Plant Aux/IAA Gene Family: Significance in Growth, Development and Stress Responses
Abstract
1. Introduction
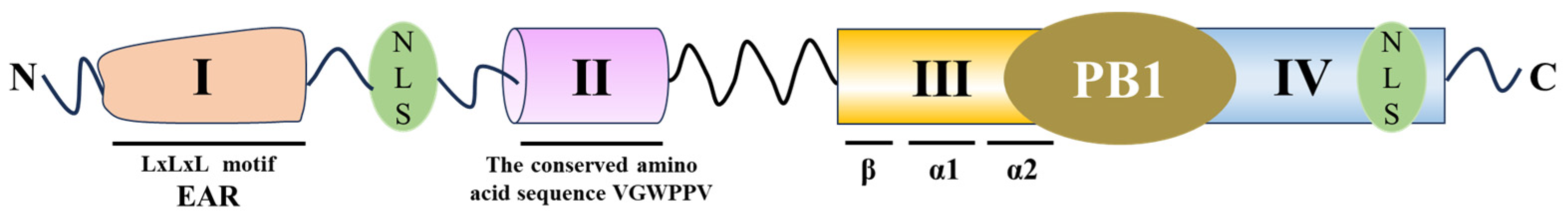
2. Structural Characteristics of Aux/IAA Proteins
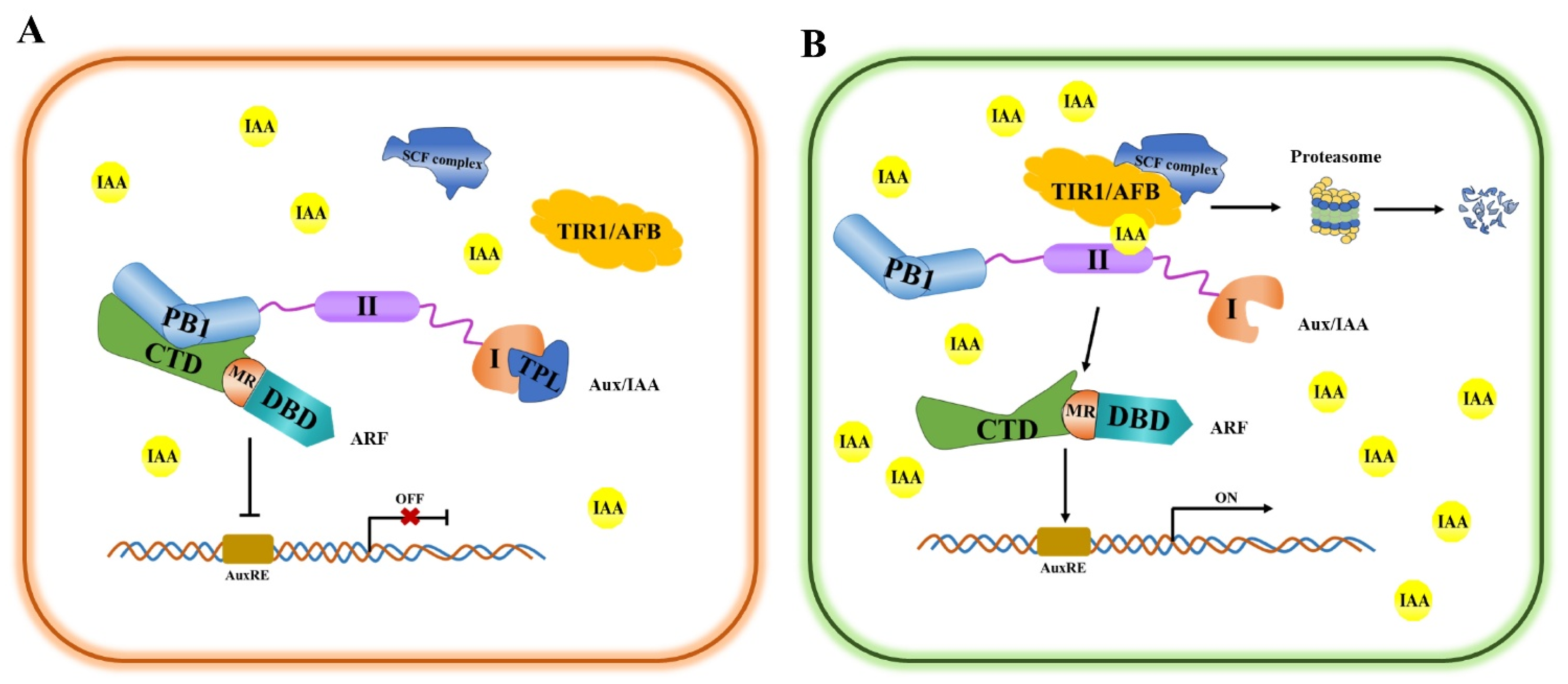
3. Auxin Signal Transduction Pathway
4. Research on the Function of Aux/IAA Genes in Arabidopsis
4.1. Regulation of Aux/IAA Genes on the Growth and Development of Arabidopsis
4.2. The Impact of Aux/IAA Genes on Abiotic Stress in Arabidopsis
5. Research on the Functions of Aux/IAA Genes in Rice
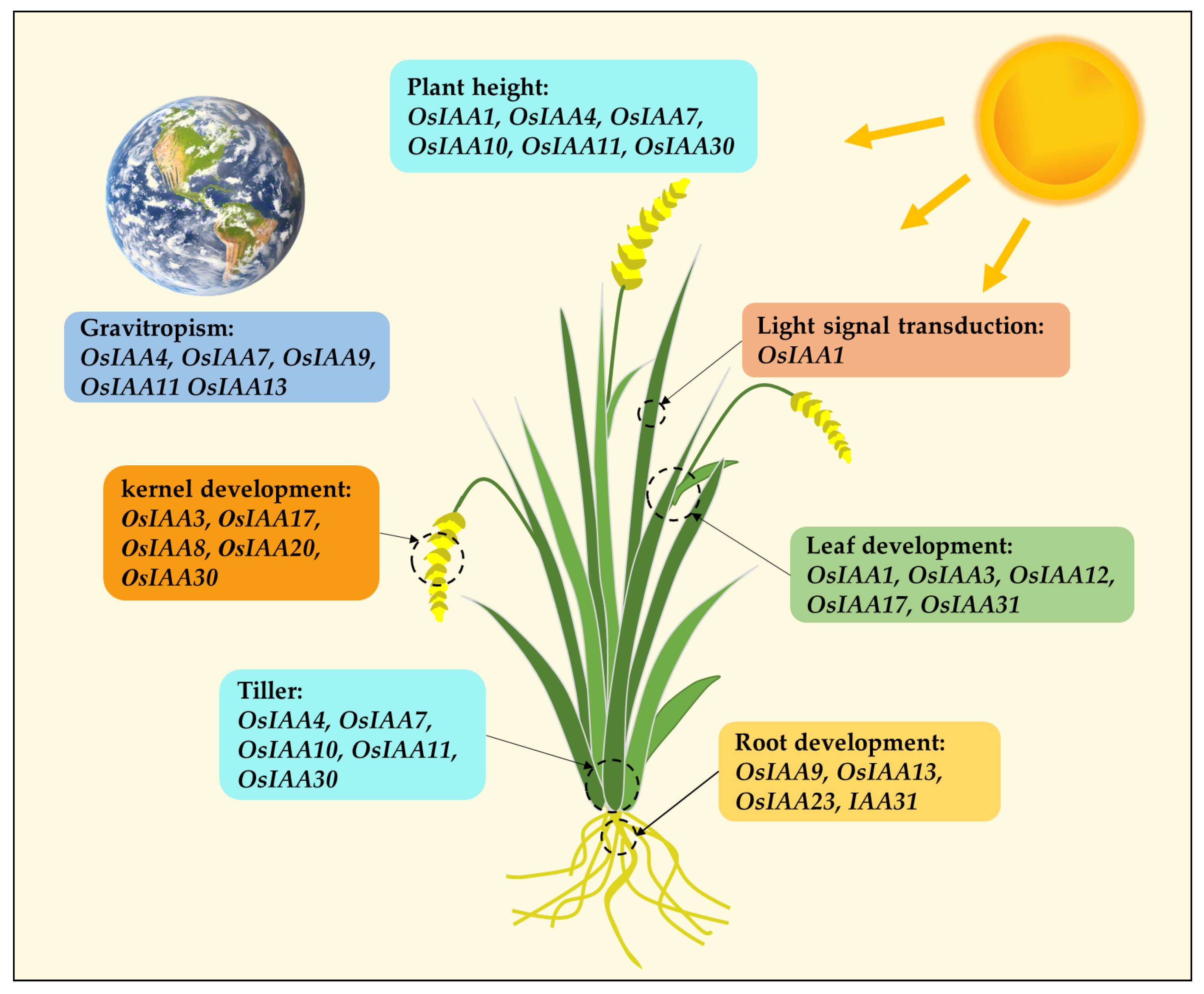
5.1. Regulation of Aux/IAA Genes in Rice Growth and Development
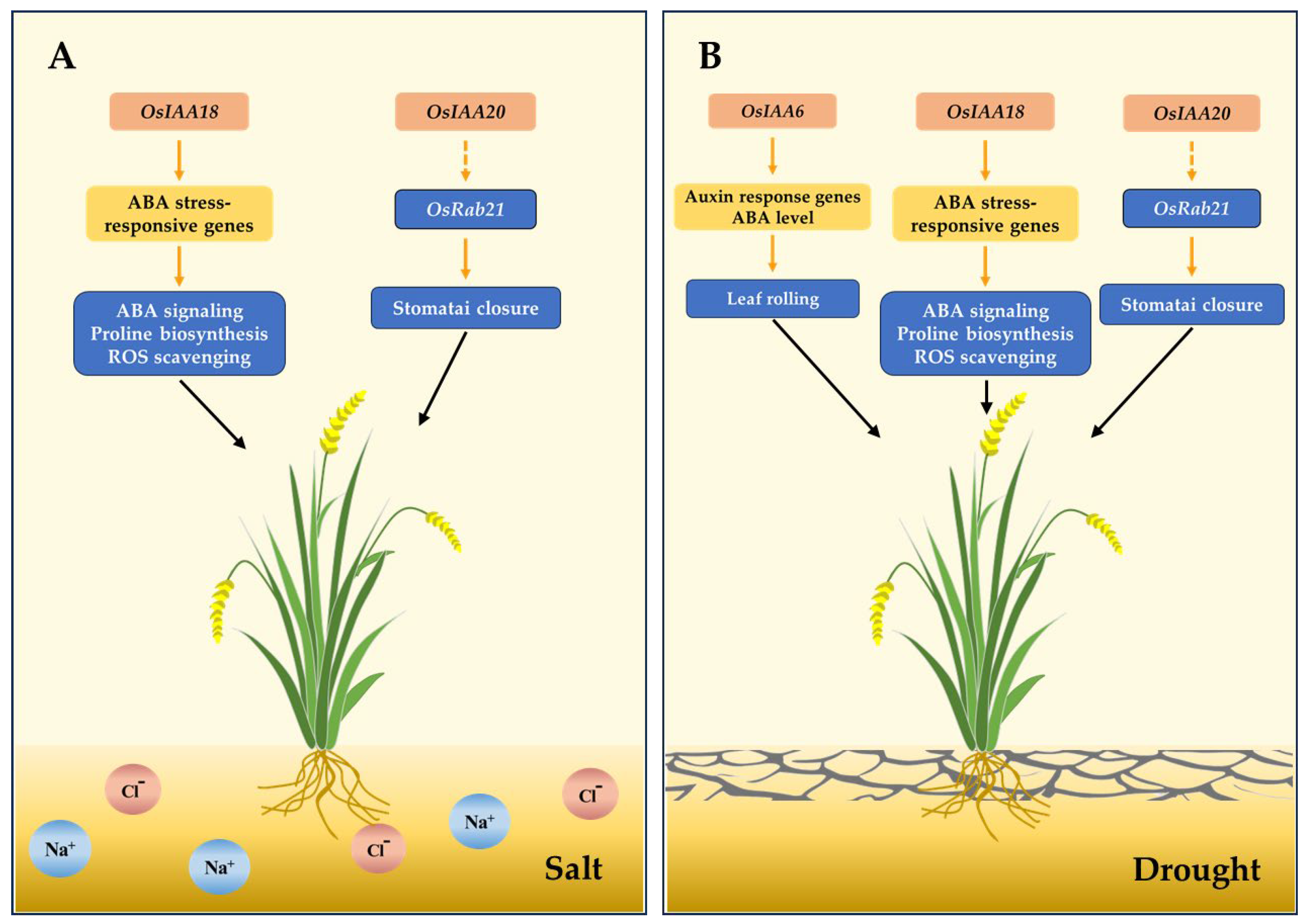
5.2. The Impact of Aux/IAA Genes on Abiotic Stress in Rice
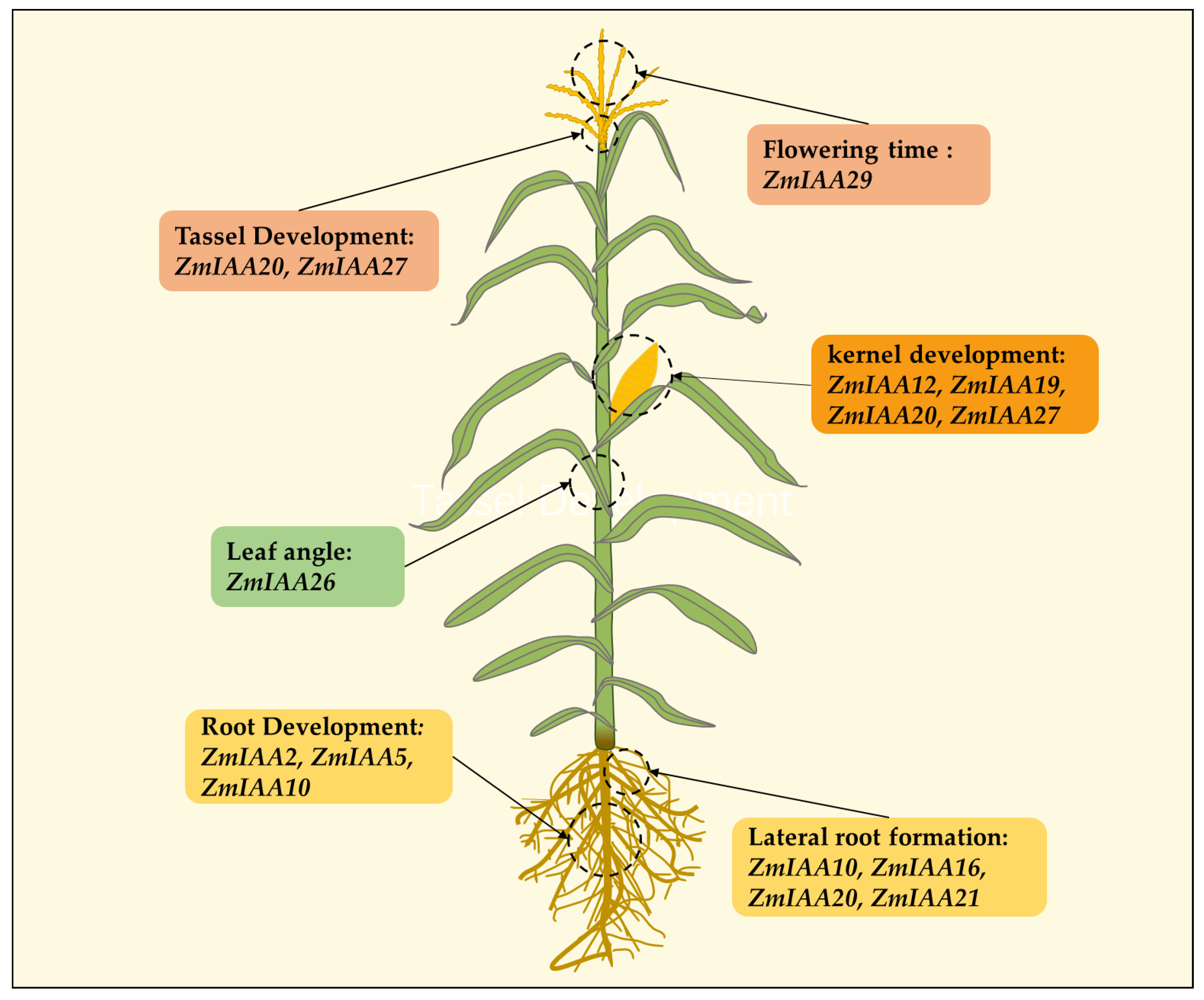
6. Research on the Functions of Aux/IAA Genes in Maize
6.1. Regulation of Aux/IAA Genes in Maize Growth and Development
6.2. The Impact of Aux/IAA Genes on Abiotic Stress in Maize
7. Research Progress on the Aux/IAA Family in Other Plants
8. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Leyser, O. The Power of Auxin in Plants. Plant Physiol. 2010, 154, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Larsson, E.; Vivian-Smith, A.; Offringa, R.; Sundberg, E. Auxin Homeostasis in Arabidopsis Ovules Is Anther-Dependent at Maturation and Changes Dynamically upon Fertilization. Front. Plant Sci. 2017, 8, 1735. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-X.; Zhang, L.-F.; Li, W.-F.; Qi, L.-W.; Han, S.-Y. MIR166a Affects the Germination of Somatic Embryos in Larixleptolepis by Modulating IAA Biosynthesis and Signaling Genes. J. Plant Growth Regul. 2017, 36, 889–896. [Google Scholar] [CrossRef]
- Park, S.-H.; Elhiti, M.; Wang, H.; Xu, A.; Brown, D.; Wang, A. Adventitious root formation of in vitro peach shoots is regulated by auxin and ethylene. Sci. Hortic. 2017, 226, 250–260. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Schuller, A.; Kehr, J.; Ludwig-Müller, J. Laser Microdissection Coupled to Transcriptional Profiling of Arabidopsis Roots Inoculated by Plasmodiophora brassicae Indicates a Role for Brassinosteroids in Clubroot Formation. Plant Cell Physiol. 2013, 55, 392–411. [Google Scholar] [CrossRef]
- Devos, S.; Laukens, K.; Deckers, P.; Van Der Straeten, D.; Beeckman, T.; Inzé, D.; Van Onckelen, H.; Witters, E.; Prinsen, E. A Hormone and Proteome Approach to Picturing the Initial Metabolic Events During Plasmodiophora brassicae Infection on Arabidopsis. Mol. Plant-Microbe Interact.® 2006, 19, 1431–1443. [Google Scholar] [CrossRef]
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef]
- Guilfoyle, T.J. The PB1 Domain in Auxin Response Factor and Aux/IAA Proteins: A Versatile Protein Interaction Module in the Auxin Response. Plant Cell 2015, 27, 33–43. [Google Scholar] [CrossRef]
- Bao, D.; Chang, S.; Li, X.; Qi, Y. Advances in the study of auxin early response genes: Aux/IAA, GH3, and SAUR. Crop J. 2024, 12, 964–978. [Google Scholar] [CrossRef]
- Reed, J.W. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 2001, 6, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhou, J.-J.; Zhang, J.-Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Qiao, L.; Zhang, X.; Han, X.; Zhang, L.; Li, X.; Zhan, H.; Ma, J.; Luo, P.; Zhang, W.; Cui, L.; et al. A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.). Front. Plant Sci. 2015, 6, 770. [Google Scholar] [CrossRef]
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. TOPLESS Regulates Apical Embryonic Fate in Arabidopsis. Science 2006, 312, 1520–1523. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of Auxin-Regulated Gene Expression in Plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Getting a grasp on domain III/IV responsible for Auxin Response Factor–IAA protein interactions. Plant Sci. 2012, 190, 82–88. [Google Scholar] [CrossRef]
- Cui, X.; Wang, J.; Li, K.; Lv, B.; Hou, B.; Ding, Z. Protein post-translational modifications in auxin signaling. J. Genet. Genom. 2024, 51, 279–291. [Google Scholar] [CrossRef]
- Tang, W.; Yu, Y.; Xu, T. The interplay between extracellular and intracellular auxin signaling in plants. J. Genet. Genom. 2025, 52, 14–23. [Google Scholar] [CrossRef]
- Liu, L.; Yahaya, B.S.; Li, J.; Wu, F. Enigmatic role of auxin response factors in plant growth and stress tolerance. Front. Plant Sci. 2024, 15, 1398818. [Google Scholar] [CrossRef]
- Wang, R.; Estelle, M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 2014, 21, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; Park, J.W.; Lee, H.W.; Kim, J. Genome-wide analysis of the auxin-responsive transcriptome downstream of iaa1 and its expression analysis reveal the diversity and complexity of auxin-regulated gene expression. J. Exp. Bot. 2009, 60, 3935–3957. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The Roles of Auxin Response Factor Domains in Auxin-Responsive Transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Li, S.-B.; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef]
- Leyser, O. Auxin Signaling. Plant Physiol. 2017, 176, 465–479. [Google Scholar] [CrossRef]
- Cho, M.; Cho, H. The function of ABCB transporters in auxin transport. Plant Signal. Behav. 2013, 8, e22990. [Google Scholar] [CrossRef]
- Dreher, K.A.; Brown, J.; Saw, R.E.; Callis, J. The Arabidopsis Aux/IAA Protein Family Has Diversified in Degradation and Auxin Responsiveness. Plant Cell 2006, 18, 699–714. [Google Scholar] [CrossRef]
- Chen, H.; Ma, B.; Zhou, Y.; He, S.-J.; Tang, S.-Y.; Lu, X.; Xie, Q.; Chen, S.-Y.; Zhang, J.-S. E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. Proc. Natl. Acad. Sci. USA 2018, 115, 4513–4518. [Google Scholar] [CrossRef]
- Cao, M.; Chen, R.; Li, P.; Yu, Y.; Zheng, R.; Ge, D.; Zheng, W.; Wang, X.; Gu, Y.; Gelová, Z.; et al. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 2019, 568, 240–243. [Google Scholar] [CrossRef]
- Lv, B.; Yu, Q.; Liu, J.; Wen, X.; Yan, Z.; Hu, K.; Li, H.; Kong, X.; Li, C.; Tian, H.; et al. Non-canonical AUX/IAA protein IAA33 competes with canonical AUX/IAA repressor IAA5 to negatively regulate auxin signaling. EMBO J. 2020, 39, e101515. [Google Scholar] [CrossRef]
- Yu, Y.; Tang, W.; Lin, W.; Li, W.; Zhou, X.; Li, Y.; Chen, R.; Zheng, R.; Qin, G.; Cao, W.; et al. ABLs and TMKs are co-receptors for extracellular auxin. Cell 2023, 186, 5457–5471.e5417. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lee, S.; So, J.-h.; Dharmasiri, S.; Dharmasiri, N.; Ge, L.; Jensen, C.; Hangarter, R.; Hobbie, L.; Estelle, M. The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J. 2004, 40, 772–782. [Google Scholar] [CrossRef]
- Ku, S.-J.; Park, J.-Y.; Ha, S.-B.; Kim, J. Overexpression of IAA1 with domain II mutation impairs cell elongation and cell division in inflorescences and leaves of Arabidopsis. J. Plant Physiol. 2009, 166, 548–553. [Google Scholar] [CrossRef]
- Tian, Q.; Reed, J.W. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 1999, 126, 711–721. [Google Scholar] [CrossRef]
- Arase, F.; Nishitani, H.; Egusa, M.; Nishimoto, N.; Sakurai, S.; Sakamoto, N.; Kaminaka, H. IAA8 Involved in Lateral Root Formation Interacts with the TIR1 Auxin Receptor and ARF Transcription Factors in Arabidopsis. PLoS ONE 2012, 7, e43414. [Google Scholar] [CrossRef]
- Fukaki, H.; Nakao, Y.; Okushima, Y.; Theologis, A.; Tasaka, M. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 2005, 44, 382–395. [Google Scholar] [CrossRef]
- Fukaki, H.; Tameda, S.; Masuda, H.; Tasaka, M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002, 29, 153–168. [Google Scholar] [CrossRef]
- Vanneste, S.; De Rybel, B.; Beemster, G.T.S.; Ljung, K.; De Smet, I.; Van Isterdael, G.; Naudts, M.; Iida, R.; Gruissem, W.; Tasaka, M.; et al. Cell Cycle Progression in the Pericycle Is Not Sufficient for SOLITARY ROOT/IAA14-Mediated Lateral Root Initiation in Arabidopsis thaliana. Plant Cell 2005, 17, 3035–3050. [Google Scholar] [CrossRef]
- Wójcikowska, B.; Belaidi, S.; Robert, H.S. Game of thrones among AUXIN RESPONSE FACTORs—Over 30 years of MONOPTEROS research. J. Exp. Bot. 2023, 74, 6904–6921. [Google Scholar] [CrossRef]
- Uehara, T.; Okushima, Y.; Mimura, T.; Tasaka, M.; Fukaki, H. Domain II Mutations in CRANE/IAA18 Suppress Lateral Root Formation and Affect Shoot Development in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T.; Benkova, E.; Bäurle, I.; Kientz, M.; Jürgens, G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002, 16, 1610–1615. [Google Scholar] [CrossRef]
- Rinaldi, M.A.; Liu, J.; Enders, T.A.; Bartel, B.; Strader, L.C. A gain-of-function mutation in IAA16 confers reduced responses to auxin and abscisic acid and impedes plant growth and fertility. Plant Mol. Biol. 2012, 79, 359–373. [Google Scholar] [CrossRef]
- Shahzad, Z.; Eaglesfield, R.; Carr, C.; Amtmann, A. Cryptic variation in RNA-directed DNA-methylation controls lateral root development when auxin signalling is perturbed. Nat. Commun. 2020, 11, 218. [Google Scholar] [CrossRef]
- Rogg, L.E.; Lasswell, J.; Bartel, B. A Gain-of-Function Mutation in IAA28 Suppresses Lateral Root Development. Plant Cell 2001, 13, 465–480. [Google Scholar] [CrossRef]
- De Rybel, B.; Vassileva, V.; Parizot, B.; Demeulenaere, M.; Grunewald, W.; Audenaert, D.; Van Campenhout, J.; Overvoorde, P.; Jansen, L.; Vanneste, S.; et al. A Novel Aux/IAA28 Signaling Cascade Activates GATA23-Dependent Specification of Lateral Root Founder Cell Identity. Curr. Biol. 2010, 20, 1697–1706. [Google Scholar] [CrossRef]
- Timpte, C.; Wilson, A.K.; Estelle, M. The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 1994, 138, 1239–1249. [Google Scholar] [CrossRef]
- Sato, A.; Sasaki, S.; Matsuzaki, J.; Yamamoto, K.T. Light-dependent gravitropism and negative phototropism of inflorescence stems in a dominant Aux/IAA mutant of Arabidopsis thaliana, axr2. J. Plant Res. 2014, 127, 627–639. [Google Scholar] [CrossRef]
- Belin, C.; Megies, C.; Hauserová, E.; Lopez-Molina, L. Abscisic Acid Represses Growth of the Arabidopsis Embryonic Axis after Germination by Enhancing Auxin Signaling. Plant Cell 2009, 21, 2253–2268. [Google Scholar] [CrossRef]
- Rouse, D.; Mackay, P.; Stirnberg, P.; Estelle, M.; Leyser, O. Changes in Auxin Response from Mutations in an AUX/IAA Gene. Science 1998, 279, 1371–1373. [Google Scholar] [CrossRef]
- Kubalová, M.; Müller, K.; Dobrev, P.I.; Rizza, A.; Jones, A.M.; Fendrych, M. Auxin co-receptor IAA17/AXR3 controls cell elongation in Arabidopsis thaliana root solely by modulation of nuclear auxin pathway. New Phytol. 2024, 241, 2448–2463. [Google Scholar] [CrossRef] [PubMed]
- Colón-Carmona, A.n.; Chen, D.L.; Yeh, K.-C.; Abel, S. Aux/IAA Proteins Are Phosphorylated by Phytochrome in Vitro1. Plant Physiol. 2000, 124, 1728–1738. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Yamamoto, K.T. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 2008, 133, 397–405. [Google Scholar] [CrossRef]
- Müller, C.J.; Valdés, A.E.; Wang, G.; Ramachandran, P.; Beste, L.; Uddenberg, D.; Carlsbecker, A. PHABULOSA Mediates an Auxin Signaling Loop to Regulate Vascular Patterning in Arabidopsis. Plant Physiol. 2016, 170, 956–970. [Google Scholar] [CrossRef]
- Kirungu, J.N.; Magwanga, R.O.; Lu, P.; Cai, X.; Zhou, Z.; Wang, X.; Peng, R.; Wang, K.; Liu, F. Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genet. 2019, 20, 62. [Google Scholar] [CrossRef]
- Ma, J.; Wang, L.-y.; Dai, J.-x.; Wang, Y.; Lin, D. The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 11. [Google Scholar] [CrossRef]
- Lin, X.; Li, R.; Zhou, Y.; Tang, F.; Wang, Y.; Lu, X.; Wang, S.; Yao, Y.; Liu, J.; Hu, X.; et al. Overexpression of Cassava MeAnn2 Enhances the Salt and IAA Tolerance of Transgenic Arabidopsis. Plants 2021, 10, 941. [Google Scholar] [CrossRef]
- Nam, H.; Han, S.; Lee, S.; Nam, H.; Lim, H.; Lee, G.; Cho, H.S.; Dang, T.V.T.; Choi, S.; Lee, M.M.; et al. CPR5-mediated nucleo-cytoplasmic localization of IAA12 and IAA19 controls lateral root development during abiotic stress. Proc. Natl. Acad. Sci. USA 2023, 120, e2209781120. [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10, 4021. [Google Scholar] [CrossRef]
- López-Bucio, J.; Ortiz-Castro, R.; Ruíz-Herrera, L.F.; Juárez, C.V.; Hernández-Madrigal, F.; Carreón-Abud, Y.; Martínez-Trujillo, M. Chromate induces adventitious root formation via auxin signalling and SOLITARY-ROOT/IAA14 gene function in Arabidopsis thaliana. BioMetals 2015, 28, 353–365. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. OsIAA1, an Aux/IAA cDNA from Rice, and Changes in Its Expression as Influenced by Auxin and Light. DNA Res. 2001, 8, 193–203. [Google Scholar] [CrossRef]
- Song, Y.; You, J.; Xiong, L. Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol. Biol. 2009, 70, 297–309. [Google Scholar] [CrossRef]
- Xian, F.; Liu, S.; Huang, J.; Xie, B.; Zhu, L.; Zhang, Q.; Lv, C.; Xu, Y.; Zhang, X.; Hu, J. The OsIAA3-OsARF16-OsBUL1 auxin signaling module regulates grain size in rice. Plant Physiol. 2025, 197, kiaf122. [Google Scholar] [CrossRef]
- Zhang, Z. Cloning and Molecular Mechanism Analysis of Yield Related Gene Gnp4 in Rice. Ph.D. Thesis, China Agricultural University, Beijing, China, 2015. [Google Scholar]
- Zhang, Z.; Li, J.; Tang, Z.; Sun, X.; Zhang, H.; Yu, J.; Yao, G.; Li, G.; Guo, H.; Li, J.; et al. Gnp4/LAX2, a RAWUL protein, interferes with the OsIAA3–OsARF25 interaction to regulate grain length via the auxin signaling pathway in rice. J. Exp. Bot. 2018, 69, 4723–4737. [Google Scholar] [CrossRef]
- Qiao, J.; Jiang, H.; Lin, Y.; Shang, L.; Wang, M.; Li, D.; Fu, X.; Geisler, M.; Qi, Y.; Gao, Z.; et al. A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 2021, 14, 1683–1698. [Google Scholar] [CrossRef]
- Song, Y.; Xu, Z.-F. Ectopic Overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) Gene OsIAA4 in Rice Induces Morphological Changes and Reduces Responsiveness to Auxin. Int. J. Mol. Sci. 2013, 14, 13645–13656. [Google Scholar] [CrossRef]
- Jung, H.; Lee, D.-K.; Choi, Y.D.; Kim, J.-K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 2015, 236, 304–312. [Google Scholar] [CrossRef]
- Han, S.; Hwang, I. Integration of multiple signaling pathways shapes the auxin response. J. Exp. Bot. 2017, 69, 189–200. [Google Scholar] [CrossRef]
- Hu, Y.; Xue, D.; Wang, S.; Zhang, Q.; Zhang, X.; Yang, J.; Lv, Y.; Yan, B.; Yin, Y.; Cui, Z.; et al. An auxin response factor regulates tiller angle and shoot gravitropism by directly activating related gene expression in rice. J. Adv. Res. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Zhu, Z.-X.; Liu, Y.; Liu, S.-J.; Mao, C.-Z.; Wu, Y.-R.; Wu, P. A Gain-of-Function Mutation in OsIAA11 Affects Lateral Root Development in Rice. Mol. Plant 2012, 5, 154–161. [Google Scholar] [CrossRef]
- Kang, B.; Zhang, Z.; Wang, L.; Zheng, L.; Mao, W.; Li, M.; Wu, Y.; Wu, P.; Mo, X. OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J. 2013, 74, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Q.; Sang, S.; Wei, Z.; Wang, P. Rice Inositol Polyphosphate Kinase (OsIPK2) Directly Interacts with OsIAA11 to Regulate Lateral Root Formation. Plant Cell Physiol. 2017, 58, 1891–1900. [Google Scholar] [CrossRef]
- Guo, F.; Zhang, H.; Liu, W.; Hu, X.; Han, N.; Qian, Q.; Xu, L.; Bian, H. Callus Initiation from Root Explants Employs Different Strategies in Rice and Arabidopsis. Plant Cell Physiol. 2018, 59, 1782–1789. [Google Scholar] [CrossRef]
- Yu, E.; Yamaji, N.; Mao, C.; Wang, H.; Ma, J.F. Lateral roots but not root hairs contribute to high uptake of manganese and cadmium in rice. J. Exp. Bot. 2021, 72, 7219–7228. [Google Scholar] [CrossRef]
- Chen, S.-H.; Zhou, L.-J.; Xu, P.; Xue, H.-W. SPOC domain-containing protein Leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PLoS Genet. 2018, 14, e1007829. [Google Scholar] [CrossRef]
- Luo, S.; Li, Q.; Liu, S.; Pinas, N.M.; Tian, H.; Wang, S. Constitutive Expression of OsIAA9 Affects Starch Granules Accumulation and Root Gravitropic Response in Arabidopsis. Front. Plant Sci. 2015, 6, 1156. [Google Scholar] [CrossRef]
- Kitomi, Y.; Inahashi, H.; Takehisa, H.; Sato, Y.; Inukai, Y. OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci. 2012, 190, 116–122. [Google Scholar] [CrossRef]
- Jun, N.; Gaohang, W.; Zhenxing, Z.; Huanhuan, Z.; Yunrong, W.; Ping, W. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 2011, 68, 433–442. [Google Scholar] [CrossRef]
- Nakamura, A.; Umemura, I.; Gomi, K.; Hasegawa, Y.; Kitano, H.; Sazuka, T.; Matsuoka, M. Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 2006, 46, 297–306. [Google Scholar] [CrossRef]
- Jain, M.; Khurana, J.P. Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 2009, 276, 3148–3162. [Google Scholar] [CrossRef]
- Zhang, A.; Yang, X.; Lu, J.; Song, F.; Sun, J.; Wang, C.; Lian, J.; Zhao, L.; Zhao, B. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Niu, H.; Xin, D.; Long, Y.; Wang, G.; Liu, Z.; Li, G.; Zhang, F.; Qi, M.; Ye, Y.; et al. OsIAA18, an Aux/IAA Transcription Factor Gene, Is Involved in Salt and Drought Tolerance in Rice. Front. Plant Sci. 2021, 12, 738660. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, Z.; Yu, X.; Liu, C. Bioinformatics analysis of Aux/IAA gene family in maize. Agron. J. 2021, 113, 932–942. [Google Scholar] [CrossRef]
- von Behrens, I.; Komatsu, M.; Zhang, Y.; Berendzen, K.W.; Niu, X.; Sakai, H.; Taramino, G.; Hochholdinger, F. Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J. 2011, 66, 341–353. [Google Scholar] [CrossRef]
- Zhang, Y.; Paschold, A.; Marcon, C.; Liu, S.; Tai, H.; Nestler, J.; Yeh, C.-T.; Opitz, N.; Lanz, C.; Schnable, P.S.; et al. The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J. Exp. Bot. 2014, 65, 4919–4930. [Google Scholar] [CrossRef]
- Galli, M.; Liu, Q.; Moss, B.L.; Malcomber, S.; Li, W.; Gaines, C.; Federici, S.; Roshkovan, J.; Meeley, R.; Nemhauser, J.L.; et al. Auxin signaling modules regulate maize inflorescence architecture. Proc. Natl. Acad. Sci. USA 2015, 112, 13372–13377. [Google Scholar] [CrossRef]
- Yang, F.; Shi, Y.; Zhao, M.; Cheng, B.; Li, X. ZmIAA5 regulates maize root growth and development by interacting with ZmARF5 under the specific binding of ZmTCP15/16/17. PeerJ 2022, 10, e13710. [Google Scholar] [CrossRef]
- Qing, X.; Li, J.; Lin, Z.; Wang, W.; Yi, F.; Chen, J.; Liu, Q.; Song, W.; Lai, J.; Chen, B.; et al. Maize transcription factor ZmEREB167 negatively regulates starch accumulation and kernel size. J. Genet. Genom. 2025, 52, 411–421. [Google Scholar] [CrossRef]
- Ma, C.; Dang, K.; Xie, Q.; Sahito, J.H.; Yuan, B.; Wan, J.; Qiu, X.; Zhao, J.; Lin, Y.; Meng, S.; et al. Over-Expression of ZmIAA29, an AUX/IAA Transcription Factor, Improved Maize Flowering Time. Agronomy 2023, 13, 2028. [Google Scholar] [CrossRef]
- Dou, D.; Han, S.; Cao, L.; Ku, L.; Liu, H.; Su, H.; Ren, Z.; Zhang, D.; Zeng, H.; Dong, Y.; et al. CLA4 regulates leaf angle through multiple hormone signaling pathways in maize. J. Exp. Bot. 2020, 72, 1782–1794. [Google Scholar] [CrossRef]
- Yan, Z.; Li, K.; Li, Y.; Wang, W.; Leng, B.; Yao, G.; Zhang, F.; Mu, C.; Liu, X. The ZmbHLH32-ZmIAA9-ZmARF1 module regulates salt tolerance in maize. Int. J. Biol. Macromol. 2023, 253, 126978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qu, J.; Lu, M.; Zhao, X.; Xu, Y.; Wang, L.; Liu, Z.; Shi, Y.; Liu, C.; Li, Y.; et al. The maize transcription factor CCT regulates drought tolerance by interacting with Fra a 1, E3 ligase WIPF2, and auxin response factor Aux/IAA8. J. Exp. Bot. 2023, 75, 103–122. [Google Scholar] [CrossRef]
- Zhang, M.; Lu, X.; Li, C.; Zhang, B.; Zhang, C.; Zhang, X.-s.; Ding, Z. Auxin Efflux Carrier ZmPGP1 Mediates Root Growth Inhibition under Aluminum Stress. Plant Physiol. 2018, 177, 819–832. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, J.; Wan, J.; Yao, Q.; Zhang, Y.; Mi, G.; Chen, L.; Li, Z.; Zhang, M. Auxin efflux carrier ZmPIN1a modulates auxin reallocation involved in nitrate-mediated root formation. BMC Plant Biol. 2023, 23, 74. [Google Scholar] [CrossRef]
- Chen, D.; Richardson, T.; Chai, S.; Lynne McIntyre, C.; Rae, A.L.; Xue, G.-P. Drought-Up-Regulated TaNAC69-1 is a Transcriptional Repressor of TaSHY2 and TaIAA7, and Enhances Root Length and Biomass in Wheat. Plant Cell Physiol. 2016, 57, 2076–2090. [Google Scholar] [CrossRef]
- Jia, M.; Li, Y.; Wang, Z.; Tao, S.; Sun, G.; Kong, X.; Wang, K.; Ye, X.; Liu, S.; Geng, S.; et al. TaIAA21 represses TaARF25-mediated expression of TaERFs required for grain size and weight development in wheat. Plant J. 2021, 108, 1754–1767. [Google Scholar] [CrossRef]
- Zhao, L.; Zheng, Y.; Wang, Y.; Wang, S.; Wang, T.; Wang, C.; Chen, Y.; Zhang, K.; Zhang, N.; Dong, Z.; et al. A HST1-like gene controls tiller angle through regulating endogenous auxin in common wheat. Plant Biotechnol. J. 2023, 21, 122–135. [Google Scholar] [CrossRef]
- Su, P.; Sui, C.; Li, J.; Wan, K.; Sun, H.; Wang, S.; Liu, X.; Guo, S. The Aux/IAA protein TaIAA15-1A confers drought tolerance in Brachypodium by regulating abscisic acid signal pathway. Plant Cell Rep. 2023, 42, 385–394. [Google Scholar] [CrossRef]
- Zeng, D.; Peng, J.; Zhang, L.; Hayden, M.J.; Rathjen, T.M.; Li, X.; Jiang, W.; Delhaize, E. Twisted Sister1: An agravitropic mutant of bread wheat (Triticum aestivum) with altered root and shoot architectures. Plant J. 2025, 122, e70122. [Google Scholar] [CrossRef]
- Audran-Delalande, C.; Bassa, C.; Mila, I.; Regad, F.; Zouine, M.; Bouzayen, M. Genome-Wide Identification, Functional Analysis and Expression Profiling of the Aux/IAA Gene Family in Tomato. Plant Cell Physiol. 2012, 53, 659–672. [Google Scholar] [CrossRef]
- Wu, L.; Tian, Z.; Zhang, J. Functional Dissection of Auxin Response Factors in Regulating Tomato Leaf Shape Development. Front. Plant Sci. 2018, 9, 957. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Israeli, A.; Ori, N.; Sun, T.-p. The Interaction between DELLA and ARF/IAA Mediates Crosstalk between Gibberellin and Auxin Signaling to Control Fruit Initiation in Tomato. Plant Cell 2018, 30, 1710–1728. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Yan, F.; Liu, M.; Wang, X.; Li, Z. Down-regulation of SlIAA15 in tomato altered stem xylem development and production of volatile compounds in leaf exudates. Plant Signal Behav 2012, 7, 911–913. [Google Scholar] [CrossRef]
- Su, L.; Bassa, C.; Audran, C.; Mila, I.; Cheniclet, C.; Chevalier, C.; Bouzayen, M.; Roustan, J.-P.; Chervin, C. The Auxin Sl-IAA17 Transcriptional Repressor Controls Fruit Size Via the Regulation of Endoreduplication-Related Cell Expansion. Plant Cell Physiol. 2014, 55, 1969–1976. [Google Scholar] [CrossRef]
- Bassa, C.; Mila, I.; Bouzayen, M.; Audran-Delalande, C. Phenotypes Associated with Down-Regulation of Sl-IAA27 Support Functional Diversity Among Aux/IAA Family Members in Tomato. Plant Cell Physiol. 2012, 53, 1583–1595. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, L.; Cai, Y.; Liu, Y.; Yan, X.; Liu, J.; Li, R.; Ge, S.; Wang, S.; Liu, X.; et al. A KNOTTED1-LIKE HOMEOBOX PROTEIN1–interacting transcription factor SlGATA6 maintains the auxin-response gradient to inhibit abscission. Sci. Adv. 2025, 11, eadt1891. [Google Scholar] [CrossRef]
- Gao, J.; Cao, X.; Shi, S.; Ma, Y.; Wang, K.; Liu, S.; Chen, D.; Chen, Q.; Ma, H. Genome-wide survey of Aux/IAA gene family members in potato (Solanum tuberosum): Identification, expression analysis, and evaluation of their roles in tuber development. Biochem. Biophys. Res. Commun. 2016, 471, 320–327. [Google Scholar] [CrossRef]
- Kloosterman, B.; Visser, R.G.F.; Bachem, C.W.B. Isolation and characterization of a novel potato Auxin/Indole-3-Acetic Acid family member (StIAA2) that is involved in petiole hyponasty and shoot morphogenesis. Plant Physiol. Biochem. 2006, 44, 766–775. [Google Scholar] [CrossRef]
- Wu, J.; Peng, Z.; Liu, S.; He, Y.; Cheng, L.; Kong, F.; Wang, J.; Lu, G. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol. Genet. Genom. 2012, 287, 295–311. [Google Scholar] [CrossRef]
- Jiang, M.; Hu, H.; Kai, J.; Traw, M.B.; Yang, S.; Zhang, X. Different knockout genotypes of OsIAA23 in rice using CRISPR/Cas9 generating different phenotypes. Plant Mol. Biol. 2019, 100, 467–479. [Google Scholar] [CrossRef]
- Jia, S.-S.; Ren, X.-Y.; Tong, M.-N.; Jiang, S.-Y.; Zhang, C.-Q.; Liu, Q.-Q.; Li, Q.-F. OsIAA19, an Aux/IAA Family Gene, Involved in the Regulation of Seed-Specific Traits in Rice. Plants 2024, 13, 3538. [Google Scholar] [CrossRef]
- Martin-Arevalillo, R.; Guillotin, B.; Schön, J.; Hugues, A.; Gerentes, M.-F.; Tang, K.; Lucas, J.; Thévenon, E.; Dreuillet, M.; Vissers, G.; et al. Synthetic deconvolution of an auxin-dependent transcriptional code. Cell 2025. [Google Scholar] [CrossRef]



| Species | Gene Name | Function | References |
|---|---|---|---|
| Wheat (Triticum aestivum) | TaIAA7 | Involved in regulating wheat root system development | [97] |
| TaIAA15 | Positively regulates wheat drought tolerance | [100] | |
| TaIAA17 | Involved in regulating wheat tillering pattern | [99] | |
| TaIAA19 | Involved in regulating wheat’s gravitropic response process | [101] | |
| TaIAA21 | Regulates wheat grain size and grain weight | [98] | |
| Tomato (Lycopersicon esculentum) | SlIAA3 | Involved in regulating organ abscission | [108] |
| SlIAA9 | Involved in regulating tomato leaf development and fruit formation | [103,104] | |
| SlIAA15 | Involved in regulating tomato plant leaf and lateral root development | [105] | |
| SlIAA17 | Involved in regulating tomato fruit size | [106] | |
| SlIAA27 | Involved in tomato nutrient and reproductive growth | [107] | |
| Potato (Solanum tuberosum) | StIAA2 | Involved in regulating potato plant morphology and plant height | [110] |
| StIAA9 | involved in the formation and development of potato tubers | [109] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, Z.; Bian, J.; Ren, Z.; Ta, W.; Peng, Y. Plant Aux/IAA Gene Family: Significance in Growth, Development and Stress Responses. Agronomy 2025, 15, 1228. https://doi.org/10.3390/agronomy15051228
Zhuang Z, Bian J, Ren Z, Ta W, Peng Y. Plant Aux/IAA Gene Family: Significance in Growth, Development and Stress Responses. Agronomy. 2025; 15(5):1228. https://doi.org/10.3390/agronomy15051228
Chicago/Turabian StyleZhuang, Zelong, Jianwen Bian, Zhenping Ren, Wanling Ta, and Yunling Peng. 2025. "Plant Aux/IAA Gene Family: Significance in Growth, Development and Stress Responses" Agronomy 15, no. 5: 1228. https://doi.org/10.3390/agronomy15051228
APA StyleZhuang, Z., Bian, J., Ren, Z., Ta, W., & Peng, Y. (2025). Plant Aux/IAA Gene Family: Significance in Growth, Development and Stress Responses. Agronomy, 15(5), 1228. https://doi.org/10.3390/agronomy15051228





