Comparative Transcriptome Analysis of Susceptible and Resistant Rutaceae Plants to Huanglongbing
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. RNA Sequencing and Transcriptomic Analysis
2.3. Data Processing
2.4. Differential Expression and Functional Enrichment Analysis
3. Results
3.1. Transcriptome Analysis of the Effects of Huanglongbing on Ponkan Mandarin and Punctate Wampee
3.2. Comparative Transcriptomics Reveals Key Genes Involved in the Response to Huanglongbing in Citrus
3.3. Identification of Core Genes Conferring Huanglongbing Resistance in Rutaceae Species Through WGCNA
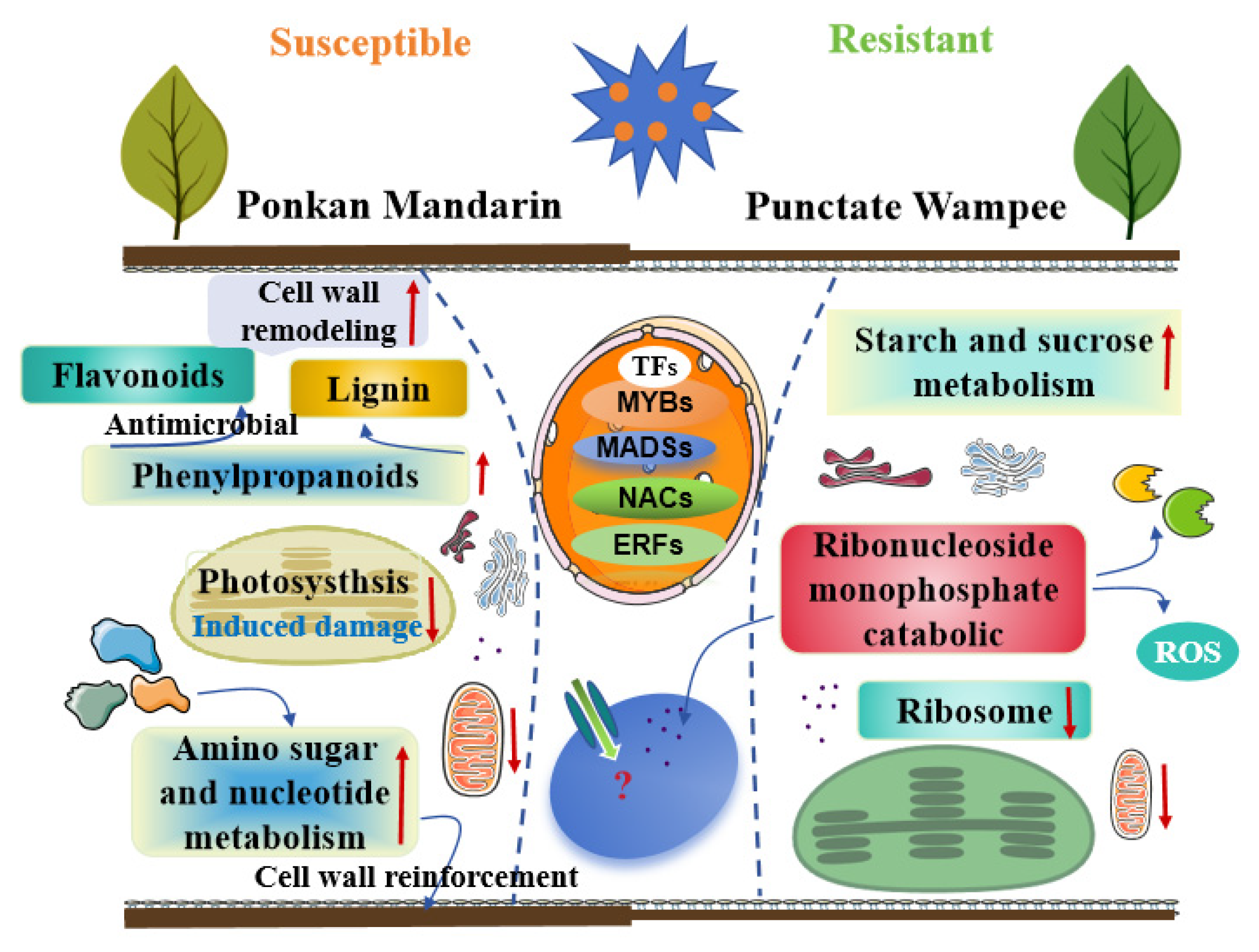
3.4. Proposed Working Model of Huanglongbing Resistance in Ponkan Mandarin and Punctate Wampee
4. Discussions
4.1. Differential Defense Strategies
4.2. Role of Transcription Factors and Transporter Proteins
4.3. Comparative Insights and Resistance Mechanisms
4.4. Implications for Breeding Programs and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus [Asia; South Africa; Brazil; Florida]. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Gottwald, T.R. Current epidemiological understanding of Citrus Huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139. [Google Scholar] [CrossRef] [PubMed]
- da Graça, J.V.; Douhan, G.W.; Halbert, S.E.; Keremane, M.L.; Lee, R.F.; Vidalakis, G.; Zhao, H. Huanglongbing: An overview of a complex pathosystem ravaging the world’s citrus. J. Integr. Plant Biol. 2016, 58, 373–387. [Google Scholar] [CrossRef]
- Graham, J.H.; Bassanezi, R.B.; Dawson, W.O.; Dantzler, R. Management of Huanglongbing of Citrus: Lessons from São Paulo and Florida. Annu. Rev. Phytopathol. 2024, 62, 243–262. [Google Scholar] [CrossRef]
- Hall, D.G.; Richardson, M.L.; Ammar, E.-D.; Halbert, S.E. Asian citrus psyllid, iaphorina citri, vector of citrus huanglongbing disease. Entomol. Exp. Appl. 2013, 146, 207–223. [Google Scholar] [CrossRef]
- Hynniewta, M.; Malik, S.K.; Rao, S.R. Genetic diversity phylogenetic analysis of Citrus (L) from north-east India as revealed by meiosis molecular analysis of internal transcribed spacer region of rDNA. Meta Gene 2014, 2, 237–251. [Google Scholar] [CrossRef]
- Albrecht, U.; Bowman, K.D. Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci. 2012, 185–186, 118–130. [Google Scholar] [CrossRef]
- Ramadugu, C.; Keremane, M.L.; Halbert, S.E.; Duan, Y.P.; Roose, M.L.; Stover, E.; Lee, R.F. Long-term field evaluation reveals Huanglongbing resistance in Citrus relatives. Plant Dis. 2016, 100, 1858–1869. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, X.; Hu, Y.; Huang, G. Analysis of huanglongbing-associated RNA-seq data reveals disturbances in biological processes within Citrus spp. triggered by Candidatus Liberibacter asiaticus infection. Front. Plant Sci. 2024, 15, 1388163. [Google Scholar] [CrossRef] [PubMed]
- Mithöfer, A.; Boland, W. Plant defense against herbivores: Chemical aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef]
- Dutt, M.; Barthe, G.; Irey, M.; Grosser, J. Transgenic Citrus expressing an Arabidopsis NPR1 gene exhibit enhanced resistance against Huanglongbing (HLB; Citrus greening). PLoS ONE 2015, 10, e0137134. [Google Scholar] [CrossRef]
- Preza-Murrieta, B.; Noa-Carrazana, J.C.; Flores-Estévez, N.; Estrella-Maldonado, H.; Santillán-Mendoza, R.; Matilde-Hernández, C.; González-Oviedo, N.A.; Saucedo-Picazo, L.E.; Flores-de la Rosa, F.R. WRKY transcription factors identified in the transcriptome of Citrus latifolia Tan. and their expression in response to Huanglongbing disease. J. General. Plant Pathol. 2024, 90, 309–321. [Google Scholar] [CrossRef]
- Dong, X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004, 7, 547–552. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Li, J.; Brader Gn Palva, E.T. The WRKY70 transcription factor: A Node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef]
- van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Weber, K.C.; Mahmoud, L.M.; Stanton, D.; Welker, S.; Qiu, W.; Grosser, J.W.; Levy, A.; Dutt, M. Insights into the mechanism of Huanglongbing tolerance in the Australian finger lime (Citrus australasica). Front. Plant Sci. 2022, 13, 1019295. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Wang, G.; Zhou, J.-M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Pierson, E.A.; Setubal, J.C.; Xu, J.; Levy, J.G.; Zhang, Y.; Li, J.; Rangel, L.T.; Martins, J. The candidatus liberibacter–host interface: Insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 2017, 55, 451–482. [Google Scholar] [CrossRef]
- Albrecht, U.; Fiehn, O.; Bowman, K.D. Metabolic variations in different citrus rootstock cultivars associated with different responses to Huanglongbing. Plant Physiol. Biochem. 2016, 107, 33–44. [Google Scholar] [CrossRef]
- Li, X.; Wang, N.; She, W.; Guo, Z.; Pan, H.; Yu, Y.; Ye, J.; Pan, D.; Pan, T. Identification and functional analysis of the CgNAC043 gene involved in lignin synthesis from Citrusgrandis “San Hong”. Plants 2022, 11, 403. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Pham-Gia, T.; Hung, T.L. The mean and median absolute deviations. Math. Comput. Model. 2001, 34, 921–936. [Google Scholar] [CrossRef]
- Hématy, K.; Cherk, C.; Somerville, S. Host–pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 2009, 12, 406–413. [Google Scholar] [CrossRef]
- Wang, L.; Chen, M.; Lam, P.-Y.; Dini-Andreote, F.; Dai, L.; Wei, Z. Multifaceted roles of flavonoids mediating plant-microbe interactions. Microbiome 2022, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Ramaroson, M.-L.; Koutouan, C.; Helesbeux, J.-J.; Le Clerc, V.; Hamama, L.; Geoffriau, E.; Briard, M. Role of phenylpropanoids and flavonoids in plant resistance to pests and diseases. Molecules 2022, 27, 8371. [Google Scholar] [CrossRef]
- Shah, A.; Smith, D.L. Flavonoids in agriculture: Chemistry and roles in, biotic and abiotic stress responses, and microbial associations. Agronomy 2020, 10, 1209. [Google Scholar] [CrossRef]
- Morkunas, I.; Ratajczak, L. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plant. 2014, 36, 1607–1619. [Google Scholar] [CrossRef]
- Seifert, G.J. Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr. Opin. Plant Biol. 2004, 7, 277–284. [Google Scholar] [CrossRef]
- Trouvelot, S.; Héloir, M.-C.; Poinssot, B.; Gauthier, A.; Paris, F.; Guillier, C.; Combier, M.; Trdá, L.; Daire, X.; Adrian, M. Carbohydrates in plant immunity and plant protection: Roles and potential application as foliar sprays. Front. Plant Sci. 2014, 5, 592. [Google Scholar] [CrossRef]
- Jeandet, P.; Formela-Luboińska, M.; Labudda, M.; Morkunas, I. The role of sugars in plant responses to stress and their regulatory function during development. Int. J. Mol. Sci. 2022, 23, 5161. [Google Scholar] [CrossRef]
- Kongala, S.I.; Kondreddy, A. A review on plant and pathogen derived carbohydrates, oligosaccharides and their role in plant’s immunity. Carbohydr. Polym. Technol. Appl. 2023, 6, 100330. [Google Scholar] [CrossRef]
- Xu, G.; Greene, G.H.; Yoo, H.; Liu, L.; Marqués, J.; Motley, J.; Dong, X. Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 2017, 545, 487–490. [Google Scholar] [CrossRef]
- Batista-Silva, W.; Heinemann, B.; Rugen, N.; Nunes-Nesi, A.; Araújo, W.L.; Braun, H.-P.; Hildebrandt, T.M. The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 2019, 42, 1630–1644. [Google Scholar] [CrossRef]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Halitschke, R.; Baldwin, I.T. Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 2003, 36, 794–807. [Google Scholar] [CrossRef] [PubMed]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef]
- Ng, D.W.; Abeysinghe, J.K.; Kamali, M. Regulating the Regulators: The control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, D. The WRKY57 transcription factor affects the expression of jasmonate ZIM-Domain genes transcriptionally to compromise botrytis cinerea resistance. Plant Physiol. 2016, 171, 2771–2782. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Bu, Q.; Jiang, H.; Li, C.-B.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008, 18, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef] [PubMed]
- Do, T.H.T.; Martinoia, E.; Lee, Y. Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar] [CrossRef]
- Kang, J.; Hwang, J.-U.; Lee, M.; Kim, Y.-Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef]
- Connorton, J.M.; Balk, J.; Rodríguez-Celma, J. Iron homeostasis in plants—A brief overview. Metallomics 2017, 9, 813–823. [Google Scholar] [CrossRef]
- Hu, Y.; Zhong, X.; Liu, X.; Lou, B.; Zhou, C.; Wang, X. Comparative transcriptome analysis unveils the tolerance mechanisms of Citrus hystrix in response to ‘Candidatus Liberibacter asiaticus’ infection. PLoS ONE 2017, 12, e0189229. [Google Scholar] [CrossRef]
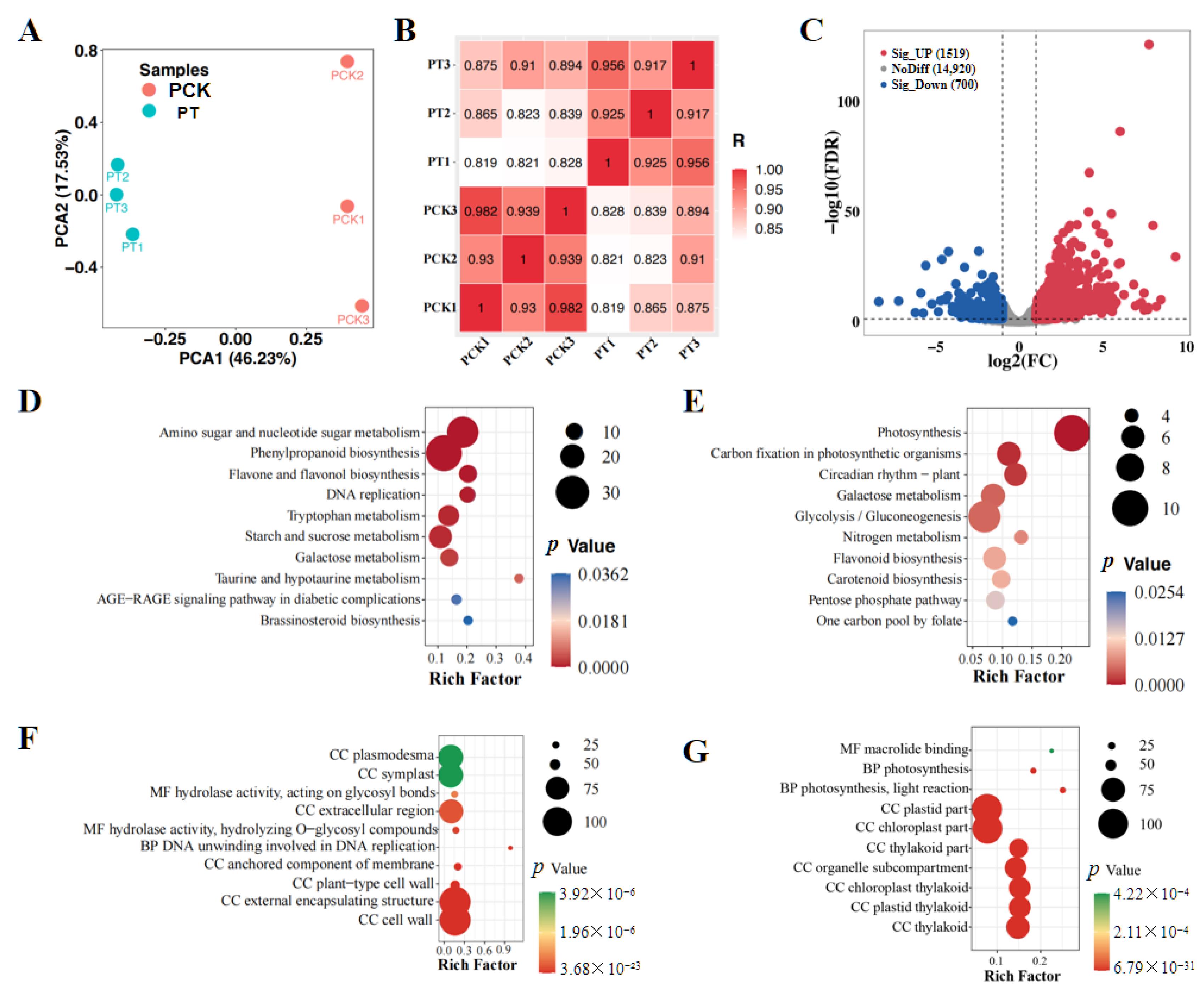
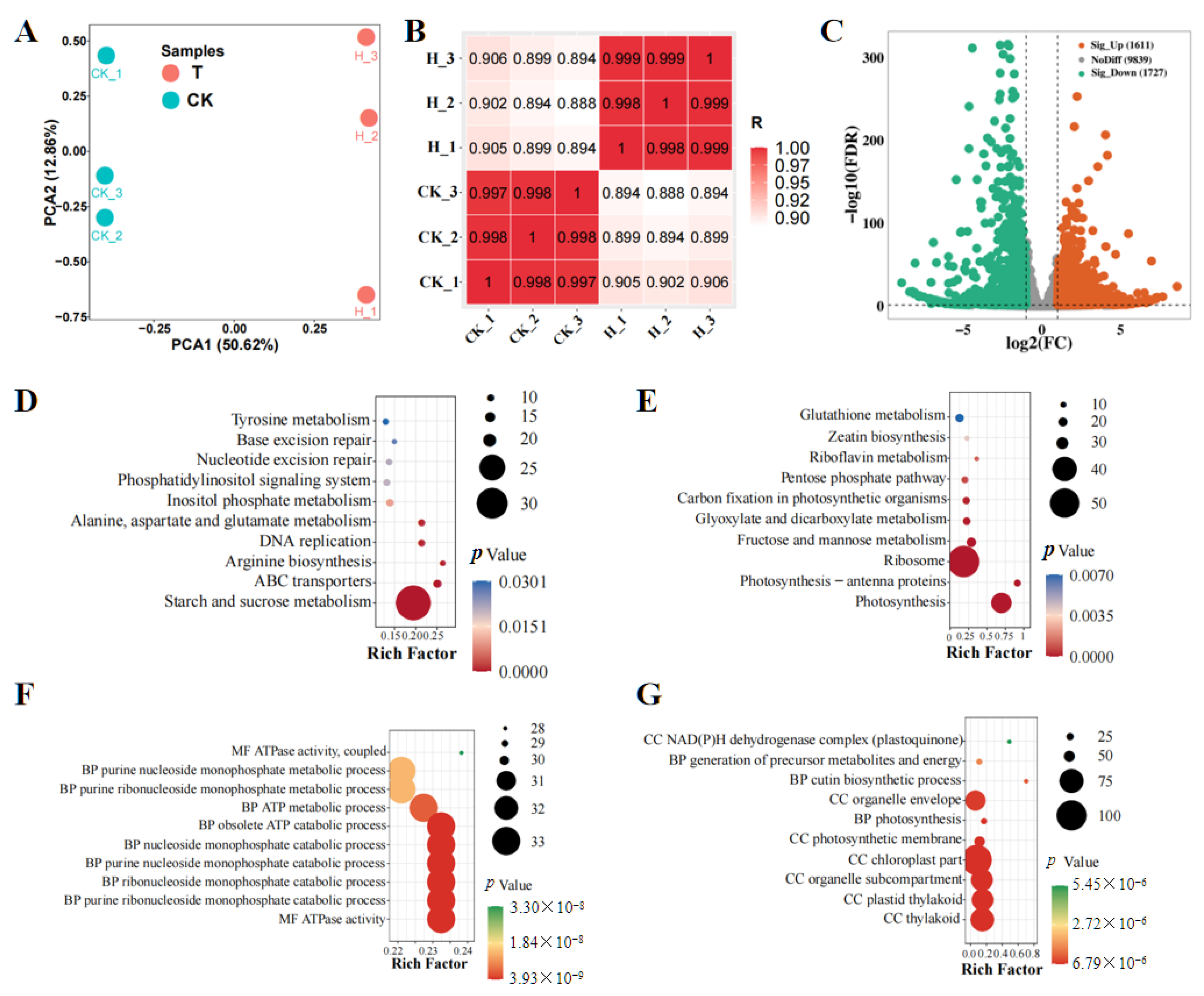
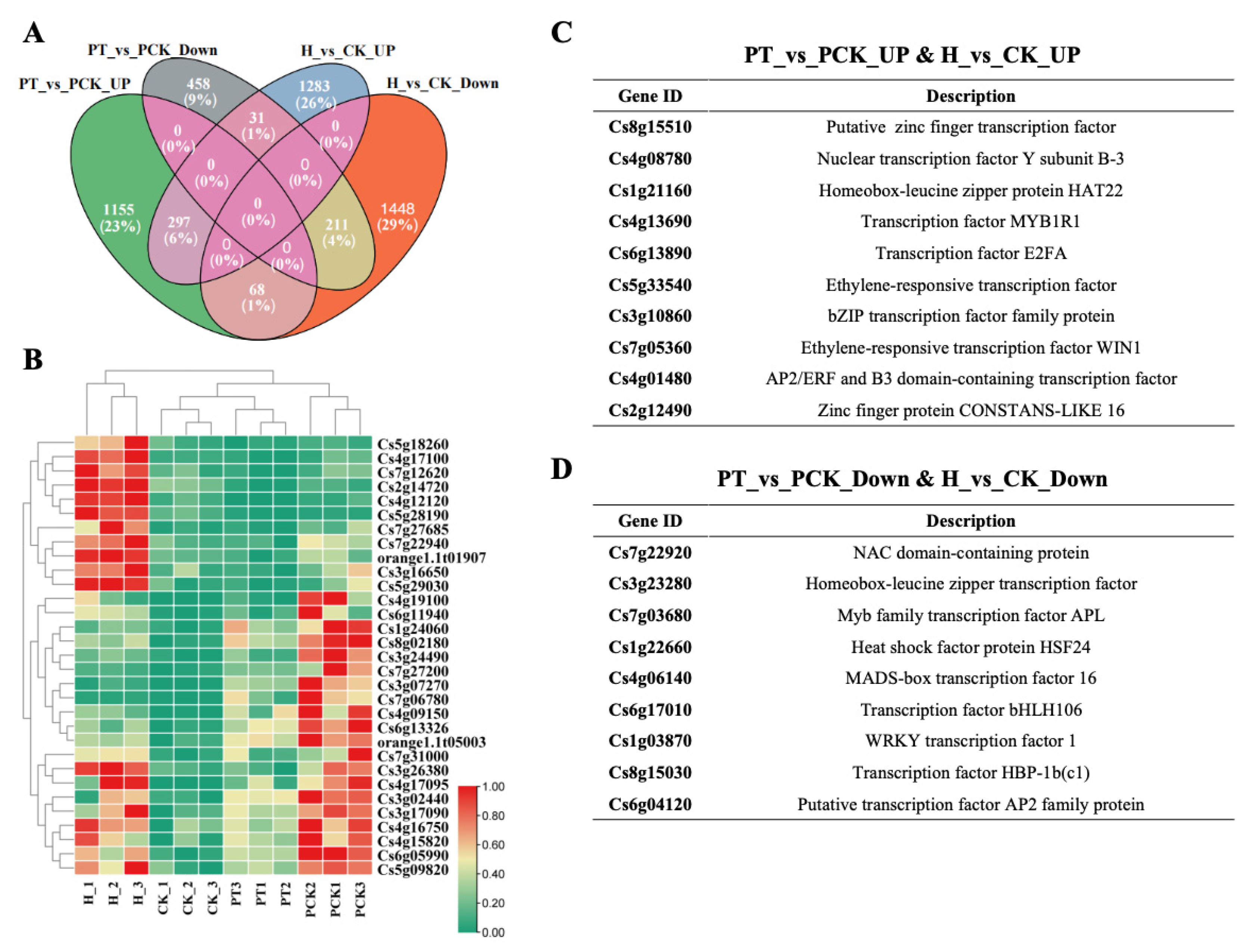

| Gene ID | PT vs. PCK (log2FC) | HT vs. HCK (log2FC) | Description |
|---|---|---|---|
| Cs7g30890 | 0.18 | 0.96 | Clathrin heavy chain 1 |
| Cs1g22900 | 0.22 | 1.02 | Putative uncharacterized protein |
| Cs1g20380 | 0.55 | 1.11 | Similarity to auxin-independent growth promoter |
| Cs6g05890 | 0.38 | 1.55 | Heat shock protein |
| Cs7g05150 | 0.13 | 1.10 | Cellulose synthase A catalytic subunit |
| Cs7g07870 | −0.03 | 0.75 | Cellulose synthase-like protein |
| Cs3g27310 | 0.49 | 0.86 | Similar to patched sphingolipid transporter |
| Cs8g14490 | 0.00 | 1.19 | Serine/threonine-protein phosphatase |
| Cs1g12370 | 0.04 | 0.93 | Serine/threonine-protein phosphatase |
| Cs5g26620 | 0.31 | 1.06 | Casein kinase I isoform delta-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, H.; Liu, F.; Wang, X.; Huang, H.; Huang, Q.; Wang, N.; Wei, C. Comparative Transcriptome Analysis of Susceptible and Resistant Rutaceae Plants to Huanglongbing. Agronomy 2025, 15, 1218. https://doi.org/10.3390/agronomy15051218
Liao H, Liu F, Wang X, Huang H, Huang Q, Wang N, Wei C. Comparative Transcriptome Analysis of Susceptible and Resistant Rutaceae Plants to Huanglongbing. Agronomy. 2025; 15(5):1218. https://doi.org/10.3390/agronomy15051218
Chicago/Turabian StyleLiao, Huihong, Fuping Liu, Xi Wang, Hongming Huang, Qichun Huang, Nina Wang, and Chizhang Wei. 2025. "Comparative Transcriptome Analysis of Susceptible and Resistant Rutaceae Plants to Huanglongbing" Agronomy 15, no. 5: 1218. https://doi.org/10.3390/agronomy15051218
APA StyleLiao, H., Liu, F., Wang, X., Huang, H., Huang, Q., Wang, N., & Wei, C. (2025). Comparative Transcriptome Analysis of Susceptible and Resistant Rutaceae Plants to Huanglongbing. Agronomy, 15(5), 1218. https://doi.org/10.3390/agronomy15051218





