Abstract
To elucidate the molecular mechanisms that underlie jujube (Ziziphus jujuba) flavor synthesis, we integrated transcriptomic and metabolomic analyses on the ‘Lingwuchangzao’ cultivar across seven developmental stages. Our multi-omics approach detected 750 metabolites, categorized into 11 primary and 35 secondary classes, with K-means clustering revealing significant stage-specific variations in sugars, alcohols, and organic acids. KEGG enrichment analysis identified differentially expressed genes (DEGs) in key metabolic pathways, including carbohydrate metabolism and plant hormone signal transduction, showing dynamic changes during development. Weighted gene co-expression network analysis (WGCNA) further pinpointed gene networks related to starch/sucrose and carbon metabolism, and eight novel genes linked to starch and fatty acid metabolism. Notably, the white ripening stage (BS) emerged as the critical phase for flavor compound accumulation, offering new molecular insights and targets for quality improvement.
1. Introduction
Fruit flavor is a crucial determinant of consumer preferences and directly influences its economic value. However, the metabolic dynamics and underlying gene regulatory networks that regulate flavor formation during fruit development remain largely unknown in many fruit species. Jujube (Ziziphus jujuba), a member of the Rhamnaceae family, has been cultivated in China for over 8000 years and holds both medicinal and culinary value. Traditionally, jujube cultivars are classified into three categories: dried, fresh, and dual-purpose. Among them, dried jujube has the highest production in China, while fresh jujube is prized for its rich nutritional content and crisp texture [1]. Jujube fruits are abundant in carbohydrates, triterpenoids, flavonoids, vitamins, and essential minerals such as phosphorus, calcium, and iron, underscoring their high nutritional and medicinal value. Understanding the dynamic changes in these compounds during fruit ripening is essential for both scientific research and the genetic improvement of jujube cultivars.
Flavor quality, a key trait that inflences consumer preference, is primarily determined by the sugar and organic acid composition [2,3]. During jujube domestication, genetic selection has shaped fruit sweetness and acidity by modulating sugar and acid accumulation [4]. Studies on sugar composition in jujube indicate that fructose and glucose predominate in the early developmental stages, whereas sucrose accumulates in later stages [5,6]. Organic acid profiling across 219 jujube germplasm accessions identified malic, quinic, and citric acids as the primary acid components [7]. These sugars and acids are metabolized through glycolysis and the tricarboxylic acid (TCA) cycle, forming key intermediates that contribute to fruit flavor. Compared to wild jujube and other fruit crops (e.g., apples, peaches, and grapes), cultivated jujube exhibits significantly higher sugar content [8]. The balance between sugar sources and sinks plays a pivotal role in determining sugar accumulation, making it essential to investigate the metabolite profiles and biosynthetic pathways regulating sugar and organic acid dynamics in jujube.
Advancements in omics technologies have facilitated the elucidation of regulatory mechanisms that govern fruit quality, including sugar metabolism, anthocyanin accumulation, and nutrient composition. Transcriptomic and metabolomic approaches have been successfully applied to study apple and jujube leaf color, fruit flavonoid biosynthesis, and nutrient accumulation [9,10,11]. In the pitaya fruit, betaine biosynthesis has been identified as a key pathway that regulates fruit peel and pulp coloration [12]. In watermelon, integrated multi-omics analysis revealed the role of UDP-glycosyltransferases in cucurbitacin glycosylation and sugar accumulation, distinguishing cultivated varieties from their wild counterparts [13,14]. Similar approaches have been used to investigate sugar, acid, and ascorbic acid accumulation in yellow kiwifruit [15] and sugar-acid metabolism during cherry ripening [16]. Despite these advancements, the complex nature of jujube flavor composition remains largely unexplored, necessitating further studies on its molecular basis.
In this study, we investigate the flavor formation mechanisms of ‘Lingwuchangzao’, a distinctive jujube cultivar from Lingwu City, Ningxia Hui Autonomous Region, recognized as a Chinese National Geographic Indication Product. This cultivar exhibits significant changes in flavor over a short period, yet the regulatory mechanisms governing its flavor development remain unclear. To address this gap, we performed a comprehensive multi-omics analysis across seven fruit developmental stages, integrating transcriptomic and metabolomic sequencing. Through this approach, we identified key metabolic pathways and genes regulating sugar and organic acid accumulation, providing novel insights into the molecular networks underlying jujube flavor formation. This is the first study to integrate transcriptomic and metabolomic data to elucidate the genetic and biochemical basis of ‘Lingwuchangzao’ flavor dynamics, offering valuable targets for future breeding and quality improvement strategies.
2. Materials and Methods
2.1. Plant Materials
Jujube fruits (Ziziphus jujuba ‘Lingwuchangzao’) were collected from an open-field orchard at Fucheng Jujube Industry, Lingwu City, Ningxia, between 7 July and 10 October 2022. This period covered seven developmental stages: the young fruit stage (YG), the hard core stage (YH), the early expansion stage (PQ), the late expansion stage (PH), the white ripening stage (BS), the half-red stage (BH), and the full-red stage (QH). At the start of the experiment, red ropes were used to mark four directions (east, south, west, and north) at a uniform height on selected trees. Each biological replicate comprised 5 trees, with 8 fruits per tree sampled across 4 canopy positions (2 fruits/position). Metabolomics used pooled extracts (equal volumes from individual fruits), while transcriptomics analyzed 4 representative single fruits per tree (one from each of the north, south, east, and west canopies). Fruits without mechanical damage that were at consistent developmental stages were randomly selected, wrapped in aluminum foil, and transported to the laboratory under controlled conditions. The samples were stored at −80 °C until further analysis. Three biological replicates were prepared for each stage, with each replicate consisting of five trees of the same age and normal growth conditions.
To quantify fruit color changes across different developmental stages, we employed the R package (version 0.1.0) for image segmentation and color classification. First, an initial segmentation was performed using the recolorization function, which assigns each pixel in the image to a distinct color category. Subsequently, color center refinement was conducted using the reclustering function, which calculates Euclidean distances between color pairs in the CIELAB color space. Hierarchical clustering was applied to group similar color centers based on a predefined cut-off threshold (50 in this study). This process merged the initial color clusters into a consensus set of representative color centers. Following clustering, the refined color centers were used to adjust the original image, generating the final segmented color categories. Each color region’s center was determined as the mean of all assigned pixel values; if no pixels were assigned, the geometric center was used. This approach provided an objective and reproducible method for quantifying fruit color variation across developmental stages [17].
2.2. Measurement of Physiological and Growth
For each developmental stage, 40 fruits were collected per replicate. Of these, 10 fruits were used to measure growth indicators, and 10–20 fruits were assessed for flesh texture. Physiological indicators, including vitamin C (Vc), organic acids, soluble solids, and soluble sugars, were measured using Solarbio reagent kits (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China), following the manufacturer’s instructions (https://www.solarbio.com/) (accessed on 30 October 2022). Growth indicators were evaluated by randomly selecting 10 fruits per stage and determining their weight using an electronic balance. The flesh hardness was measured using a TMS-Pro texture analyzer with a 2 mm probe, applying the following parameters: a test speed of 30 mm/min, a deformation percentage of 40%, a trigger force of 0.1 N, and an ascent height of 15 mm. The fruits were sliced perpendicularly at the equatorial region near the pit and placed on the texture analyzer tray for measurement. Hardness and other texture parameters were extracted from the texture profile analysis (TPA) curve.
2.3. Metabolomics Analysis
A 100 mg portion of vacuum freeze-dried biological sample powder was dissolved overnight at 4 °C in 1.2 mL of 70% methanol solution. The sample was centrifuged at 13,400× g for 10 min, and the supernatant was subjected to UPLC-ESI-MS/MS analysis. UPLC-ESI MS/MS analysis was performed on a Shimadzu Nexera X2 UPLC system (Agilent SB-C18, 1.8 μm, 2.1 × 100 mm; 40 °C; gradient: 5–95% acetonitrile/water [0.1% FA] over 9 min, 0.35 mL/min, Agilent Technologies, Santa Clara, CA, USA) coupled to an AB Sciex 4500 QTRAP MS (positive/negative ESI: 5500/4500 V, source temp 550 °C, GS1/GS2 50/60 psi, Sciex, Framingham, MA, USA). MRM transitions were optimized individually (DP 40–120 V, CE 15–35 eV) using 10/100 μmol/L polypropylene glycol calibrants, with nitrogen as the collision gas. Method validation demonstrated >85% recovery (RSD < 8%), an LOD of 0.05–0.2 ng/mL, and linearity (r2 > 0.998) across 3 orders of magnitude. Quality control samples (QC) were prepared by mixing sample extracts and were used to analyze the repeatability of samples under the same processing methods. During the instrument analysis process, the QC samples were interspersed between samples for testing to monitor the repeatability of the analysis process. The better the repeatability, the higher the stability of the detection. The metabolites were annotated using the NWDB proprietary database and quantified using multiple reaction monitoring (MRM). Data processing and normalization (log2 transformation) were performed using the Analyst 1.6.1 software. Differentially accumulated metabolites (DAMs) were identified based on log2 (fold change) ≥ 1 and variable importance in projection (VIP) > 1. Venn diagrams were generated to illustrate the distribution of the DAMs. Principal component analysis (PCA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were used to investigate metabolic pathway variations across the seven developmental stages.
2.4. Transcriptomic Analysis
The total RNA was extracted from frozen fruit samples at seven developmental stages using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA sequencing (RNA-Seq) was performed by Metware Biotechnology (Wuhan, China). The sequencing library was constructed using the NEBNext® Ultra™ RNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA). The raw sequence data from 210 RNA-seq samples were quality-filtered using fastp v0.19.3, and clean reads were aligned to the jujube reference genome (Accession number: PRJNA1032544). Raw RNA-seq reads were processed using fastp v0.19.3 to remove adapter sequences and low-quality bases (retaining reads with ≥30 bp length and Phred quality score ≥ 20 for at least 90% of bases). Quality-filtered clean data for all 21 samples exceeded 6 Gb per sample, with a Q30 base percentage of ≥91%, reflecting high sequencing fidelity. The reads were mapped to the reference genome using HISAT2 (version 2.2.1), and mapping statistics (e.g., alignment rates) are provided in Supplementary Materials. Novel genes were predicted using StringTie v1.3.4d, while gene alignment counts were calculated using FeatureCounts v1.6.2. The gene expression was quantified using the RSEM software (version 1.4.0), yielding FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values. Differentially expressed genes (DEGs) were identified using DESeq2 v1.22.1, with a significance threshold of adjusted p-value < 0.05 and |log2(fold change)| ≥ 1. Gene Ontology (GO) and KEGG pathway enrichment analyses were performed for functional annotation. Weighted gene co-expression network analysis (WGCNA) was conducted in R (version 1.72.5) to identify the co-expression modules associated with fruit flavor development.
2.5. Integrated Transcriptome-Metabolome Analysis
To integrate transcriptomic and metabolomic data, DAMs and DEGs were first enriched in shared pathways. A bidirectional orthogonal partial least squares (O2PLS) model was constructed to analyze correlations between metabolite and gene expression datasets. Correlation analysis was performed using the R package corrplot (version 0.92), with significant gene–metabolite pairs defined as having correlation coefficients of >0.8 or <−0.8 and p-values of <0.05. KEGG enrichment analysis was performed on significant gene–metabolite pairs to explore their functional associations. Enrichment significance was calculated via hypergeometric tests with FDR correction (q-value < 0.05) using the clusterProfiler package (version 4.0) in R. Additionally, selected metabolic phenotypes from the metabolome data were integrated with gene expression profiles to construct a phenotype–gene interaction network. WGCNA (version 1.72.5) was employed to identify correlation-based associations between gene modules and phenotypic traits. The modules were validated via correlation with phenotypic traits (e.g., sugar and organic acid content), and the significance was determined by permutation tests (p < 0.01). The adjacency matrix was transformed into a topological overlap matrix (TOM), and co-expressed genes were grouped into modules. Cytoscape (version 3.10.2) was used to visualize gene–metabolite interactions and the regulatory networks influencing jujube fruit flavor.
2.6. Statistical Analysis
All experiments were performed with biological replicates (n ≥ 3), and data are presented as the mean ± standard deviation (SD). Graphical visualization (e.g., heatmaps, volcano plots, and pathway diagrams) was achieved using ggplot2 (version 3.4.0) pheatmap, and Pathview in R.
3. Results
3.1. Observation and Characterization of Fruit Development and Ripening
Our experiment showed that in the seven developmental stages of Lingwu jujube, the fruit underwent significant morphological and physiological changes. The fruit shape evolved gradually (Figure 1a), and the single fruit weight reached its peak in the BH stage (Figure 1b).
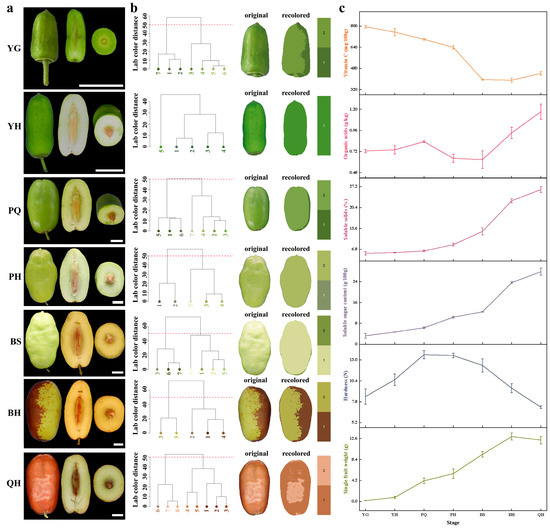
Figure 1.
Physiological and growth indicators during jujube development. (a) Fruit morphology; (b) color changes during fruit development; (c) physiological index: single fruit weight, hardness, soluble sugar content, soluble solids, organic acids, and vitamin C.
Color changes were also observed throughout the developmental stages. At the YG stage, the fruits were predominantly green due to high chlorophyll content. As development progressed to the YH and PQ stages, the green color began to fade, and a yellowish hue emerged, indicating chlorophyll degradation and the accumulation of carotenoids. By the PH and BS stages, the fruit exhibited a transition to a pale or whitish color, which marked the onset of ripening. During the BH stage, anthocyanin biosynthesis intensified, leading to the emergence of red pigmentation. Finally, in the QH stage, fruits attained a deep red color, indicating full ripeness and peak anthocyanin accumulation. These color transitions visually reflect the biochemical changes that occur during fruit maturation and serve as important indicators of ripening progress.
From the YG to PQ stages, the fruit hardness gradually increased, while from the YG to QH stages, the soluble sugar content progressively rose, peaking at 27.870 ± 1.325 g/100 g in the QH stage, indicating a relatively high sugar content in Lingwu long jujube fruits. The soluble solids content in the QH stage was 26.24%. Conversely, the organic acid content exhibited irregular fluctuations throughout these seven stages, with a peak of 1.173% in the QH stage, suggesting that ‘Lingwu long jujube’ is a variety with a sweet and sour taste, high quality, and high edibility. The vitamin C (Vc) content continuously decreased from the YG to BS stages, stabilized during the BH stage, and increased, reaching a stable level in the QH stage (Figure 1c), albeit slightly lower compared to the YG stage.
3.2. Metabolomic Analysis
To investigate the dynamic metabolic changes during jujube fruit development, we conducted a comprehensive targeted metabolomic analysis across the seven developmental stages. A total of 750 metabolites were identified, categorized into 11 primary metabolite classes and 35 secondary metabolite classes.
Specifically, we detected 48 alkaloids, 54 organic acids, 69 amino acids and derivatives, and 85 metabolites from other classes. Additionally, 115 phenolic acids, 10 lignans and coumarins, and 106 lipids were identified. Furthermore, 163 flavonoids, 52 terpenoids, eight sterols, and 40 nucleic acids and derivatives were present in the dataset.
Principal component analysis (PCA) revealed clear separation among samples from different developmental stages, with the first three principal components explaining 74.24% of the total variance (Figure 2a). Differentially accumulated metabolites (DAMs) were identified in pairwise comparisons across developmental stages, totaling 550 DAMs (Table S1). The most substantial metabolic shift occurred between the YH and PQ stages, where 281 DAMs were identified, including 201 downregulated and 80 upregulated metabolites. Similarly, 220 DAMs were detected between the BS and DLW6 stages, with 139 downregulated and 81 upregulated (Table 1). Notably, none of the DAMs were shared across all seven pairwise comparisons (Figure 2b), highlighting the dynamic and stage-specific nature of jujube fruit metabolism.
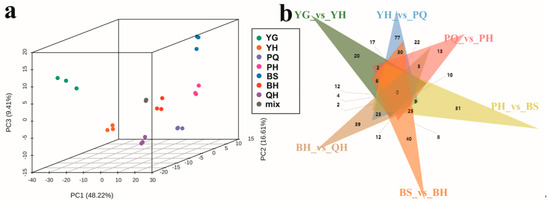
Figure 2.
Principal component analysis and Venn diagram analysis of jujube fruits at different stages. (a) Principal component analysis of jujube fruit samples; (b) metabolomics Venn diagram analysis.

Table 1.
Statistics of differential metabolite counts among groups.
3.3. DAMs Time-Series Analysis
To investigate the dynamic accumulation patterns of differentially accumulated metabolites (DAMs) across developmental stages, all identified DAMs from pairwise comparisons were standardized using z-score normalization. Subsequently, K-means clustering analysis was performed to classify metabolites based on their temporal abundance trends.
The K-means clustering results revealed distinct metabolic trends among sugars, alcohols, and organic acids (Figure 3). Class 1 consisted of 36 DAMs, including 3-dehydro-L-threo-sugar acid and D-glucuronic acid-6,3-lactone, which exhibited a significant increase from the young green (YG) to the yellow–green (YH) stage, followed by a decline during later developmental stages (Figure 3a). Class 2 comprised 152 DAMs, such as 2-aminoisobutyric acid and methylpyruvate, which showed a sharp decrease from YG to pre-quiescent (PQ), followed by a gradual decline thereafter (Figure 3b). Class 3 contained 81 DAMs, including arabinose, glucose, trehalose, and maltotriose, which demonstrated a continuous increase throughout fruit development (Figure 3c).
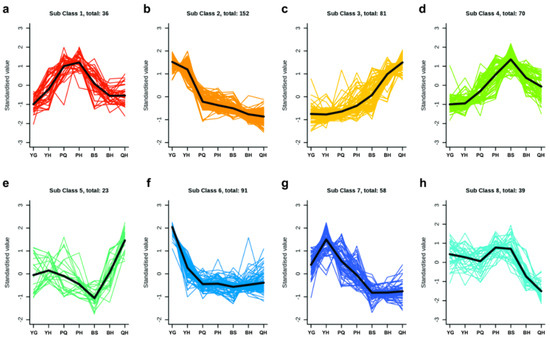
Figure 3.
Results of k-means clustering of differential metabolites. (a) Class 1 contained 36 DAMs; (b) Class 2 contained 152 DAMs; (c) Class 3 contained 81 DAMs; (d) Class 4 contained 70 DAMs; (e) Class 5 contained 23 DAMs; (f) Class 6 contained 91 DAMs; (g) Class 7 contained 58 DAMs; and (h) Class 8 contained 39 DAMs.
Class 4 included 70 DAMs, such as trans-butenedioic acid, 3-methylpropionic acid, and 4-hydroxybutyric acid, which accumulated progressively from YG to blue skin (BS), peaked at BS, and subsequently declined (Figure 3d). Class 5 consisted of 23 DAMs, with isopropylmalic acid exhibiting an increasing trend from YG to YH, declining to its lowest level at BS, and then rapidly increasing again (Figure 3e). Additional clusters included Class 6 (91 DAMs), Class 7 (58 DAMs), and Class 8 (39 DAMs) (Figure 3f–h). Notably, γ-aminobutyric acid, 2-hydroxyisobutyric acid, D-erythroascorbic acid, glutaric acid, 4-aminobutyric acid, 4-phenylbutyric acid, jasmonic acid, and argininosuccinic acid peaked at YH before rapidly decreasing until BS, where their levels stabilized (Figure 3h).
Furthermore, a total of 90 lipid compounds and 53 flavonol compounds were identified. The dynamic shifts in sugar compounds and organic acids indicate that YH and BS represent two critical transition points in jujube fruit development, reflecting significant metabolic reprogramming events that contribute to fruit flavor and quality.
3.4. Genetic Basis of Flavor-Related Metabolites Dynamics
To elucidate the molecular mechanisms underlying flavor-related metabolite changes, RNA sequencing (RNA-seq) was performed across the seven developmental stages. Differentially expressed genes (DEGs) were identified in six pairwise comparisons (Table 2), with the highest number observed in the YH_vs_PQ group (9342 DEGs, including 2724 downregulated), whereas the BS_vs_BH group exhibited the highest number of upregulated DEGs (1872) (Table S2).

Table 2.
Statistics of differential gene counts among groups.
Among all comparisons, only four DEGs were consistently shared across all stages: FEM48_Zijuj07G0138200, FEM48_Zijuj08G0156700, FEM48_Zijuj11G0017500, and novel.882. Excluding the BH_vs_QH group, 44 DEGs were commonly detected across all other comparisons (Figure 4). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses indicated that these DEGs were primarily associated with metabolic pathways and plant hormone signal transduction (Figure 5 and Figure 6). Notably, the biosynthesis of secondary metabolites and circadian rhythm regulation exhibited substantial variation across developmental stages.
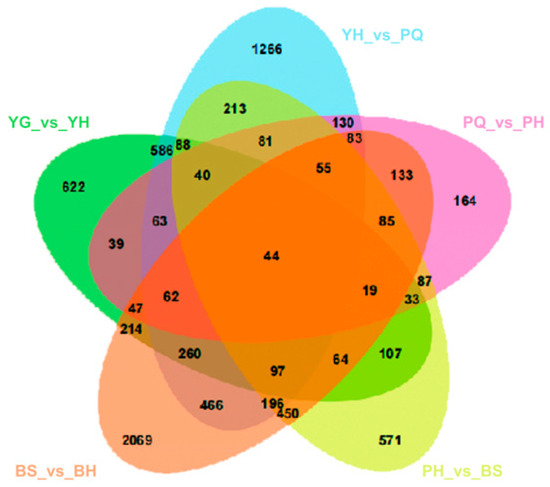
Figure 4.
Transcriptome Venn diagram analysis.
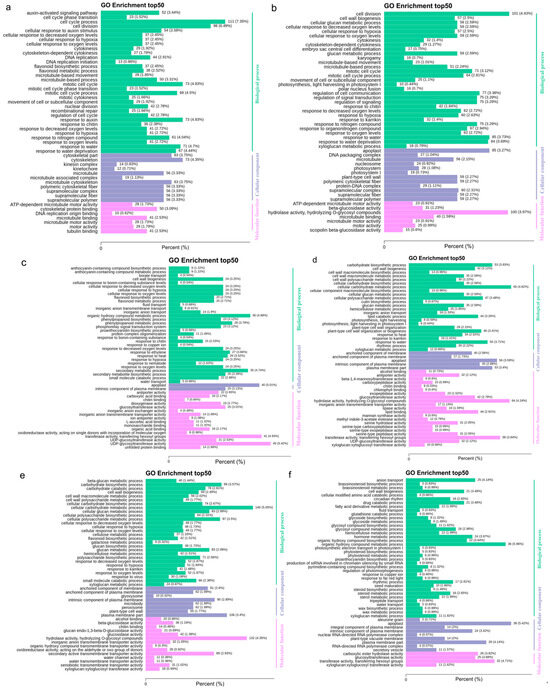
Figure 5.
Transcriptome GO classification of differentially expressed genes at different stages. (a) YG_vs_YH.go enrich; (b) YH_vs_PQ.go enrich; (c) PQ_vs_PH.go enrich; (d) PH_vs_BS.go enrich; (e) BS_vs_BH.go enrich; and (f) BH_vs_QH.go enrich.
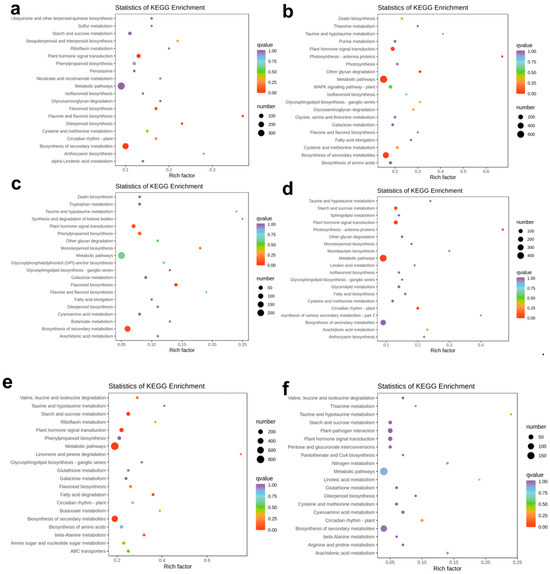
Figure 6.
KEGG analysis of differentially expressed genes at different stages. (a) BS_vs_BH.path enrichment; (b) PH_vs_BS.path enrichment; (c) PQ_vs_PH.path enrichment; (d) YH_vs_PQ.path enrichment; (e) YG_vs_YH.path enrichment; and (f) BH_vs_QH.path enrichment.
3.5. Key Pathways Underlying Flavor Changes in Jujube
To identify key pathways influencing flavor changes, an integrative transcriptome and metabolome analysis was conducted. An orthogonal partial least squares (O2PLS) model was constructed to assess the interactions between different omics datasets, directly indicating the weight of variables that influenced each other. Variables with higher weights had a greater impact on the transcriptome–metabolome relationship.
By selecting all DAMs and DEGs, key metabolic pathways were identified using the O2PLS model (Figure 7). The top 50 variables revealed significant enrichment in phenylpropanoid biosynthesis (ko00940), carbon metabolism (ko01200), starch and sucrose metabolism (ko00500), and cofactor biosynthesis (ko01240). Additionally, pathways such as ascorbate and aldarate metabolism (ko00053), the pentose phosphate pathway, and fatty acid biosynthesis were strongly influenced by metabolite changes.
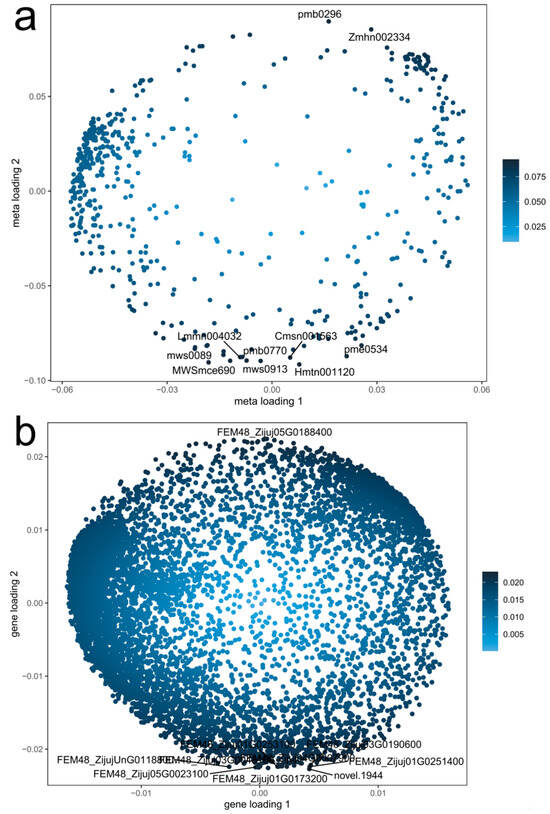
Figure 7.
Metabolite and gene loading plot. (a) Metabolite loading plot; (b) gene loading plot.
The integrated analysis revealed that starch and sucrose continuously degraded from YG to BS, leading to an enrichment of fructose at BS, contributing to increased sweetness. Organic acids accumulated from YG to PH, peaking at BS. Similarly, aroma compounds increased throughout development, reaching their highest levels at BS. These findings suggest that the optimal fruit flavor is achieved at BS, characterized by a balance of sweetness, acidity, and aroma.
3.6. WGCNA Analysis of Genes Related to Flavor Changes
To further explore the key genetic factors that regulate flavor changes, weighted gene co-expression network analysis (WGCNA) was performed on all DEGs (Figure 8a). Genes were primarily clustered into the MEturquoise and MEblue modules. A total of 277 DEGs were related to the starch and sucrose metabolism pathway, with 155 DEGs clustering in the MEturquoise module and 40 in the MEblue module. Similarly, 278 DEGs were associated with carbon metabolism, with 104 DEGs clustering in the MEturquoise module and 76 in the MEblue module (Figure 8b,c).
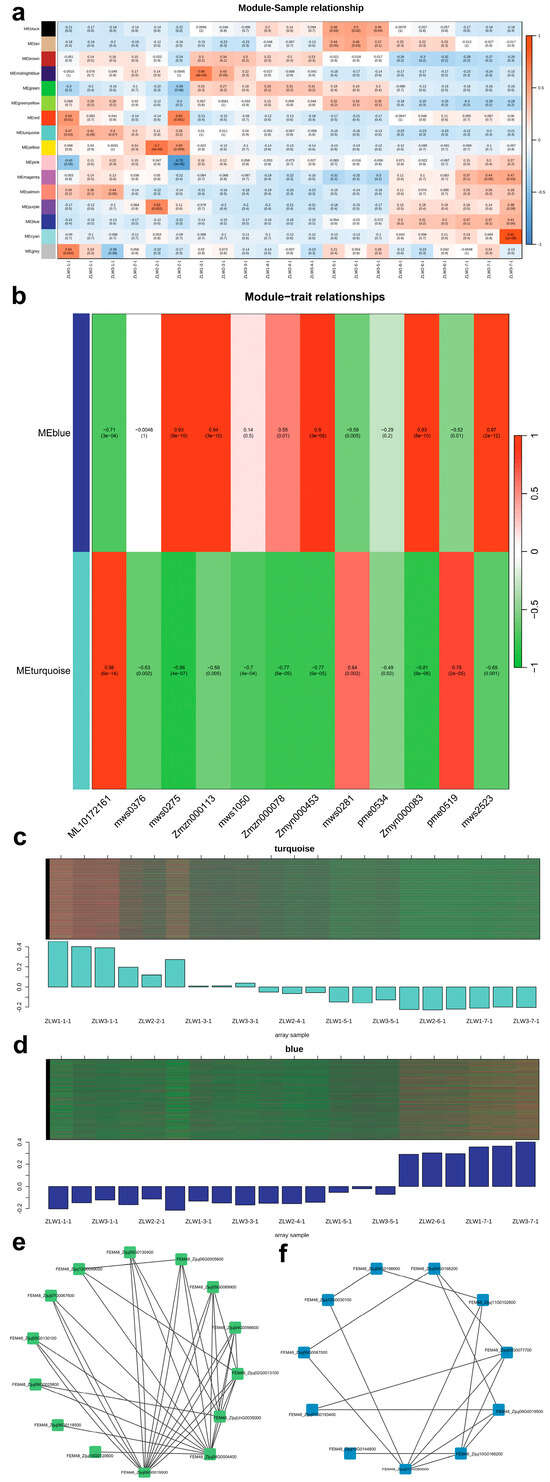
Figure 8.
Differential gene weighted co-expression network analysis (WGCNA) plot and phenotypes and the gene interaction network. (a) represents module correlation; (b) represents starch and sucrose metabolism and the carbon metabolism pathway; (c) represents the turquoise module results plot; (d) represents the blue module results plot; (e) represents gene network of the MEturquoise module; and (f) represents the gene network of the MEblue module.
Further analysis of the starch and sucrose metabolism pathway (ko00500) and the carbon metabolism pathway (ko01200) identified 155 gene–metabolite pairs from YG to BH, of which 35 pairs exhibited positive correlations and 120 pairs exhibited negative correlations (Table S3). In addition, 160 gene–metabolite pairs were identified in the carbon metabolism pathway, with 86 showing positive correlations and 74 negative correlations (Table S4). Visualization using the Cytoscape software (version 3.10.2) revealed key regulatory networks (Figure 8d–f).
Among the identified genes, eight candidate genes were found to be strongly associated with flavor changes: FEM48_Zijuj06G0019500, FEM48_Zijuj04G0004400, FEM48_Zijuj02G0013100, FEM48_ZijujunG0035000, FEM48_Zijuj01G0089000, FEM48_Zijuj10G0166200, FEM48_Ziiui03G0077700, and FEM48_Ziu09G0019500. Except for FEM48_Zijuj03G0077700, whose function remains unclear, all other genes were associated with enzymes involved in sugar metabolism (Table 3, Figure 9). These findings, based on co-expression and metabolite correlation analyses, suggest potential associations between these genes and flavor-related metabolic pathways during jujube fruit development. Due to unforeseen sample limitations, experimental validation is pending, and these genes are therefore proposed as priority candidates for future functional studies.

Table 3.
The gene functions.
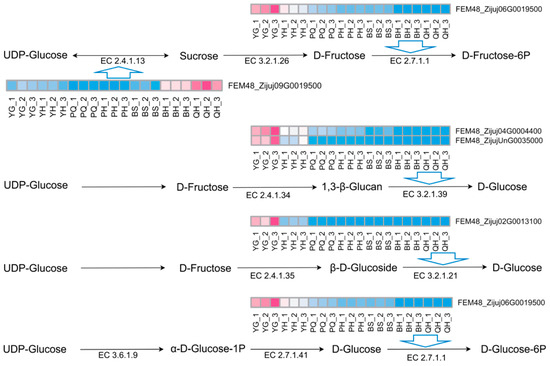
Figure 9.
The expression levels of key genes in key metabolic pathways.
4. Discussion
4.1. Dynamic Changes of Key Metabolites During Fruit Development and Their Effects on Flavor
The development of fruit flavor is primarily influenced by variations in the content and trends of sugars, organic acids, and specific lipid compounds. The metabolic pathways associated with these components, which were simultaneously enriched in DAMs and DEGs, include starch and sucrose metabolism, fructose and mannose metabolism, galactose metabolism, glycolysis/gluconeogenesis, the pentose phosphate pathway, pentose and glucuronate interconversions, the citric acid cycle, fatty acid biosynthesis, and flavonoid biosynthesis.
Through differential metabolite time-series analysis, we identified that the key DAMs were mainly concentrated in sugars, alcohols, organic acids, lipids, and flavonols. Notably, 3-dehydro-L-gulonic acid, D-glucurono-6,3-lactone, L-gulono-1,4-lactone, and gluconic acid exhibited an increasing trend from YG to the physiological maturity (PQ) stage. Sugars such as sorbitol and rhamnose increased from YG to YH and then remained stable, suggesting that the sugar content dynamically changes throughout fruit development. In general, sugar accumulation increases as the fruit matures, significantly influencing fruit taste and sweetness [18]. During fruit ripening, metabolic flux alterations lead to a decline in organic acid synthesis rates, followed by redistribution or transport to other plant tissues [19]. As the fruit matures, the demand for organic acids diminishes in favor of sucrose accumulation and seed development, rather than organic acid synthesis and accumulation [20].
Several organic acids displayed distinctive trends throughout fruit development. 2-Aminobutyric acid, 2-isopropylpropanoic acid, malic acid, 3-dehydroshikimic acid, 2-decylpentanedioic acid, adipic acid diethyl ester, and 2-isopropylhexadecanoic acid exhibited a rapid decline from YG to PQ, followed by a stable decrease. In contrast, γ-aminobutyric acid, 2-isopropylbutanoic acid, D-ketoglutaric acid, adipic acid, 4-ethylaminobutyric acid, 4-phenylbutanoic acid, jasmonic acid, and argininosuccinic acid increased from YG to YH, peaked at YH, then rapidly declined toward the breaking stage (BS) and eventually stabilized. Citric acid and 2,2-dimethylsuccinic acid decreased from YG to PQ, gradually increased from PQ to BS, and then sharply declined from BS to over-ripening (QH).
Lipids play a crucial role in fruit development beyond their contribution to the cuticle structure. They provide energy, influence fruit texture and flavor, and act as precursors for aromatic compounds that modify fruit aroma [21]. In this study, we identified 90 lipid compounds. During early fruit development, proanthocyanidins and quercetin were the predominant flavonoids, but their levels decreased significantly as maturation progressed. Conversely, anthocyanins accumulated substantially in the later stages, emerging as the dominant flavonoids in mature fruit [22,23]. In total, we identified 53 flavonols and 31 flavones. These differential metabolites provide important insights into the metabolic shifts that occur during jujube fruit development, though further experimental validation is necessary.
4.2. Enrichment of Primary and Secondary Metabolic Pathways and Their Central Roles in Fruit Flavor Formation
Plant metabolism is categorized into primary and secondary metabolism. Primary metabolism includes essential biochemical reactions, while secondary metabolism plays a critical role in growth, development, and plant–environment interactions. Primary metabolites from glycolysis, the tricarboxylic acid (TCA) cycle, and the shikimate pathway serve as precursors for a vast array of secondary metabolites [24]. Unlike the highly conserved primary metabolism, secondary metabolic pathways exhibit species-specific, organ-specific, and even developmental-stage-specific variations [25]. Mapping DAMs and DEGs onto KEGG pathway diagrams revealed that key pathways enriched during fruit maturation included linoleic acid metabolism, flavonoid biosynthesis, and flavone and flavonol biosynthesis [9]. Additionally, α-linolenic acid metabolism, phenylalanine metabolism, purine metabolism, linoleic acid metabolism, and isoquinoline alkaloid biosynthesis were consistently enriched across all developmental stages. These pathways significantly influence fruit flavor, as enzyme-mediated metabolic activities generate key compounds that contribute to fruit aroma [26,27].
The metabolism of starch, sucrose, and other intermediates plays a crucial role in fruit flavor development. Their levels are determined by primary metabolic pathways, such as glycolysis, the TCA cycle, and respiration [28]. Throughout jujube fruit development, we observed significant shifts in key metabolites. From YG to YH, sucrose and raffinose contents decreased, while S-malic acid and fumaric acid levels increased (citric acid cycle, ko00020). Between YH and PQ, D-gluconate (the pentose phosphate pathway, ko00030) and α-D-glucose 1,6-bisphosphate levels increased, fumaric acid continued to rise, while S-malic acid remained stable, and glutaric acid declined. From PQ to PH, L-rhamnose (fructose and mannose metabolism, ko00051) and α-D-glucose 1,6-bisphosphate increased, along with continued fumaric acid accumulation. During PH to BS, raffinose levels declined (galactose metabolism), while D-fructose 6-phosphate, trehalose 6-phosphate, and α-D-glucose 1,6-bisphosphate levels rose. Fumaric acid levels stabilized, whereas malonic acid and glutaric acid decreased. Notably, fatty acid biosynthesis (ko00061) was associated with enhanced fruit aroma through increased levels of lauric acid, myristic acid, and palmitoleic acid [29].
The primary organic acids in jujube fruit include citric acid, malic acid, and quinic acid, with lower concentrations of other TCA intermediates such as oxalic acid, succinic acid, isocitric acid, fumaric acid, and cis-aconitic acid. During BS to QH, trehalose 6-phosphate, lauric acid, and myristic acid continued to accumulate, while fumaric acid and citric acid levels declined (citric acid cycle). These findings confirm that starch and sugar metabolism, fatty acid metabolism, and organic acid metabolism are the key pathways that affect jujube fruit flavor.
4.3. Evolutionary Conservation and Lineage-Specific Divergence in Jujube Fruit Flavor Metabolism: Integrative Multi-Omics Insights from Cross-Species Comparisons
Our multi-omics analysis identified several key enriched pathways in jujube fruit flavor development, which align with and diverge from metabolic networks reported in other fruit species, providing the basis for the conserved and unique mechanisms of flavor formation. For instance, the enrichment of sugar metabolism pathways (e.g., sucrose synthesis, glycolysis) mirrors findings in apple (Malus domestica) and grape (Vitis vinifera), where genes encoding sucrose synthase (SS) and invertase (INV) play central roles in sugar partitioning and accumulation during ripening [29,30,31]. In jujube, the co-expression of SS homologs (e.g., FEM48_Zijuj06G0019500) with hexose transporters echoes similar regulatory modules in apple, suggesting a conserved strategy for coordinating sugar biosynthesis and transport. However, jujube-specific expansions in the glucosidase gene family—highly correlated with glucose content (Figure 9)—distinguish it from model fruits like apple, pear, and tomato (Solanum lycopersicum), where sucrose biosynthesis dominates secondary metabolism [32,33]. This divergence may underlie the unique flavor of jujube, highlighting species-specific adaptations in flavor-related secondary metabolism.
4.4. Screening of Key Genes Involved in Carbon and Sugar Metabolism Based on WGCNA and Their Functional Prediction
Through weighted gene co-expression network analysis (WGCNA), we identified key genes associated with carbon and sugar metabolism, including FEM48_Zijuj06G0019500, FEM48_Zijuj04G0004400, FEM48_Zijuj02G0013100, FEM48_ZijujunG0035000, FEM48_Zijuj01G0089000, FEM48_Zijuj10G0166200, FEM48_Ziiui03G0077700, and FEM48_Ziiu09G0019500. Future studies will focus on the functional validation of these genes to further elucidate their roles in flavor development in jujube fruit.
5. Conclusions
In this study, transcriptome and metabolite analyses were conducted on jujube fruits to elucidate the dynamic changes in genes and metabolites during fruit development and to explore the regulatory gene networks that influence flavor formation. Our findings indicate that the biosynthesis of secondary metabolites, metabolic pathways, phenylpropanoid metabolism, and linoleic acid metabolism play crucial roles in flavor synthesis and actively participate in fruit development. Among these pathways, starch and sugar metabolism, along with carbon metabolism, exhibited the highest number of differentially expressed genes, highlighting their significance in determining fruit flavor. Additionally, through co-expression pattern analysis, we identified eight gene networks associated with jujube flavor modulation. These genes represent novel discoveries, as their roles in jujube fruit development and flavor formation have not been previously reported. Further functional validation of these genes will provide deeper insights into their contributions to jujube fruit quality and metabolic regulation.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/agronomy15061337/s1, Figure S1. plants of Z. jujuba ‘Lingwuchangzao’. A, Photo was taken on 25 May 2024; B. Photo was taken on 25 August 2024; C. Photo was taken on 20 October 2024. Figure S2. CV_ECDF of all samples. Note: The abscissa represents the CV value, and the ordinate indicates the proportion of the number of substances with CV values less than the corresponding CV value to the total number of substances. Different colors represent different grouped samples, where “mix” denotes QC (Quality Control) samples. The two reference lines perpendicular to the X-axis correspond to CV values of 0.3 and 0.5, while the two reference lines parallel to the X-axis correspond to proportions of 75% and 85% of the total number of substances. Figure S3. PCA analysis of all samples Table S1: Total DAMs during the developmental stage of Ziziphus jujuba ‘Lingwuchangzao’; Table S2: Total DEGs during the developmental stage of Ziziphus jujuba ‘Lingwuchangzao’; Table S3: 155 gene–metabolite pairs in starch and sucrose metabolism pathway and the carbon metabolism pathway; Table S4: 160 gene–metabolite pairs identified in the carbon metabolism pathway; Table S5: quality control of transcriptome.
Author Contributions
Conceptualization, X.Z. and J.Z.; methodology, T.W.; software, Z.B.; validation, J.Z. and X.Z.; formal analysis, Y.R.; investigation, X.W., W.X., and H.L.; resources, T.W. and J.Z.; data curation, X.Z.; writing—original draft preparation, X.Z.; writing—review and editing, Z.B.; visualization, Z.B.; supervision, J.Z. and T.W.; project administration, Y.R., J.Z., and T.W.; funding acquisition, J.Z. and T.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Research Initiation Project of North Minzu University, grant numbers 2023QNPY25 and 2023QNPY26, Breeding of New Jujube Varieties (Lines) and Research with Integrated Demonstration of High-Quality and High-Efficiency Cultivation Techniques for ‘Lingwu Changzao’ of Ningxia Academy of Agricultural and Forestry Sciences (No. NGSB-2021-1-02), the Innovation Team for Genetic Improvement of Economic Forests (No. 2022QCXTD04) and the Baijitan Scientific Research Project of North Minzu University (No. SKBJT202204, SKBJT202208).
Data Availability Statement
The original contributions presented in this study are included in the article and Supplementary Material. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Li, B.; Muhammad, N.; Zhang, S.; Lan, Y.; Yang, Y.; Han, S.; Liu, M.; Yang, M. Multiple-genome-based simple sequence repeat is an efficient and successful method in genotyping and classifying different Jujube germplasm resources. Plants 2023, 12, 2885. [Google Scholar] [CrossRef] [PubMed]
- Barrett, D.M.; Beaulieu, J.C.; Shewfelt, R. Color, Flavor, Texture, and Nutritional Quality of Fresh-Cut Fruits and Vegetables: Desirable Levels, Instrumental and Sensory Measurement, and the Effects of Processing. Crit. Rev. Food Sci. Nutr. 2010, 50, 369–389. [Google Scholar] [CrossRef]
- Goldenberg, L.; Yaniv, Y.; Porat, R.; Carmi, N. Mandarin fruit quality: A review. J. Sci. Food Agric. 2018, 98, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, C.; Zhao, X.; Fei, Z.; Wan, K.; Zhang, Z.; Pang, X.; Yin, X.; Bai, Y.; Sun, X.; et al. The Jujube Genome Provides Insights into Genome Evolution and the Domestication of Sweetness/Acidity Taste in Fruit Trees. PLoS Genet. 2016, 12, e1006433. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Jin, J.; Li, M.; Kong, D.; Cao, M.; Wang, X.; Li, Y.; Chen, X.; Zhang, X.; Pang, X.; et al. The Key Metabolic Network and Genes Regulating the Fresh Fruit Texture of Jujube (Ziziphus jujuba Mill.) Revealed via Metabolomic and Transcriptomic Analysis. Plants 2023, 12, 2087. [Google Scholar] [CrossRef]
- Zhao, A.; Xue, X.; Wang, Y.; Sui, C.; Ren, H.; Li, D. The Sugars and Organic Acids Composition in Fruits of Different Chinese Jujube Cultivars of Different Development Stages. Acta Hortic. Sin. 2016, 43, 1175–1185. [Google Scholar]
- Zhao, A.; Xue, X.; Ren, H.; Wang, Y.; Li, D.; Li, Y. Analysis of Composition and Content Characteristics of Organic Acids in Jujube Germplasm. Acat Agric. Boreali-Occident. Sin. 2021, 30, 1185–1198. [Google Scholar]
- Huang, J.; Chen, X.; He, A.; Ma, Z.; Gong, T.; Xu, K.; Chen, R. Integrative Morphological, Physiological, Proteomics Analyses of Jujube Fruit Development Provide Insights into Fruit Quality Domestication from Wild Jujube to Cultivated Jujube. Front. Plant Sci. 2021, 12, 773825. [Google Scholar] [CrossRef]
- Li, X.; Jiang, J.; Zhang, L.; Yu, Y.; Ye, Z.; Wang, X.; Zhou, J.; Chai, M.; Zhang, H.; Arús, P.; et al. Identification of volatile and softening-related genes using digital gene expression profiles in melting peach. Tree Genet. Genomes 2015, 11, 71. [Google Scholar] [CrossRef]
- Shi, Q.Q.; Du, J.T.; Zhu, D.J.; Li, X.; Li, X.G. Metabolomic and Transcriptomic Analyses of Anthocyanin Biosynthesis Mechanisms in the Color Mutant Ziziphus jujuba cv. Tailihong. J. Agric. Food Chem 2020, 68, 15186–15198. [Google Scholar] [CrossRef]
- Xu, J.; Yan, J.; Li, W.; Wang, Q.; Wang, C.; Guo, J.; Geng, D.; Guan, Q.; Ma, F. Integrative analyses of widely targeted metabolic profiling and transcriptome data reveals molecular insight into metabolomic variations during apple (Malus domestica) fruit development and ripening. Int. J. Mol. Sci. 2020, 21, 4797. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gao, H.; Ming, J.; Ding, Z.; Lin, X.; Zhan, R. Combined Transcriptome and Metabolome analysis of Pitaya fruit unveiled the mechanisms underlying Peel and pulp color formation. BMC Genom. 2020, 21, 734. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Zhu, H.; Lu, X.; Yang, D.; Zhao, S.; Umer, M.J.; He, N.; Yuan, P.; Anees, M.; Diao, W.; et al. An integrated transcriptome and metabolome approach reveals the accumulation of taste-related metabolites and gene regulatory networks during watermelon fruit development. Planta 2021, 254, 35. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Wang, S.; Zhang, R.; Yin, M.; Yang, X.; Shi, Q. Integrative analysis of transcriptomic and metabolomic profiles reveals new insights into the molecular foundation of fruit quality formation in Citrullus lanatus (Thunb.). Food Qual. Saf. 2022, 6, fyac015. [Google Scholar] [CrossRef]
- Xiong, Y.; Yan, P.; Du, K.; Li, M.Z.; Xie, Y.; Gao, P. Nutritional component analyses of kiwifruit in different development stages by metabolomic and transcriptomic approaches. J. Sci. Food Agric. 2020, 100, 2399–2409. [Google Scholar] [CrossRef]
- Yang, H.; Tian, C.; Ji, S.; Ni, F.; Fan, X.; Yang, Y.; Sun, C.; Gong, H.; Zhang, A. Integrative analyses of metabolome and transcriptome reveals metabolomic variations and candidate genes involved in sweet cherry (Prunus avium L.) fruit quality during development and ripening. PLoS ONE 2021, 16, e0260004. [Google Scholar] [CrossRef]
- Weller, H.I.; Hiller, A.E.; Lord, N.P.; Van Belleghem, S.M. Recolorize: An R package for flexible colour segmentation of biological images. Ecol. Lett. 2024, 27, e14378. [Google Scholar] [CrossRef]
- Kanayama, Y. Sugar Metabolism and Fruit Development in the Tomato. Hortic. J. 2017, 86, 417–425. [Google Scholar] [CrossRef]
- Sarahuldhat, P. Organic Acid Metabolism and Accumulation During Pineapple Fruit Growth and Development; University of Hawaii at Manoa: Honolulu, HI, USA, 2005. [Google Scholar]
- Albertini, M.-V.; Carcouet, E.; Pailly, O.; Gambotti, C.; Luro, F.; Berti, L. Changes in organic acids and sugars during early stages of development of acidic and acidless citrus fruit. J. Agric. Food Chem. 2006, 54, 8335–8339. [Google Scholar] [CrossRef]
- Shahidi, F.; Hossain, A. Role of Lipids in Food Flavor Generation. Molecules 2022, 27, 5014. [Google Scholar] [CrossRef]
- Jaakola, L.; Maatta, K.; Pirttila, A.M.; Torronen, R.; Karenlampi, S.; Hohtola, A. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 2002, 130, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Kosma, D.K.; Parsons, E.P.; Isaacson, T.; Lü, S.Y.; Rose, J.K.C.; Jenks, M.A. Fruit cuticle lipid composition during development in tomato ripening mutants. Physiol. Plant. 2010, 139, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Kroymann, J. Natural diversity and adaptation in plant secondary metabolism. Curr. Opin. Plant Biol. 2011, 14, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Izmailov, S.F.; Nikitin, A.V.; Rodionov, V.A. Nitrate Signaling in Plants: Introduction to the Problem. Russ. J. Plant Physiol. 2018, 65, 477–489. [Google Scholar] [CrossRef]
- Jia, Z.; Wang, Y.; Wang, L.; Zheng, Y.; Jin, P. Amino acid metabolomic analysis involved in flavor quality and cold tolerance in peach fruit treated with exogenous glycine betaine. Food Res. Int. 2022, 157, 111204. [Google Scholar] [CrossRef]
- Saladié, M.; Wright, L.P.; Garcia-Mas, J.; Rodriguez-Concepcion, M.; Phillips, M.A. The 2-C-methylerythritol 4-phosphate pathway in melon is regulated by specialized isoforms for the first and last steps. J. Exp. Bot. 2014, 65, 5077–5092. [Google Scholar] [CrossRef]
- Quan, Q.; Liu, W.; Zuo, M.; Zhang, J. Advances in the flavor of fruit and vegetable juices fermented by lactic acid bacteria. Food Ferment. Ind. 2022, 48, 315–323. [Google Scholar]
- Grechkin, A. Recent developments in biochemistry of the plant lipoxygenase pathway. Prog. Lipid Res. 1998, 37, 317–352. [Google Scholar] [CrossRef]
- Li, M.; Feng, F.; Cheng, L. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS ONE 2012, 7, e33055. [Google Scholar] [CrossRef]
- Tong, X.; Wang, Z.; Ma, B.; Zhang, C.; Zhu, L.; Ma, F.; Li, M. Structure and expression analysis of the sucrose synthase gene family in apple. J. Integr. Agric. 2018, 17, 847–856. [Google Scholar] [CrossRef]
- Wang, X.Q.; Li, L.M.; Yang, P.P.; Gong, C.L. The role of hexokinases from grape berries (Vitis vinifera L.) in regulating the expression of cell wall invertase and sucrose synthase genes. Plant Cell Rep. 2014, 33, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Feng, X.; Peng, F.; Muhammad, A.; Zhang, Y.; Zhao, Y.; Han, W.; Lu, J.; Cao, Y.; et al. Analysis of the β-Glucosidase family reveals genes involved in the lignification of stone cells in Chinese White Pear (Pyrus bretschneideri Rehd.). Front. Plant Sci. 2022, 13, 852001. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).