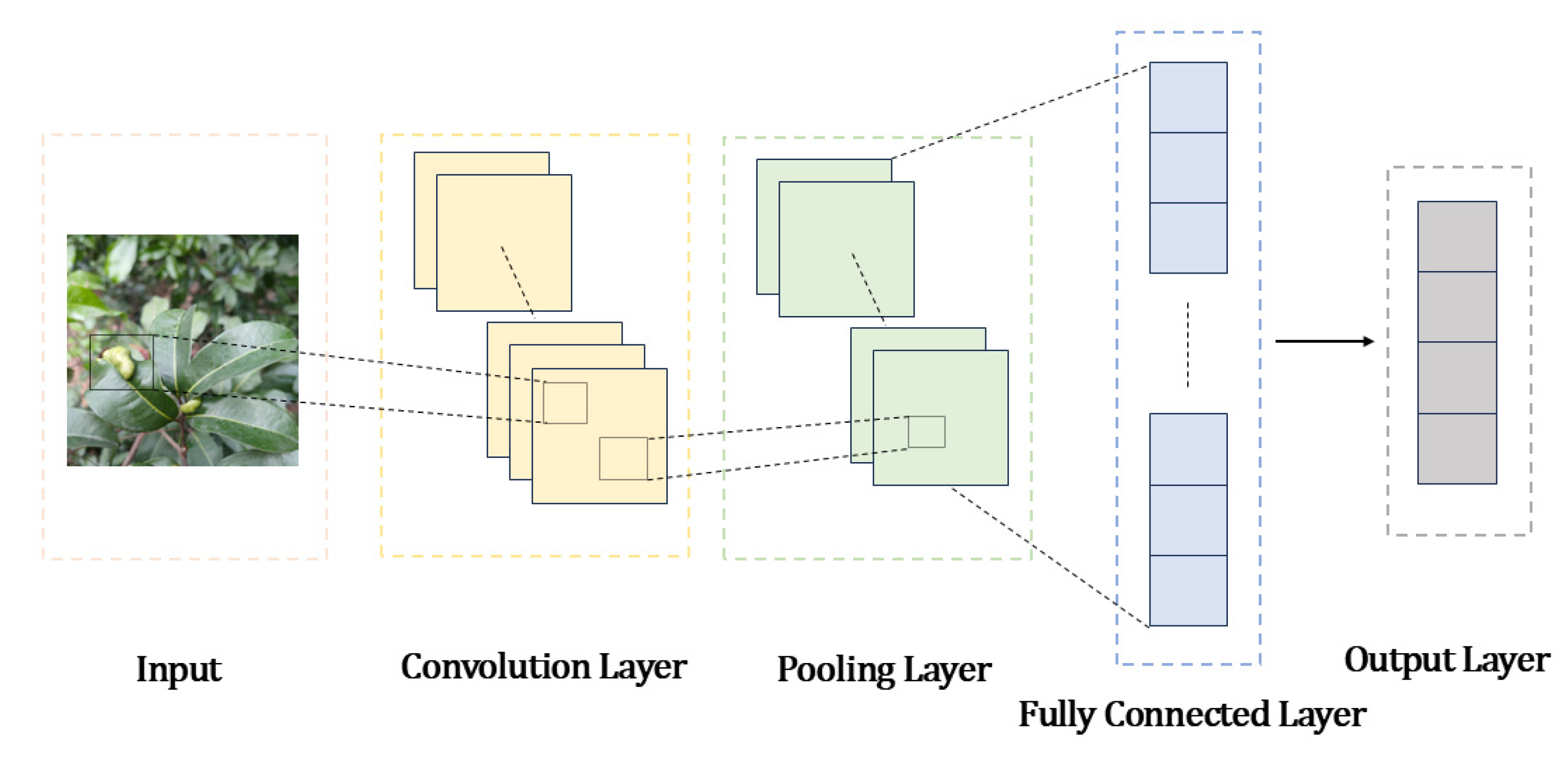
3.1. CNN
3.1.1. Image Classification Algorithms
AlexNet
Visual Geometry Group (VGG)
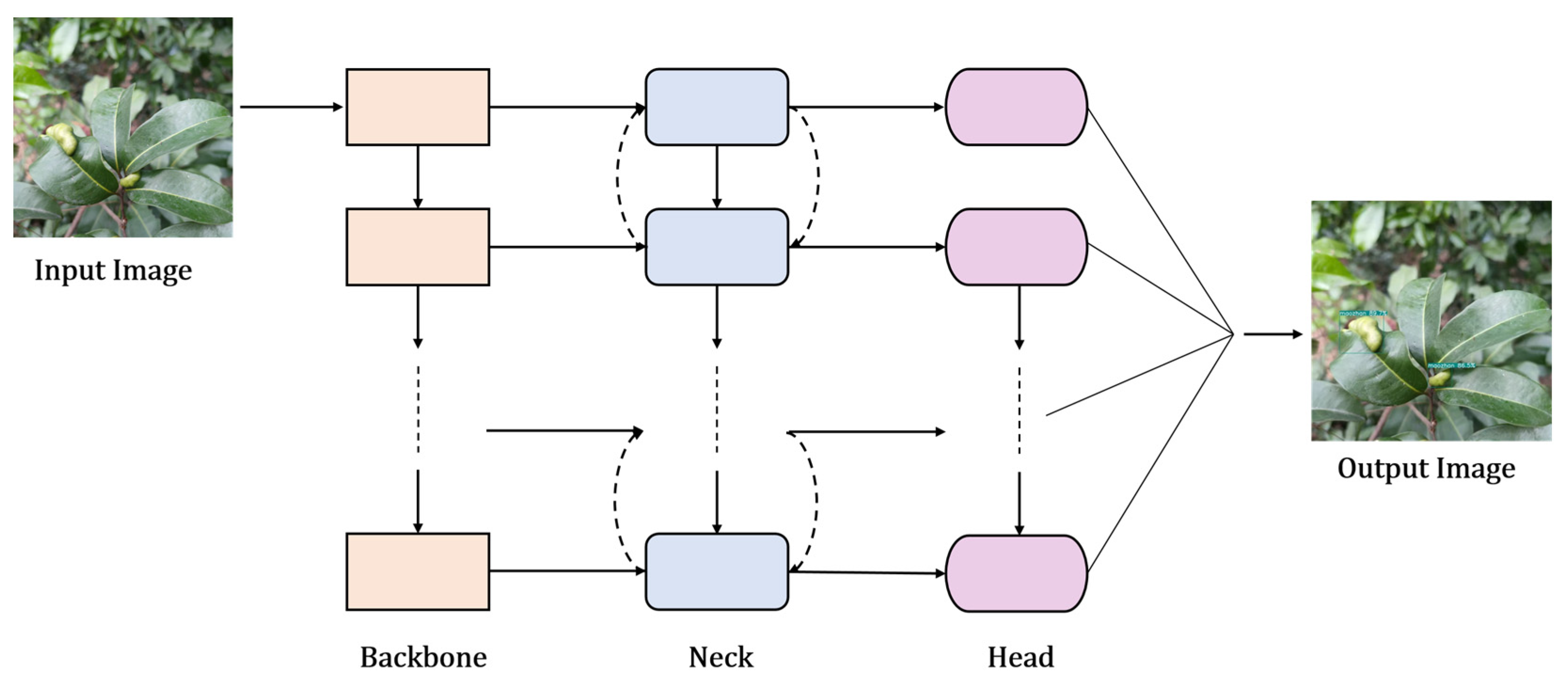
3.1.2. Object Detection Algorithms
R-CNN Series
You Only Look Once (YOLO) Series
Detection Transformer (DETR)
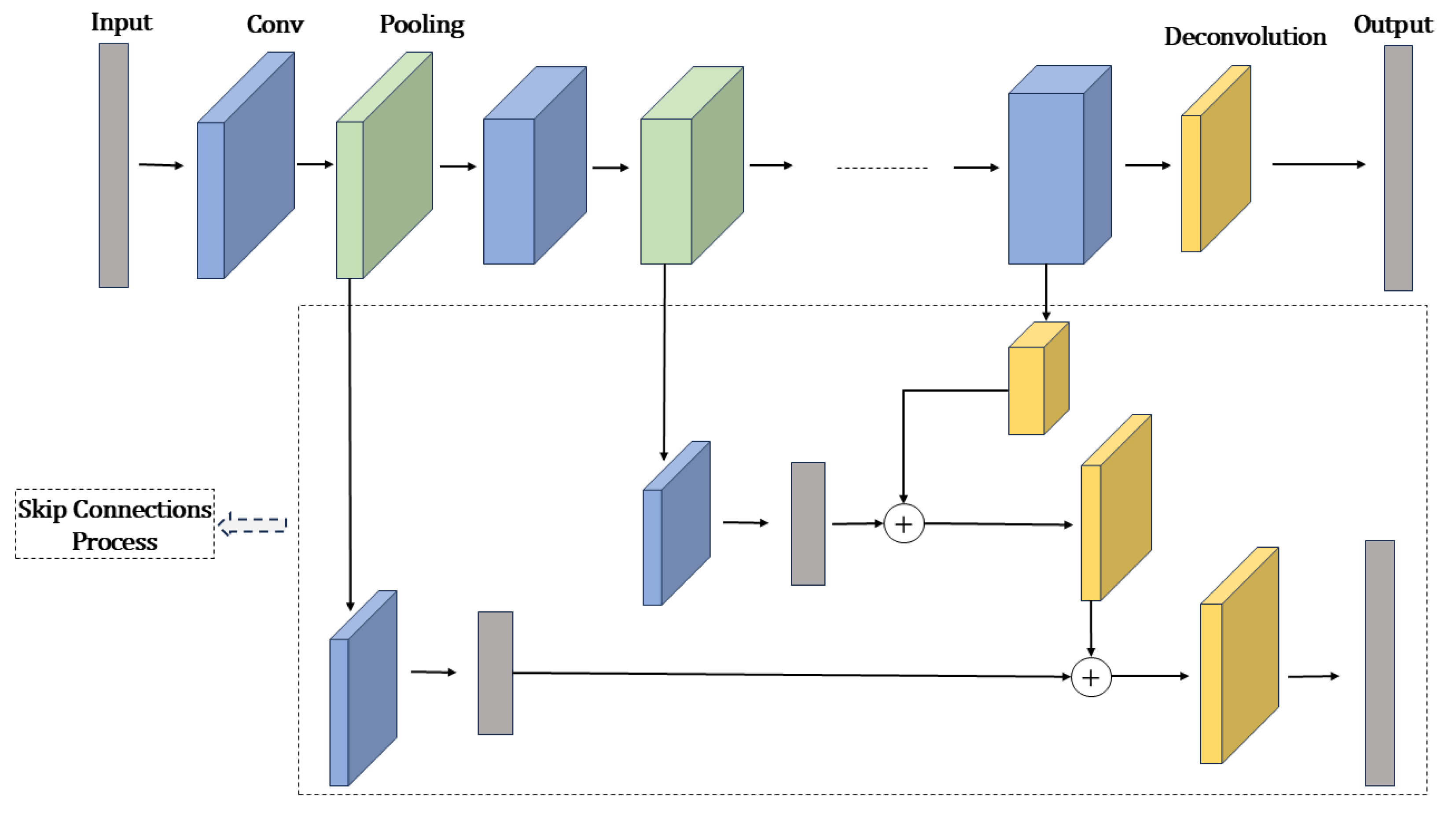
3.1.3. Image Segmentation Algorithms
Fully Convolutional Networks (FCNs)
Mask R-CNN
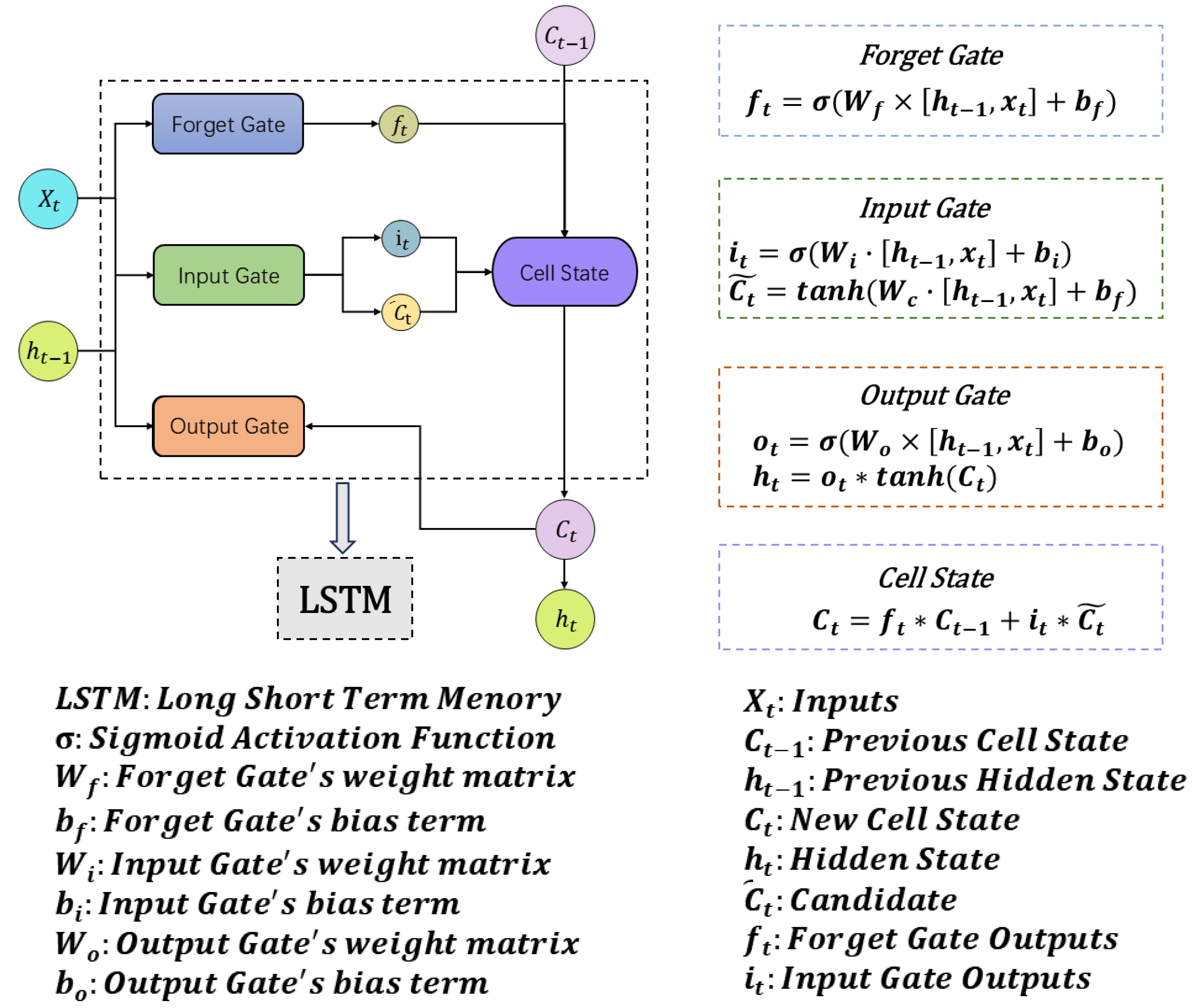
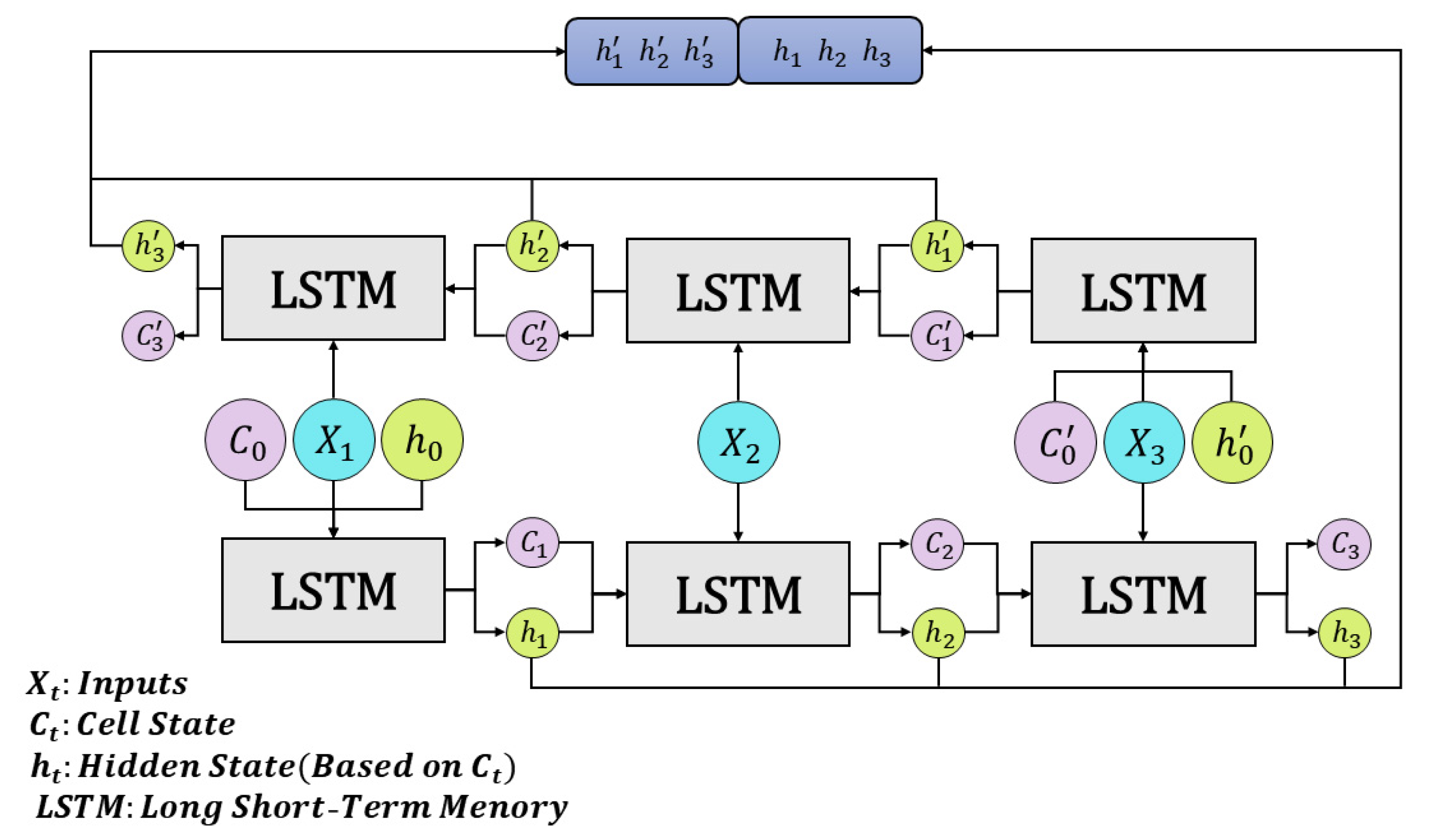
3.2. RNN
LSTM
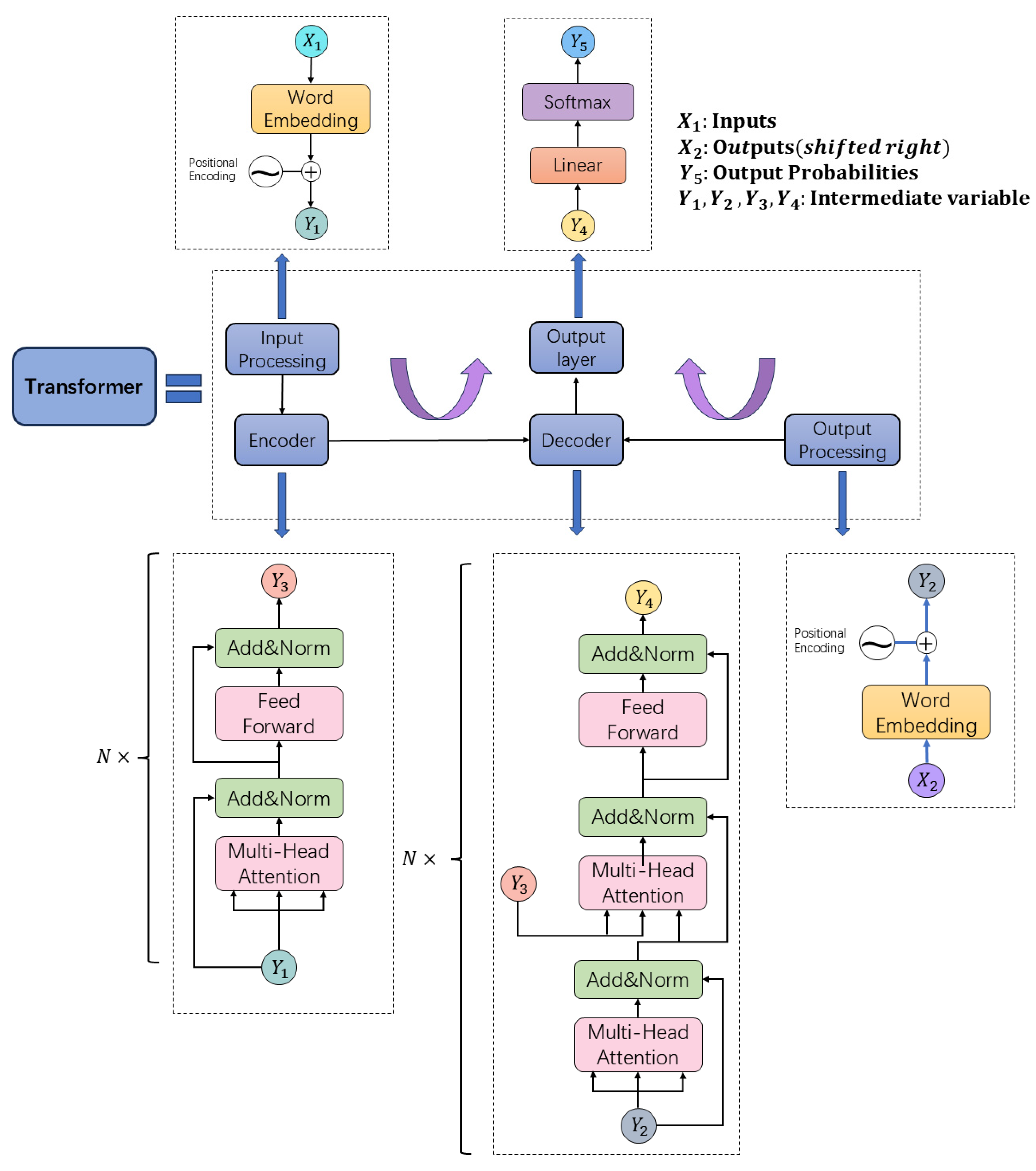
4. Large Language Models
4.1. Semantic Large Language Models
4.2. Vision–Language Models
5. Discussion and Conclusions
5.1. Discussion
5.2. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Population Growth and Sustainable Development|Policy Commons. Available online: https://policycommons.net/artifacts/8983020/global-population-growth-and-sustainable-development/9868536/ (accessed on 19 March 2025).
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The Global Burden of Pathogens and Pests on Major Food Crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Researchers Helping Protect Crops From Pests|NIFA. Available online: https://www.nifa.usda.gov/about-nifa/blogs/researchers-helping-protect-crops-pests (accessed on 20 March 2025).
- Abubakar, M.; Koul, B.; Chandrashekar, K.; Raut, A.; Yadav, D. Whitefly (Bemisia tabaci) Management (WFM) Strategies for Sustainable Agriculture: A Review. Agriculture 2022, 12, 1317. [Google Scholar] [CrossRef]
- Lidwell-Durnin, J.; Lapthorn, A. The Threat to Global Food Security from Wheat Rust: Ethical and Historical Issues in Fighting Crop Diseases and Preserving Genetic Diversity. Glob. Food Secur. 2020, 26, 100446. [Google Scholar] [CrossRef]
- Azfar, S.; Nadeem, A.; Alkhodre, A.B.; Ahsan, K.; Mehmood, N.; Alghmdi, T.; Alsaawy, Y. Monitoring, Detection and Control Techniques of Agriculture Pests and Diseases Using Wireless Sensor Network: A Review. Int. J. Adv. Comput. Sci. Appl. 2018, 9, 424–433. [Google Scholar] [CrossRef]
- Domingues, T.; Brandão, T.; Ferreira, J.C. Machine Learning for Detection and Prediction of Crop Diseases and Pests: A Comprehensive Survey. Agriculture 2022, 12, 1350. [Google Scholar] [CrossRef]
- Rwakipamba, E.; Sseremba, G.; Byalebeka, J.; Ssekandi, J.; Mwine, J. Over Reliance on Pesticides and Poor Handling Practices Characterize Intensive Vegetable Farming: Case of Selected Smallholders in Southwestern Uganda. Preprints 2020, 1–36. [Google Scholar] [CrossRef]
- Kartikeyan, P.; Shrivastava, G. Review on Emerging Trends in Detection of Plant Diseases Using Image Processing with Machine Learning. Int. J. Comput. Appl. 2021, 174, 39–48. [Google Scholar] [CrossRef]
- Cornuéjols, A.; Moulet, M. Machine Learning: A Survey. In Knowledge Based Systems. Advanced Concepts, Techniques and Applications; Tzafestas, S.G., Ed.; World Scientific: Singapore, 1997; pp. 61–86. [Google Scholar]
- Rajan, P.; Radhakrishnan, B.; Suresh, L.P. Detection and Classification of Pests from Crop Images Using Support Vector Machine. In Proceedings of the 2016 International Conference on Emerging Technological Trends (ICETT), Kollam, India, 21–22 October 2016; pp. 1–6. [Google Scholar]
- Pattnaik, G.; Parvathi, K. Machine Learning-Based Approaches for Tomato Pest Classification. TELKOMNIKA Telecommun. Comput. Electron. Control. 2022, 20, 321–328. [Google Scholar] [CrossRef]
- Reddy, D.T.K.; Ramesh, S. Identification of the Pest Detection Using Random Forest Algorithm and Support Vector Machine with Improved Accuracy. AIP Conf. Proc. 2024, 3193, 020185. [Google Scholar] [CrossRef]
- Revathy, R.; Lawrance, R. Classifying Crop Pest Data Using C4.5 Algorithm. In Proceedings of the 2017 IEEE International Conference on Intelligent Techniques in Control, Optimization and Signal Processing (INCOS), Krishnankoil, India, 23–25 March 2017; pp. 1–6. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, X.; Liu, Z.; Liao, F.; Wang, W.; Li, J. Detection of Litchi Leaf Diseases and Insect Pests Based on Improved FCOS. Agronomy 2023, 13, 1314. [Google Scholar] [CrossRef]
- Liu, S.; Qiao, Y.; Li, J.; Zhang, H.; Zhang, M.; Wang, M. An Improved Lightweight Network for Real-Time Detection of Apple Leaf Diseases in Natural Scenes. Agronomy 2022, 12, 2363. [Google Scholar] [CrossRef]
- Pest Identification and Counting of Yellow Plate in Field Based on Improved Mask R-CNN-Rong-2022-Discrete Dynamics in Nature and Society—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/full/10.1155/2022/1913577 (accessed on 20 March 2025).
- Roumeliotis, K.I.; Tselikas, N.D. ChatGPT and Open-AI Models: A Preliminary Review. Future Internet 2023, 15, 192. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, B.; Ji, M.; Wang, X.; Yan, Y.; Zhang, J.; Liu, S.; Ye, M.; Lv, C. Implementation of Large Language Models and Agricultural Knowledge Graphs for Efficient Plant Disease Detection. Agriculture 2024, 14, 1359. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, Q.; Chen, X.; Li, M.; Zhao, Z.; Li, F.; Guo, L. IPM-AgriGPT: A Large Language Model for Pest and Disease Management with a G-EA Framework and Agricultural Contextual Reasoning. Mathematics 2025, 13, 566. [Google Scholar] [CrossRef]
- Mahesh, B. Machine Learning Algorithms—A Review. Int. J. Sci. Res. 2020, 9, 381–386. [Google Scholar] [CrossRef]
- Sharma, R. Study of Supervised Learning and Unsupervised Learning. Int. J. Res. Appl. Sci. Eng. Technol. 2020, 8, 588–593. [Google Scholar] [CrossRef]
- Nasteski, V. An Overview of the Supervised Machine Learning Methods. Horizons. B 2017, 4, 51–62. [Google Scholar] [CrossRef]
- Singh, A.; Thakur, N.; Sharma, A. A Review of Supervised Machine Learning Algorithms. In Proceedings of the 2016 3rd International Conference on Computing for Sustainable Global Development (INDIACom), New Delhi, India, 16–18 March 2016; pp. 1310–1315. [Google Scholar]
- Naeem, S.; Ali, A.; Anam, S.; Ahmed, M. An Unsupervised Machine Learning Algorithms: Comprehensive Review. IJCDS J. 2023, 13, 911–921. [Google Scholar] [CrossRef]
- Song, Y.; Lu, Y. Decision Tree Methods: Applications for Classification and Prediction. Shanghai Arch. Psychiatry 2015, 27, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, J.R. Induction of Decision Trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Elsevier: Amsterdam, The Netherlands, 2014; ISBN 978-0-08-050058-4. [Google Scholar]
- Breiman, L.; Friedman, J.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Chapman and Hall/CRC: New York, NY, USA, 2017; ISBN 978-1-315-13947-0. [Google Scholar]
- Kass, G.V. An Exploratory Technique for Investigating Large Quantities of Categorical Data. J. R. Stat. Soc. Ser. C Appl. Stat. 1980, 29, 119–127. [Google Scholar] [CrossRef]
- Dewanto, S.; Lukas, J. Expert System For Diagnosis Pest And Disease In Fruit Plants. EPJ Web Conf. 2014, 68, 00024. [Google Scholar] [CrossRef]
- Carisse, O.; Fall, M.L. Decision Trees to Forecast Risks of Strawberry Powdery Mildew Caused by Podosphaera aphanis. Agriculture 2021, 11, 29. [Google Scholar] [CrossRef]
- Pratheepa, M.; Meena, K.; Subramaniam, K.R.; Venugopalan, R.; Bheemanna, H. A Decision Tree Analysis for Predicting the Occurrence of the Pest, Helicoverpa Armigera and Its Natural Enemies on Cotton Based on Economic Threshold Level. Curr. Sci. 2011, 100, 238–246. [Google Scholar]
- Vapnik, V.N.; Chervonenkis, A.Y. On the Uniform Convergence of Relative Frequencies of Events to Their Probabilities. In Measures of Complexity: Festschrift for Alexey Chervonenkis; Vovk, V., Papadopoulos, H., Gammerman, A., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 11–30. ISBN 978-3-319-21852-6. [Google Scholar]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A Training Algorithm for Optimal Margin Classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 1 July 1992; Association for Computing Machinery: New York, NY, USA, 1992; pp. 144–152. [Google Scholar]
- Mokhtar, U.; El Bendary, N.; Hassenian, A.E.; Emary, E.; Mahmoud, M.A.; Hefny, H.; Tolba, M.F. SVM-Based Detection of Tomato Leaves Diseases. In Proceedings of the Intelligent Systems’2014; Filev, D., Jabłkowski, J., Kacprzyk, J., Krawczak, M., Popchev, I., Rutkowski, L., Sgurev, V., Sotirova, E., Szynkarczyk, P., Zadrozny, S., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 641–652. [Google Scholar]
- Ebrahimi, M.A.; Khoshtaghaza, M.H.; Minaei, S.; Jamshidi, B. Vision-Based Pest Detection Based on SVM Classification Method. Comput. Electron. Agric. 2017, 137, 52–58. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, M.; Wan, X.; Zhao, N.; Xiong, W. Image Recognition of Rapeseed Pests Based on Random Forest Classifier. Int. J. Inf. Technol. Web Eng. IJITWE 2017, 12, 1–10. [Google Scholar] [CrossRef]
- Thirumoorthy, S.; Govindarajan, L.; Kesavan, M.; Kumar, T.R. Detection of Pest and Disease in Banana Leaf Using Convolution Random Forest. Test Eng. Manag. 2020, 83, 3727–3735. [Google Scholar]
- Resti, Y.; Irsan, C.; Latif, J.F.; Yani, I.; Dewi, N.R. A Bootstrap-Aggregating in Random Forest Model for Classification of Corn Plant Diseases and Pests|Science and Technology Indonesia. Available online: https://sciencetechindonesia.com/index.php/jsti/article/view/695 (accessed on 20 March 2025).
- Ahmed, M.; Seraj, R.; Islam, S.M.S. The k-means Algorithm: A Comprehensive Survey and Performance Evaluation. Electronics 2020, 9, 1295. [Google Scholar] [CrossRef]
- Faithpraise, F.; Birch, P.; Young, R.; Obu, J.; Faithpraise, B.; Chatwin, C. Automatic Plant Pest Detection and Recognition Using k-Means Clustering Algorithm and Correspondence Filters. Int. J. Adv. Biotechnol. Res. 2013, 4, 189–199. [Google Scholar]
- Xia, J.; Yang, Y.; Cao, H.; Ke, Y.; Ge, D.; Zhang, W.; Ge, S.; Chen, G. Performance Analysis of Clustering Method Based on Crop Pest Spectrum. Eng. Agric. Environ. Food 2018, 11, 84–89. [Google Scholar] [CrossRef]
- Sutton, R.S.; Barto, A. Reinforcement Learning: An Introduction; Adaptive computation and machine learning; Nachdruck; The MIT Press: Cambridge, MA, USA, 2014; ISBN 978-0-262-19398-6. [Google Scholar]
- Lu, Y.; Yu, X.; Hu, Z.; Wang, X. Convolutional Neural Network Combined with Reinforcement Learning-Based Dual-Mode Grey Wolf Optimizer to Identify Crop Diseases and Pests. Swarm Evol. Comput. 2025, 94, 101874. [Google Scholar] [CrossRef]
- Fu, H.; Li, Z.; Zhang, W.; Feng, Y.; Zhu, L.; Fang, X.; Li, J. Research on Path Planning of Agricultural UAV Based on Improved Deep Reinforcement Learning. Agronomy 2024, 14, 2669. [Google Scholar] [CrossRef]
- Kim, K.G. Book Review: Deep Learning. Healthc. Inform. Res. 2016, 22, 351. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of Deep Learning: Concepts, CNN Architectures, Challenges, Applications, Future Directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Bhatt, D.; Patel, C.; Talsania, H.; Patel, J.; Vaghela, R.; Pandya, S.; Modi, K.; Ghayvat, H. CNN Variants for Computer Vision: History, Architecture, Application, Challenges and Future Scope. Electronics 2021, 10, 2470. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Volume 25. [Google Scholar]
- Morankar, D.; Shinde, D.; Pawar, S.; Sabri, M. Identification of Pests and Diseases Using Alex-Net. SSRN Electron. J. 2020, 7, 53–62. [Google Scholar]
- Qiu, J.; Lu, X.; Wang, X.; Chen, C.; Chen, Y.; Yang, Y. Research on Image Recognition of Tomato Leaf Diseases Based on Improved AlexNet Model. Heliyon 2024, 10. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Paymode, A.S.; Malode, V.B. Transfer Learning for Multi-Crop Leaf Disease Image Classification Using Convolutional Neural Network VGG. Artif. Intell. Agric. 2022, 6, 23–33. [Google Scholar] [CrossRef]
- Swasono, D.I.; Tjandrasa, H.; Fathicah, C. Classification of Tobacco Leaf Pests Using VGG16 Transfer Learning. In Proceedings of the 2019 12th International Conference on Information & Communication Technology and System (ICTS), Surabaya, Indonesia, 18 July 2019; pp. 176–181. [Google Scholar]
- Ye, H.; Han, H.; Zhu, L.; Duan, Q. Vegetable Pest Image Recognition Method Based on Improved VGG Convolution Neural Network. J. Phys. Conf. Ser. 2019, 1237, 032018. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich Feature Hierarchies for Accurate Object Detection and Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Girshick, R. Fast R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2015; Volume 28. [Google Scholar]
- Patel, D.; Bhatt, N. Improved Accuracy of Pest Detection Using Augmentation Approach with Faster R-CNN. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1042, 012020. [Google Scholar] [CrossRef]
- Nieuwenhuizen, A.; Hemming, J.; Suh, H.K. Detection and Classification of Insects on Stick-Traps in a Tomato Crop Using Faster R-CNN. In Proceedings of the The Netherlands Conference on Computer Vision, Eindhoven, The Netherlands, 26–27 September 2018. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Terven, J.; Córdova-Esparza, D.-M.; Romero-González, J.-A. A Comprehensive Review of YOLO Architectures in Computer Vision: From YOLOv1 to YOLOv8 and YOLO-NAS. Mach. Learn. Knowl. Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- Diwan, T.; Anirudh, G.; Tembhurne, J.V. Object Detection Using YOLO: Challenges, Architectural Successors, Datasets and Applications|Multimedia Tools and Applications. Multimed. Tools Appl. 2023, 82, 9243–9275. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Tomato Diseases and Pests Detection Based on Improved Yolo V3 Convolutional Neural Network. Front. Plant Sci. 2020, 11, 898. [Google Scholar] [CrossRef]
- Sun, D.; Zhang, K.; Zhong, H.; Xie, J.; Xue, X.; Yan, M.; Wu, W.; Li, J. Efficient Tobacco Pest Detection in Complex Environments Using an Enhanced YOLOv8 Model. Agriculture 2024, 14, 353. [Google Scholar] [CrossRef]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. In Proceedings of the Computer Vision—ECCV 2020; Vedaldi, A., Bischof, H., Brox, T., Frahm, J.-M., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 213–229. [Google Scholar]
- Wu, J.; Wen, C.; Chen, H.; Ma, Z.; Zhang, T.; Su, H.; Yang, C. DS-DETR: A Model for Tomato Leaf Disease Segmentation and Damage Evaluation. Agronomy 2022, 12, 2023. [Google Scholar] [CrossRef]
- Liu, B.; Jia, Y.; Liu, L.; Dang, Y.; Song, S. Skip DETR: End-to-End Skip Connection Model for Small Object Detection in Forestry Pest Dataset. Front. Plant Sci. 2023, 14, 1219474. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Gong, H.; Liu, T.; Luo, T.; Guo, J.; Feng, R.; Li, J.; Ma, X.; Mu, Y.; Hu, T.; Sun, Y.; et al. Based on FCN and DenseNet Framework for the Research of Rice Pest Identification Methods. Agronomy 2023, 13, 410. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Z.; Zhang, S. Segmenting Crop Disease Leaf Image by Modified Fully-Convolutional Networks. In Proceedings of the Intelligent Computing Theories and Application; Huang, D.-S., Bevilacqua, V., Premaratne, P., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 646–652. [Google Scholar]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Lin, T.-L.; Chang, H.-Y.; Chen, K.-H. The Pest and Disease Identification in the Growth of Sweet Peppers Using Faster R-CNN and Mask R-CNN. J. Internet Technol. 2020, 21, 605–614. [Google Scholar]
- Kasinathan, T.; Uyyala, S.R. Detection of Fall Armyworm (Spodoptera Frugiperda) in Field Crops Based on Mask R-CNN. Signal Image Video Process. 2023, 17, 2689–2695. [Google Scholar] [CrossRef]
- Survey on Research of RNN-Based Spatio-Temporal Sequence Prediction Algorithms—ProQuest. Available online: https://www.proquest.com/openview/9ebe553918e3e43d67209a82d3243534/1?cbl=4585453&pq-origsite=gscholar (accessed on 20 March 2025).
- Graves, A.; Schmidhuber, J. Framewise Phoneme Classification with Bidirectional LSTM and Other Neural Network Architectures. Neural Netw. 2005, 18, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Li, W.; Kai, Y.; Chen, P.; Zhang, J.; Wang, B. Occurrence Prediction of Pests and Diseases in Cotton on the Basis of Weather Factors by Long Short Term Memory Network. BMC Bioinform. 2019, 20, 688. [Google Scholar] [CrossRef]
- Chen, C.-J.; Li, Y.-S.; Tai, C.-Y.; Chen, Y.-C.; Huang, Y.-M. Pest Incidence Forecasting Based on Internet of Things and Long Short-Term Memory Network. Appl. Soft Comput. 2022, 124, 108895. [Google Scholar] [CrossRef]
- Wahyono, T.; Heryadi, Y.; Soeparno, H.; Abbas, B.S. Crop Pest Prediction Using Climate Anomaly Model Based on Deep-LSTM Method. ICIC Express letters. Part B Appl. Int. J. Res. Surv. 2021, 12, 395–401. [Google Scholar]
- Fanni, S.C.; Febi, M.; Aghakhanyan, G.; Neri, E. Natural Language Processing. In Introduction to Artificial Intelligence; Klontzas, M.E., Fanni, S.C., Neri, E., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 87–99. ISBN 978-3-031-25928-9. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-Training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; Burstein, J., Doran, C., Solorio, T., Eds.; Association for Computational Linguistics: Minneapolis, MN, USA, 2019; Volume 1, (Long and Short Papers). pp. 4171–4186. [Google Scholar]
- Radford, A.; Narasimhan, K.; Salimans, T.; Sutskever, I. Improving Language Understanding by Generative Pre-Training. 2018. Available online: https://www.mikecaptain.com/resources/pdf/GPT-1.pdf (accessed on 20 March 2025).
- Gururangan, S.; Marasović, A.; Swayamdipta, S.; Lo, K.; Beltagy, I.; Downey, D.; Smith, N.A. Don’t Stop Pretraining: Adapt Language Models to Domains and Tasks. arXiv 2020, arXiv:2004.10964. [Google Scholar]
- Bi, X.; Chen, D.; Chen, G.; Chen, S.; Dai, D.; Deng, C.; Ding, H.; Dong, K.; Du, Q.; Fu, Z.; et al. DeepSeek LLM: Scaling Open-Source Language Models with Longtermism. arXiv 2024, arXiv:2401.02954. [Google Scholar]
- Bai, J.; Bai, S.; Chu, Y.; Cui, Z.; Dang, K.; Deng, X.; Fan, Y.; Ge, W.; Han, Y.; Huang, F.; et al. Qwen Technical Report. arXiv 2023, arXiv:2309.16609. [Google Scholar]
- OpenAI; Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L.; Almeida, D.; Altenschmidt, J.; Altman, S.; et al. GPT-4 Technical Report. arXiv 2023, arXiv:2303.08774. [Google Scholar]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models Are Few-Shot Learners. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 1877–1901. [Google Scholar]
- Touvron, H.; Martin, L.; Stone, K.; Albert, P.; Almahairi, A.; Babaei, Y.; Bashlykov, N.; Batra, S.; Bhargava, P.; Bhosale, S.; et al. Llama 2: Open Foundation and Fine-Tuned Chat Models. arXiv 2023, arXiv:2307.09288. [Google Scholar]
- Grattafiori, A.; Dubey, A.; Jauhri, A.; Pandey, A.; Kadian, A.; Al-Dahle, A.; Letman, A.; Mathur, A.; Schelten, A.; Vaughan, A.; et al. The Llama 3 Herd of Models. arXiv 2024, arXiv:2407.21783. [Google Scholar]
- Scheepens, D.; Millard, J.; Farrell, M.; Newbold, T. Large Language Models Help Facilitate the Automated Synthesis of Information on Potential Pest Controllers. Methods Ecol. Evol. 2024, 15, 1261–1273. [Google Scholar] [CrossRef]
- Tzachor, A.; Devare, M.; Richards, C.; Pypers, P.; Ghosh, A.; Koo, J.; Johal, S.; King, B. Large Language Models and Agricultural Extension Services. Nat. Food 2023, 4, 941–948. [Google Scholar] [CrossRef]
- Yuan, Z.; Liu, K.; Peng, R.; Li, S.; Leybourne, D.; Musa, N.; Huang, H.; Yang, P. PestGPT: Leveraging Large Language Models and IoT for Timely and Customized Recommendation Generation in Sustainable Pest Management. IEEE Internet Things Mag. 2025, 8, 26–33. [Google Scholar] [CrossRef]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning Transferable Visual Models From Natural Language Supervision. In Proceedings of the Proceedings of the 38th International Conference on Machine Learning; Virtual, 8–24 July 2021, PMLR; pp. 8748–8763.
- Liu, J.; Xing, J.; Zhou, G.; Wang, J.; Sun, L.; Chen, X. Transfer Large Models to Crop Pest Recognition—a Cross-Modal Unified Framework for Parameters Efficient Fine-Tuning. 2024. Available online: https://papers.ssrn.com/abstract=4999751 (accessed on 20 March 2025).
- Wang, L.; Jin, T.; Yang, J.; Leonardis, A.; Wang, F.; Zheng, F. Agri-LLaVA: Knowledge-Infused Large Multimodal Assistant on Agricultural Pests and Diseases. arXiv 2024, arXiv:2412.02158. [Google Scholar]
- Li, J.; Chen, D.; Qi, X.; Li, Z.; Huang, Y.; Morris, D.; Tan, X. Label-Efficient Learning in Agriculture: A Comprehensive Review. Comput. Electron. Agric. 2023, 215, 108412. [Google Scholar] [CrossRef]
- Sapkota, R.; Qureshi, R.; Hassan, S.Z.; Shutske, J.; Shoman, M.; Sajjad, M.; Dharejo, F.A.; Paudel, A.; Li, J.; Meng, Z.; et al. Multi-Modal LLMs in Agriculture: A Comprehensive Review. TechRxiv 2024, 1–28. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Jin, Z.; Müller, C.; Pugh, T.A.M.; Chen, A.; Wang, T.; Huang, L.; Zhang, Y.; Li, L.X.Z.; et al. Occurrence of Crop Pests and Diseases Has Largely Increased in China since 1970. Nat. Food 2022, 3, 57–65. [Google Scholar] [CrossRef]
- Zhao, W.X.; Zhou, K.; Li, J.; Tang, T.; Wang, X.; Hou, Y.; Min, Y.; Zhang, B.; Zhang, J.; Dong, Z.; et al. A Survey of Large Language Models. arXiv 2025, arXiv:2303.18223. [Google Scholar]
| Category | Supervised Learning | Unsupervised Learning |
|---|---|---|
| Data Labeling | Requires labeled data | No labeled data required |
| Task Objectives | Prediction, Classification | Discovering intrinsic data structures or patterns |
| Common Tasks | Classification, Regression | Clustering, Dimensionality Reduction |
| Classic Algorithms | Decision Trees, Support Vector Machines, Random Forests | K-means Clustering |
| Algorithm | Feature Selection Criterion | Tree Structure | Handling Continuous Features | Pruning Strategy | Reference |
|---|---|---|---|---|---|
| ID3 | Information Gain | Multi-way Tree | Manual Discretization Required | none | [29] |
| C4.5 | Gain Ratio | Multi-way Tree | Automatic Binary Splitting | Pessimistic Error Postpruning | [30] |
| CART | Gini Index | Binary Tree | Automatically Finds Optimal Binary Split | Cost Complexity Postpruning | [31] |
| CHAID | Chi-square, F-Test | Multi-way Tree | Manual Binning and Interval Merging | Significance-based Pre-pruning | [32] |
| Algorithm | Region Proposal Method | Feature Extraction Strategy |
|---|---|---|
| R-CNN | Selective Search | Extract features independently for each region |
| Fast R-CNN | Selective Search | Share feature maps |
| Fast R-CNN | Region Proposal Network | Share feature maps |
| Model | Research Institution | Core Strengths | Potential Agricultural Applications | References |
|---|---|---|---|---|
| DeepSeek | DeepSeek | Strong Chinese–English reasoning capability, deep integration of agricultural knowledge | Cross-regional pest warning, precision agriculture decision support | [89] |
| Qwen | Alibaba | Strong Chinese adaptation, supports localized deployment | Agricultural technology dissemination, farmer training | [90] |
| Chat-GPT | OpenAI | Powerful multimodal reasoning, extensive knowledge base | Global agricultural knowledge integration, intelligent agricultural Q&A | [91,92] |
| Llama | Meta | Open-source with customizable local fine-tuning, suitable for agricultural optimization | Agricultural monitoring and customized model optimization | [93,94] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, J.; Lu, M.; Gao, Q.; Chen, L.; Zou, Y.; Wu, J.; Cao, Y.; Xu, N.; Wang, W.; Li, J. Intelligent Detection and Control of Crop Pests and Diseases: Current Status and Future Prospects. Agronomy 2025, 15, 1416. https://doi.org/10.3390/agronomy15061416
Xie J, Lu M, Gao Q, Chen L, Zou Y, Wu J, Cao Y, Xu N, Wang W, Li J. Intelligent Detection and Control of Crop Pests and Diseases: Current Status and Future Prospects. Agronomy. 2025; 15(6):1416. https://doi.org/10.3390/agronomy15061416
Chicago/Turabian StyleXie, Jiaxing, Meiyi Lu, Qunpeng Gao, Liye Chen, Yingxin Zou, Jiatao Wu, Yue Cao, Niechong Xu, Weixing Wang, and Jun Li. 2025. "Intelligent Detection and Control of Crop Pests and Diseases: Current Status and Future Prospects" Agronomy 15, no. 6: 1416. https://doi.org/10.3390/agronomy15061416
APA StyleXie, J., Lu, M., Gao, Q., Chen, L., Zou, Y., Wu, J., Cao, Y., Xu, N., Wang, W., & Li, J. (2025). Intelligent Detection and Control of Crop Pests and Diseases: Current Status and Future Prospects. Agronomy, 15(6), 1416. https://doi.org/10.3390/agronomy15061416







