Genome-Wide Dissection of Shade Tolerance in Soybean at Seedling Stage
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials, Design, and Phenotypic Measurements
2.2. Statistical Analysis of Phenotype Data
2.3. SNP Linkage Disequilibrium (LD), PCA, and Kinship
2.4. Genome-Wide Association Analysis and Candidate Genes Identification
3. Results
3.1. Descriptive Statistics of Shade-Tolerance-Related Traits
3.2. LD, PCA, and Kinship
3.3. Light Intensity Effects on Soybean Plant Height in Control Group
3.4. Finding Superior Shade-Tolerant Soybean Germplasm
3.5. GWAS and Proposed QTLs and Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “green revolution” for soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Assefa, Y.; Carter, P.; Hinds, M.; Bhalla, G.; Schon, R.; Jeschke, M.; Paszkiewicz, S.; Smith, S.; Ciampitti, I.A. Analysis of long term study indicates both agronomic optimal plant density and increase maize yield per plant contributed to yield gain. Sci. Rep. 2018, 8, 4937. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.-Y.; Li, Y.-H.; Zhang, J.-W.; Peng, L.; Bin, Z.; Dong, S.-T. Increased plant density and reduced N rate lead to more grain yield and higher resource utilization in summer maize. J. Integr. Agric. 2016, 15, 2515–2528. [Google Scholar] [CrossRef]
- Du, X.; Wang, Z.; Lei, W.; Kong, L. Increased planting density combined with reduced nitrogen rate to achieve high yield in maize. Sci. Rep. 2021, 11, 358. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Liang, Y.; Liu, Q.; Zeng, J.; Ren, Q.; Guo, J.; Xiong, F.; Lu, D. Enhancing production efficiency through optimizing plant density in maize–soybean strip intercropping. Front. Plant Sci. 2024, 15, 1473786. [Google Scholar] [CrossRef]
- Finlayson, S.A.; Krishnareddy, S.R.; Kebrom, T.H.; Casal, J.J. Phytochrome regulation of branching in Arabidopsis. Plant Physiol. 2010, 152, 1914–1927. [Google Scholar] [CrossRef]
- Franklin, K.A.; Whitelam, G.C. Phytochromes and shade-avoidance responses in plants. Ann. Bot. 2005, 96, 169–175. [Google Scholar] [CrossRef]
- Reed, J.W.; Nagpal, P.; Poole, D.S.; Furuya, M.; Chory, J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 1993, 5, 147–157. [Google Scholar] [CrossRef]
- Casal, J.J. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef]
- Chen, M.; Chory, J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011, 21, 664–671. [Google Scholar] [CrossRef]
- Suetsugu, N.; Wada, M. Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: Phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol. 2013, 54, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Lin, C. Plant blue-light receptors. Trends Plant Sci. 2000, 5, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, G.I. The UV-B photoreceptor UVR8: From structure to physiology. Plant Cell 2014, 26, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Shui, Z.; Xu, L.; Yang, Y.; Li, Y.; Yuan, X.; Shang, J.; Asghar, M.A.; Wu, X.; Yu, L. Gibberellins modulate shade-induced soybean hypocotyl elongation downstream of the mutual promotion of auxin and brassinosteroids. Plant Physiol. Biochem. 2020, 150, 209–221. [Google Scholar] [CrossRef]
- Morelli, G.; Ruberti, I. Shade avoidance responses. Driving auxin along lateral routes. Plant Physiol. 2000, 122, 621–626. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Gao, Y.; Guo, Y.; Zheng, N.; Xu, X.; Xu, M.; Wang, W.; Liu, C.; Liu, W. Genome-wide identification of SMXL Gene family in soybean and expression analysis of GmSMXLs under Shade stress. Plants 2022, 11, 2410. [Google Scholar] [CrossRef]
- Jia, Q.; Hu, S.; Li, X.; Wei, L.; Wang, Q.; Zhang, W.; Zhang, H.; Liu, X.; Chen, X.; Wang, X. Identification of candidate genes and development of KASP markers for soybean shade-tolerance using GWAS. Front. Plant Sci. 2024, 15, 1479536. [Google Scholar] [CrossRef]
- Su, Y.; Hao, X.; Zeng, W.; Lai, Z.; Pan, Y.; Wang, C.; Guo, P.; Zhang, Z.; He, J.; Xing, G. Genome-wide association with transcriptomics reveals a shade-tolerance gene network in soybean. Crop J. 2024, 12, 232–243. [Google Scholar] [CrossRef]
- Su, Y.; Zhang, Z.; He, J.; Zeng, W.; Cai, Z.; Lai, Z.; Pan, Y.; Hao, X.; Xing, G.; Wang, W. Gene–allele system of shade tolerance in southern China soybean germplasm revealed by genome-wide association study using gene–allele sequence as markers. Theor. Appl. Genet. 2023, 136, 152. [Google Scholar] [CrossRef]
- Lyu, X.; Cheng, Q.; Qin, C.; Li, Y.; Xu, X.; Ji, R.; Mu, R.; Li, H.; Zhao, T.; Liu, J. GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 2021, 14, 298–314. [Google Scholar] [CrossRef]
- Qin, C.; Li, Y.-h.; Li, D.; Zhang, X.; Kong, L.; Zhou, Y.; Lyu, X.; Ji, R.; Wei, X.; Cheng, Q. PH13 improves soybean shade traits and enhances yield for high-density planting at high latitudes. Nat. Commun. 2023, 14, 6813. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Meng, S.; Zhao, T.; Xing, G.; Yang, S.; Li, Y.; Guan, R.; Lu, J.; Wang, Y.; Xia, Q. An innovative procedure of genome-wide association analysis fits studies on germplasm population and plant breeding. Theor. Appl. Genet. 2017, 130, 2327–2343. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Kolde, R. Package ‘pheatmap’. R Package 2015, 1, 790. [Google Scholar]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef]
- Wen, Z.; Tan, R.; Yuan, J.; Bales, C.; Du, W.; Zhang, S.; Chilvers, M.I.; Schmidt, C.; Song, Q.; Cregan, P.B. Genome-wide association mapping of quantitative resistance to sudden death syndrome in soybean. BMC Genom. 2014, 15, 809. [Google Scholar] [CrossRef]
- Wang, K.; Guo, H.; Yin, Y. AP2/ERF transcription factors and their functions in Arabidopsis responses to abiotic stresses. Environ. Exp. Bot. 2024, 222, 105763. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, X.; Chen, L.; Chen, Q.; Tian, X.; Ai, L.; Zhao, H.; Yang, C.; Yan, L.; Zhang, M. Genetic and transcriptome analyses reveal the candidate genes and pathways involved in the inactive shade-avoidance response enabling high-density planting of soybean. Front. Plant Sci. 2022, 13, 973643. [Google Scholar] [CrossRef]
- Cheng, B.; Wang, L.; Liu, R.; Wang, W.; Yu, R.; Zhou, T.; Ahmad, I.; Raza, A.; Jiang, S.; Xu, M. Shade-tolerant soybean reduces yield loss by regulating its canopy structure and stem characteristics in the maize–soybean strip intercropping system. Front. Plant Sci. 2022, 13, 848893. [Google Scholar] [CrossRef] [PubMed]
- Jiang, A.; Liu, J.; Gao, W.; Ma, R.; Zhang, J.; Zhang, X.; Du, C.; Yi, Z.; Fang, X.; Zhang, J. Transcriptomic and metabolomic analyses reveal the key genes related to shade tolerance in soybean. Int. J. Mol. Sci. 2023, 24, 14230. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; He, J.; Zhao, T.; Xing, G.; Wang, Y.; Yu, D.; Chen, S.; Gai, J. Efficient QTL detection of flowering date in a soybean RIL population using the novel restricted two-stage multi-locus GWAS procedure. Theor. Appl. Genet. 2018, 131, 2581–2599. [Google Scholar] [CrossRef] [PubMed]
- Gai, J.-Y.; He, J.-B. Major characteristics, often-raised queries and potential usefulness of the restricted two-stage multi-locus genome-wide association analysis. Sci. Agric. Sin. 2020, 53, 1699–1703. [Google Scholar] [CrossRef]
- DeMers, L.C.; Redekar, N.R.; Kachroo, A.; Tolin, S.A.; Li, S.; Saghai Maroof, M.A. A transcriptional regulatory network of Rsv3-mediated extreme resistance against soybean mosaic virus. PLoS ONE 2020, 15, e0231658. [Google Scholar] [CrossRef]
- Li, L.; Li, X.; Yang, C.; Cheng, Y.; Cai, Z.; Nian, H.; Ma, Q. GsERF1 enhances Arabidopsis thaliana aluminum tolerance through an ethylene-mediated pathway. BMC Plant Biol. 2022, 22, 258. [Google Scholar] [CrossRef]
- Jhan, L.-H.; Yang, C.-Y.; Huang, C.-M.; Lai, M.-C.; Huang, Y.-H.; Baiya, S.; Kao, C.-F. Integrative pathway and network analysis provide insights on flooding-tolerance genes in soybean. Sci. Rep. 2023, 13, 1980. [Google Scholar] [CrossRef]
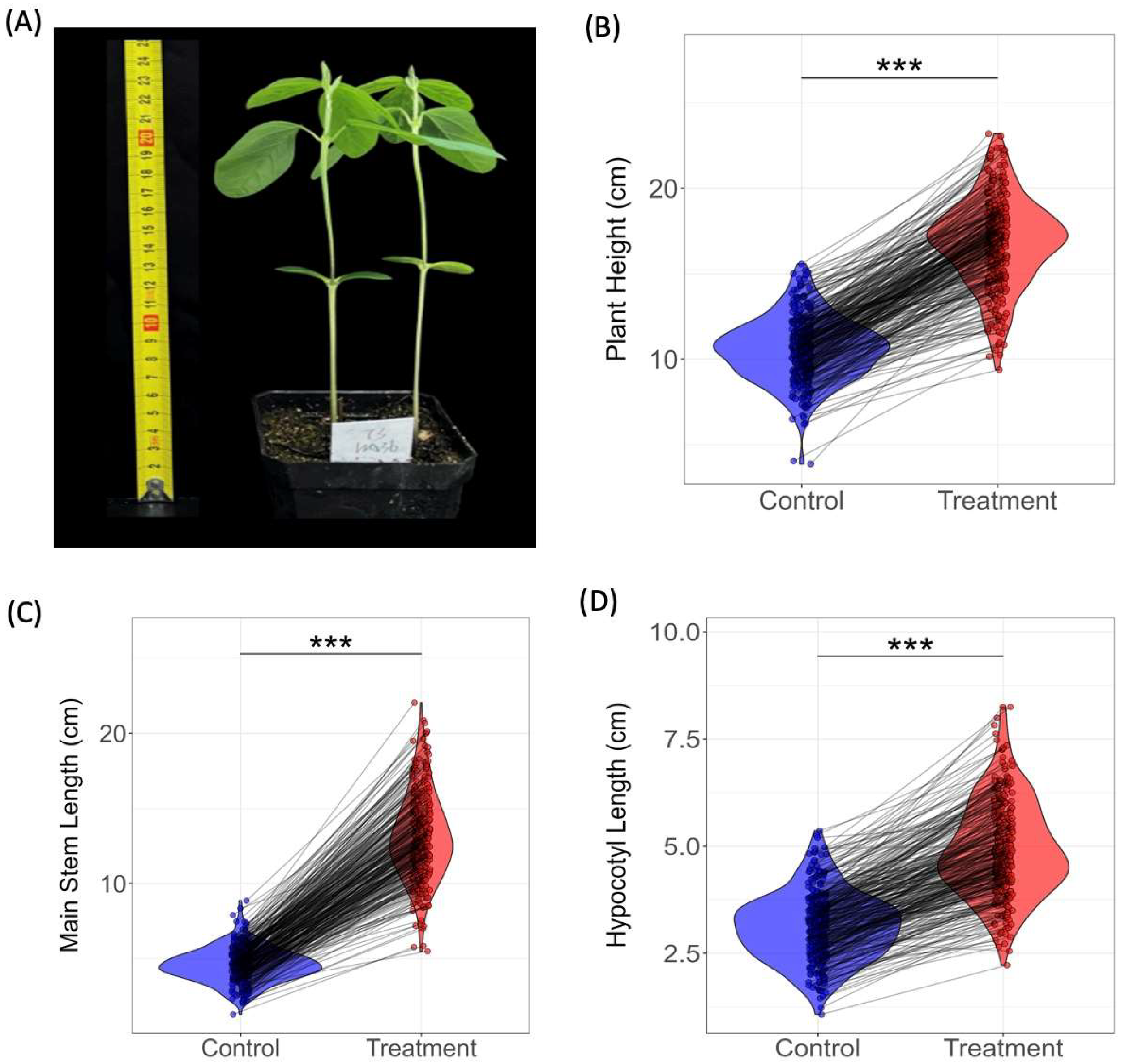

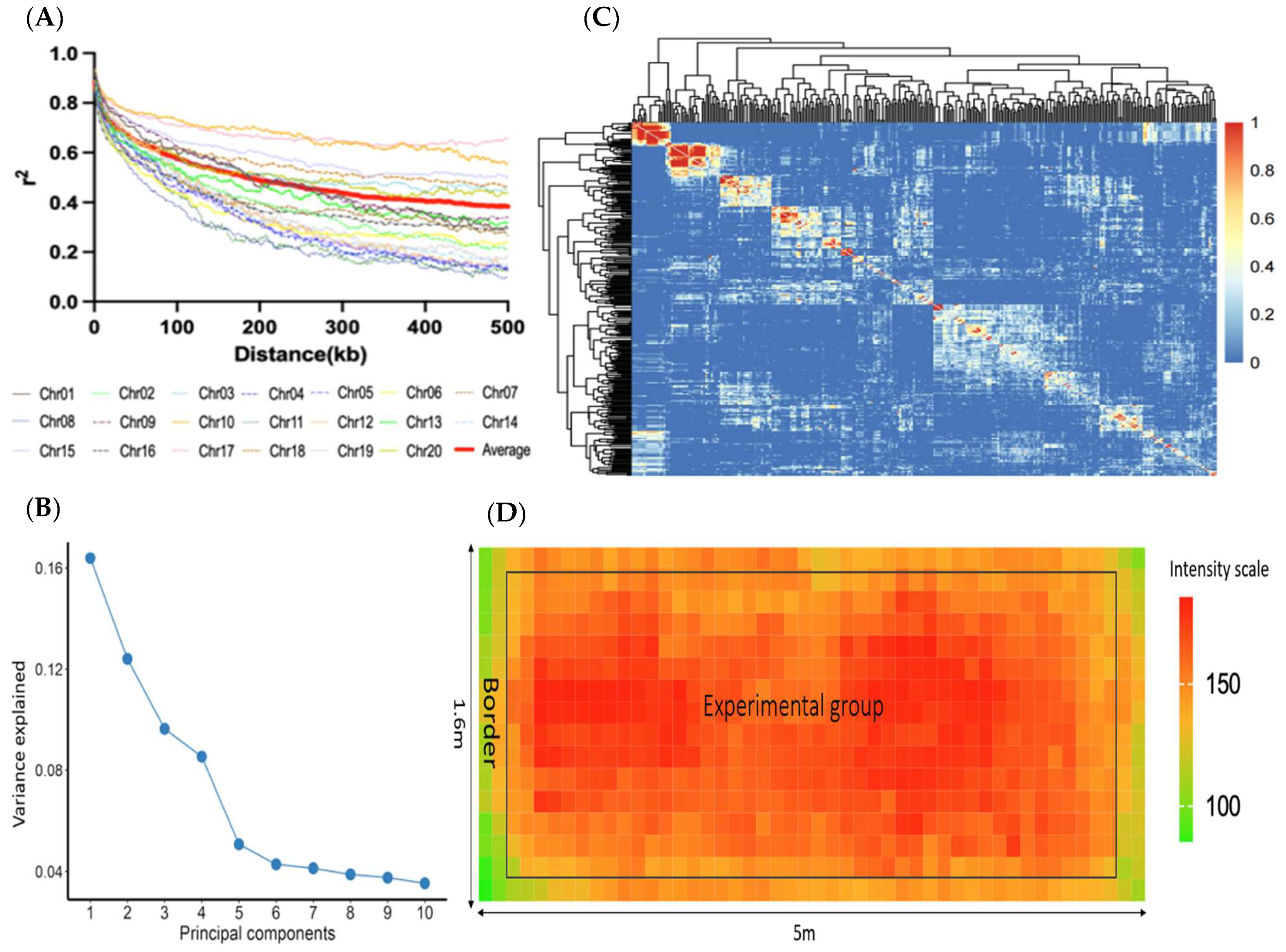

| Trait | Min | Max | Mean | 1 SD | 2 PCV (%) | 3 H2 (%) |
|---|---|---|---|---|---|---|
| PH_0 | 3.32 | 12.32 | 7.66 | 1.76 | 22.97 | 83.81 |
| PH_75 | 8.8 | 27.32 | 18.08 | 4.94 | 27.33 | 77.13 |
| PH_r | 1.3 | 5.41 | 2.43 | 0.75 | 30.71 | 57.49 |
| MSL_0 | 1.3 | 8.86 | 4.56 | 1.23 | 26.99 | 80.06 |
| MSL_75 | 5.5 | 24.05 | 13.18 | 4.19 | 31.82 | 73.29 |
| MSL_r | 1.01 | 8.37 | 3.01 | 1.08 | 35.79 | 50 |
| HL_0 | 0.68 | 5.36 | 3.10 | 0.88 | 28.48 | 73.88 |
| HL_75 | 2.22 | 8.25 | 4.92 | 1.56 | 31.77 | 67.75 |
| HL_r | 0.66 | 4.73 | 1.66 | 0.61 | 36.85 | 31.66 |
| Source of Variation | DF | MSE | F | p | η2 |
|---|---|---|---|---|---|
| Intercept | 1 | 464.79 | 73.94 | 0 | 0.21 |
| Light | 1 | 1.91 | 0.31 | 0.58 | 0.001 |
| Accession | 306 | 18.22 | 2.90 | 0 | 0.76 |
| Error | 284 | 6.29 | |||
| Total | 592 |
| Trait | QTLs | SNPLDBs | Chr. | Model p Value | Allele No. | SNP No. | QTL Main Effect | Candidate Genes | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| −log10(p) | R2 (%) | ZH13 Gene Number | W82.a2 Gene Number | Description | |||||||
| PH_r | |||||||||||
| PH_r_shade.1.1 | Block_1_11646785_11746784 | 1 | 8.23 × 10−5 | 11 | 320 | 3.66 | 3.78 | SoyZH13_01G064100 | Glyma.01G068600 | Transcription factor bHLH49 | |
| PH_r_shade.3.1 | Block_3_3114852_3115149 | 3 | 1.38 × 10−11 | 4 | 7 | 7.25 | 5.76 | ||||
| PH_r_shade.10.1 | Block_10_4801122_4860661 | 10 | 0.001718 | 8 | 219 | 2.68 | 2.39 | SoyZH13_10G051100 | Glyma.10G053500 | Auxin response factor 16 | |
| PH_r_shade.12.1 | Block_12_37743951_37744542 | 12 | 0.000161 | 2 | 4 | 3.63 | 1.47 | ||||
| PH_r_shade.15.1 | Block_15_23412759_23416902 | 15 | 3.06 × 10−6 | 5 | 56 | 4.52 | 3.25 | ||||
| PH_r_shade.16.1 | Block_16_33893242_33893951 | 16 | 3.29 × 10−18 | 6 | 24 | 10.89 | 10.18 | ||||
| PH_r_shade.16.2 | Block_16_35747737_35749257 | 16 | 4.49 × 10−9 | 8 | 12 | 6.04 | 5.62 | ||||
| PH_r_shade.16.3 | Chr16_36081826 | 16 | 0.000499 | 2 | 2 | 3.3 | 1.25 | ||||
| PH_r_shade.17.1 | Block_17_7316304_7397881 | 17 | 2.52 × 10−5 | 5 | 333 | 3.91 | 2.77 | ||||
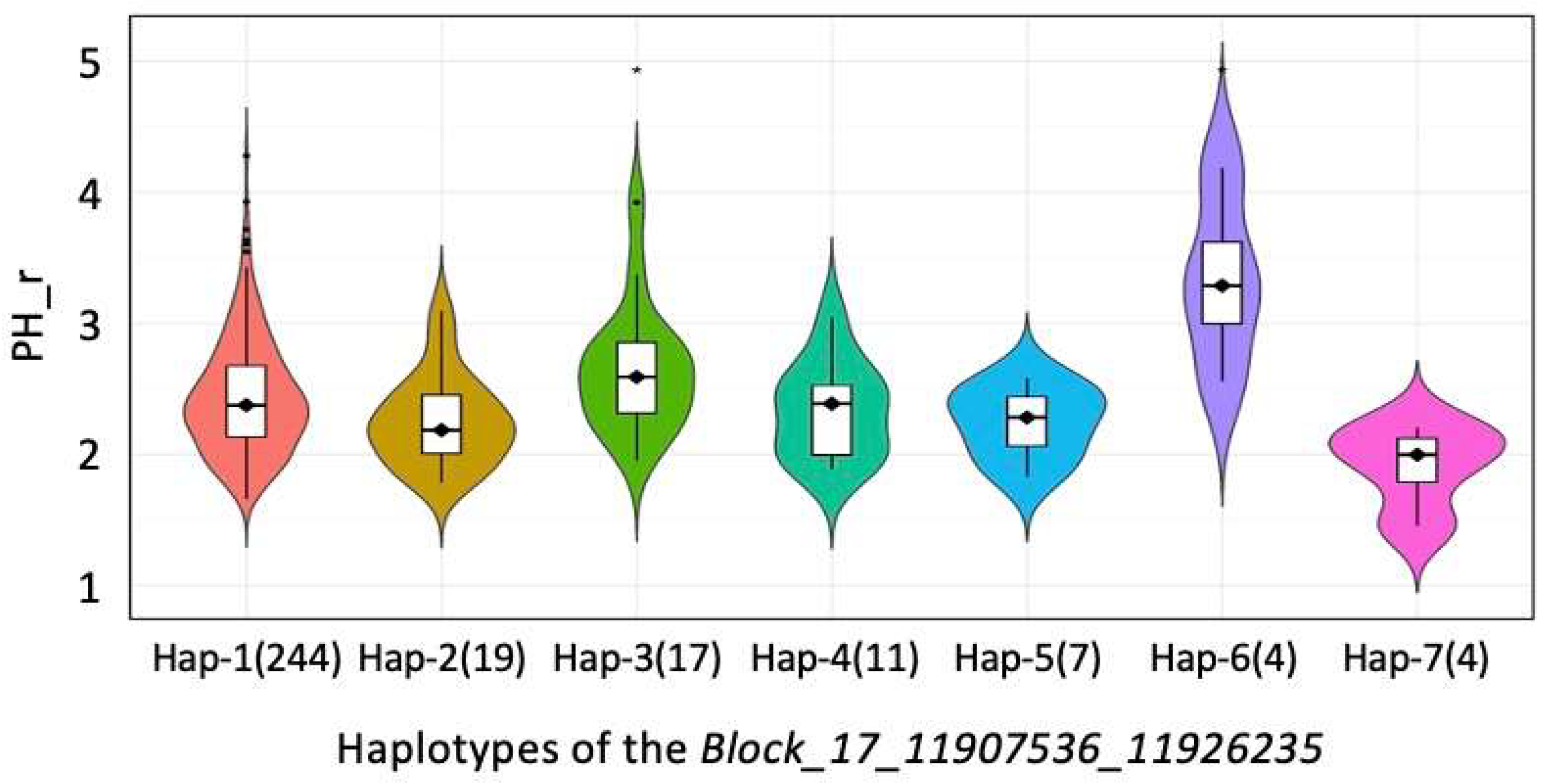
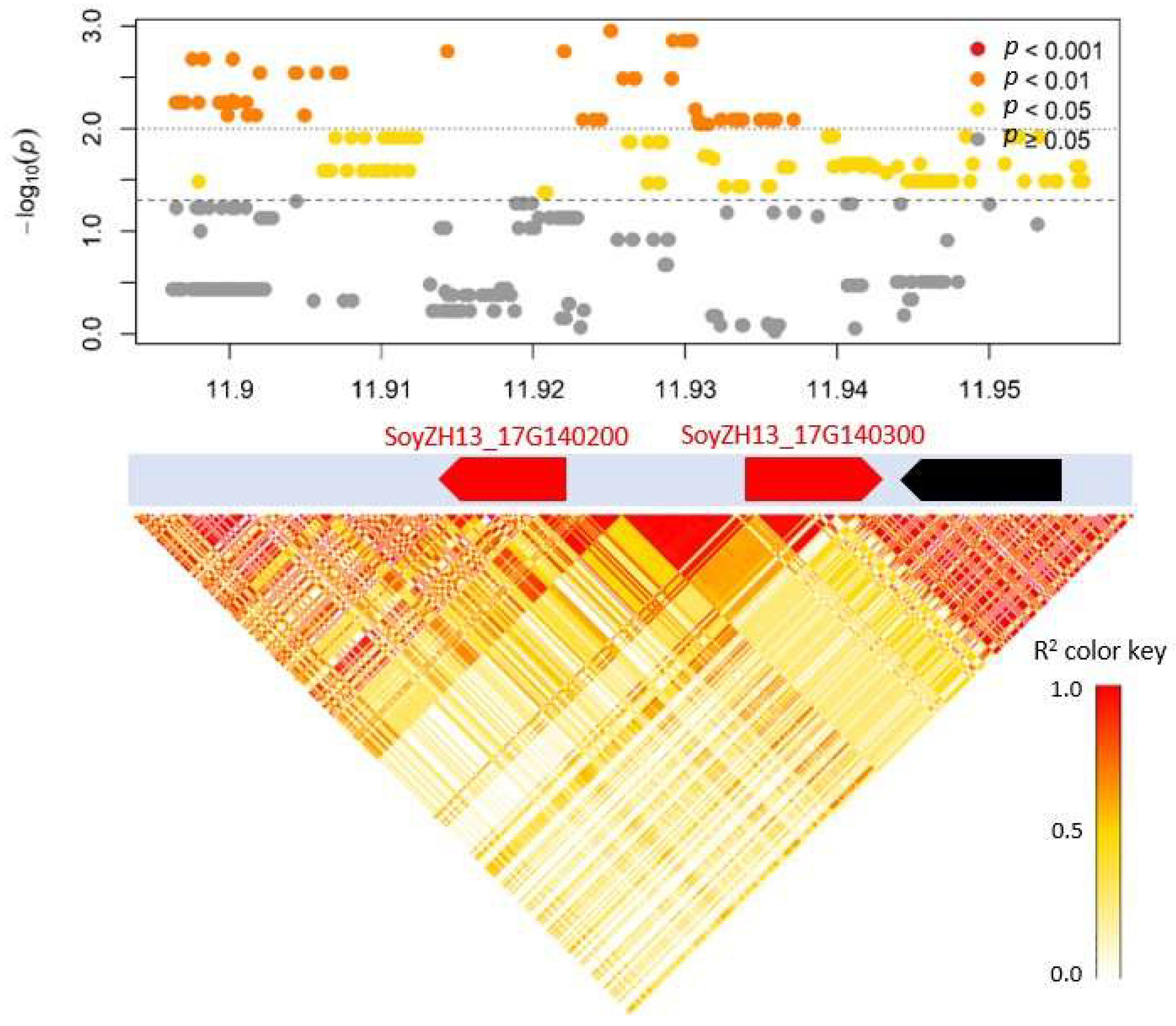
| PH_r_shade.17.2 | Block_17_11907536_11926235 | 17 | 2.2 × 10−7 | 7 | 134 | 5.12 | 4.4 | SoyZH13_17G140200 | Glyma.17G145300 | Ethylene-responsive transcription factor 5 | |
| SoyZH13_17G140300 | Glyma.17G145400 | Ethylene-responsive transcription factor 1A | |||||||||
| PH_r_shade.17.3 | Block_17_11933118_12033106 | 17 | 1.04 × 10−10 | 11 | 507 | 6.56 | 7.37 | ||||
| PH_r_shade.19.1 | Chr19_478140 | 19 | 9.47 × 10−5 | 2 | 2 | 3.63 | 1.58 | ||||
| MSL_r | |||||||||||
| MSL_r_shade.3.1 | Block_3_3253737_3254065 | 3 | 1.38 × 10−10 | 4 | 5 | 7.34 | 6.36 | ||||
| MSL_r_shade.4.1 | Block_4_45771799_45788317 | 4 | 1.10 × 10−12 | 3 | 147 | 8.57 | 7.21 | ||||
| MSL_r_shade.8.1 | Block_8_15201156_15201636 | 8 | 2.33 × 10−5 | 3 | 3 | 4.20 | 2.71 | ||||
| MSL_r_shade.11.1 | Block_11_4331465_4356051 | 11 | 5.16 × 10−9 | 6 | 62 | 6.86 | 6.12 | ||||
| MSL_r_shade.12.1 | Chr12_1168266 | 12 | 9.72 × 10−7 | 2 | 2 | 5.35 | 3.05 | ||||
| MSL_r_shade.16.1 | Chr16_34243799 | 16 | 0.000268 | 2 | 2 | 3.42 | 1.68 | ||||
| MSL_r_shade.17.1 | Block_17_11907536_11926235 | 17 | 5.3 × 10−9 | 7 | 134 | 5.12 | 4.4 | SoyZH13_17G140200 | Glyma.17G145300 | Ethylene-responsive transcription factor 5 | |
| SoyZH13_17G140300 | Glyma.17G145400 | Ethylene-responsive transcription factor 1A | |||||||||
| MSL_r_shade.18.1 | Chr18_55533676 | 18 | 0.000368 | 2 | 2 | 3.35 | 1.60 | SoyZH13_18G217700 | Glyma.18G246000 | Transcription factor bHLH25 | |
| MSL_r_shade.19.1 | Block_19_476384_476514 | 19 | 5.79 × 10−5 | 3 | 4 | 3.95 | 2.47 | ||||
| MSL_r_shade.20.1 | Block_20_1172558_1173696 | 20 | 0.00082 | 7 | 10 | 3.09 | 2.90 | SoyZH13_20G012300 | Glyma.20G013200 | U-box domain-containing protein 10 | |
| HL_r | |||||||||||
| HL_r_shade.1.1 | Block_1_55630414_55715065 | 1 | 4.04 × 10−11 | 11 | 224 | 8.09 | 10.38 | ||||
| HL_r_shade.7.1 | Block_7_6887632_6888597 | 7 | 4.32 × 10−5 | 3 | 13 | 4.36 | 2.86 | ||||
| HL_r_shade.7.2 | Block_7_27001385_27047263 | 7 | 6.66 × 10−6 | 7 | 44 | 4.97 | 4.88 | ||||
| HL_r_shade.10.1 | Block_10_8406436_8478177 | 10 | 1.21 × 10−6 | 9 | 370 | 5.10 | 6.10 | ||||
| HL_r_shade.13.1 | Block_13_35550086_35550267 | 13 | 5.67 × 10−6 | 2 | 2 | 4.89 | 2.93 | SoyZH13_13G214700 | Glyma.13G236500 | Ethylene-responsive transcription factor 9 | |
| HL_r_shade.20.1 | Chr20_18734811 | 20 | 1.36 × 10−6 | 2 | 2 | 5.07 | 3.33 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Arshad, K.; Zheng, M.; Ou, R.; Song, Y.; Xie, M.; Wei, Y.; Ling, L.; Zeng, W.; Zhang, J. Genome-Wide Dissection of Shade Tolerance in Soybean at Seedling Stage. Agronomy 2025, 15, 1382. https://doi.org/10.3390/agronomy15061382
Hu L, Arshad K, Zheng M, Ou R, Song Y, Xie M, Wei Y, Ling L, Zeng W, Zhang J. Genome-Wide Dissection of Shade Tolerance in Soybean at Seedling Stage. Agronomy. 2025; 15(6):1382. https://doi.org/10.3390/agronomy15061382
Chicago/Turabian StyleHu, Linfang, Kamran Arshad, Meiying Zheng, Ran Ou, Yinmeng Song, Mengyan Xie, Yazhi Wei, Luyi Ling, Weiying Zeng, and Jiaoping Zhang. 2025. "Genome-Wide Dissection of Shade Tolerance in Soybean at Seedling Stage" Agronomy 15, no. 6: 1382. https://doi.org/10.3390/agronomy15061382
APA StyleHu, L., Arshad, K., Zheng, M., Ou, R., Song, Y., Xie, M., Wei, Y., Ling, L., Zeng, W., & Zhang, J. (2025). Genome-Wide Dissection of Shade Tolerance in Soybean at Seedling Stage. Agronomy, 15(6), 1382. https://doi.org/10.3390/agronomy15061382





