Transcriptomic Analysis for Key Periods of Radicle Development in Contrasting Soybean Varieties HN75 and HN76
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Phenotypic Observations and Trait Measurement
2.3. Transcriptome Sequencing and Data Analysis
2.4. Bioinformatics and Functional Enrichment
2.5. qRT-PCR Validation
2.6. Statistical Analysis
3. Results
3.1. Differences in Root-Related Traits Between Soybeans HN75 and HN76
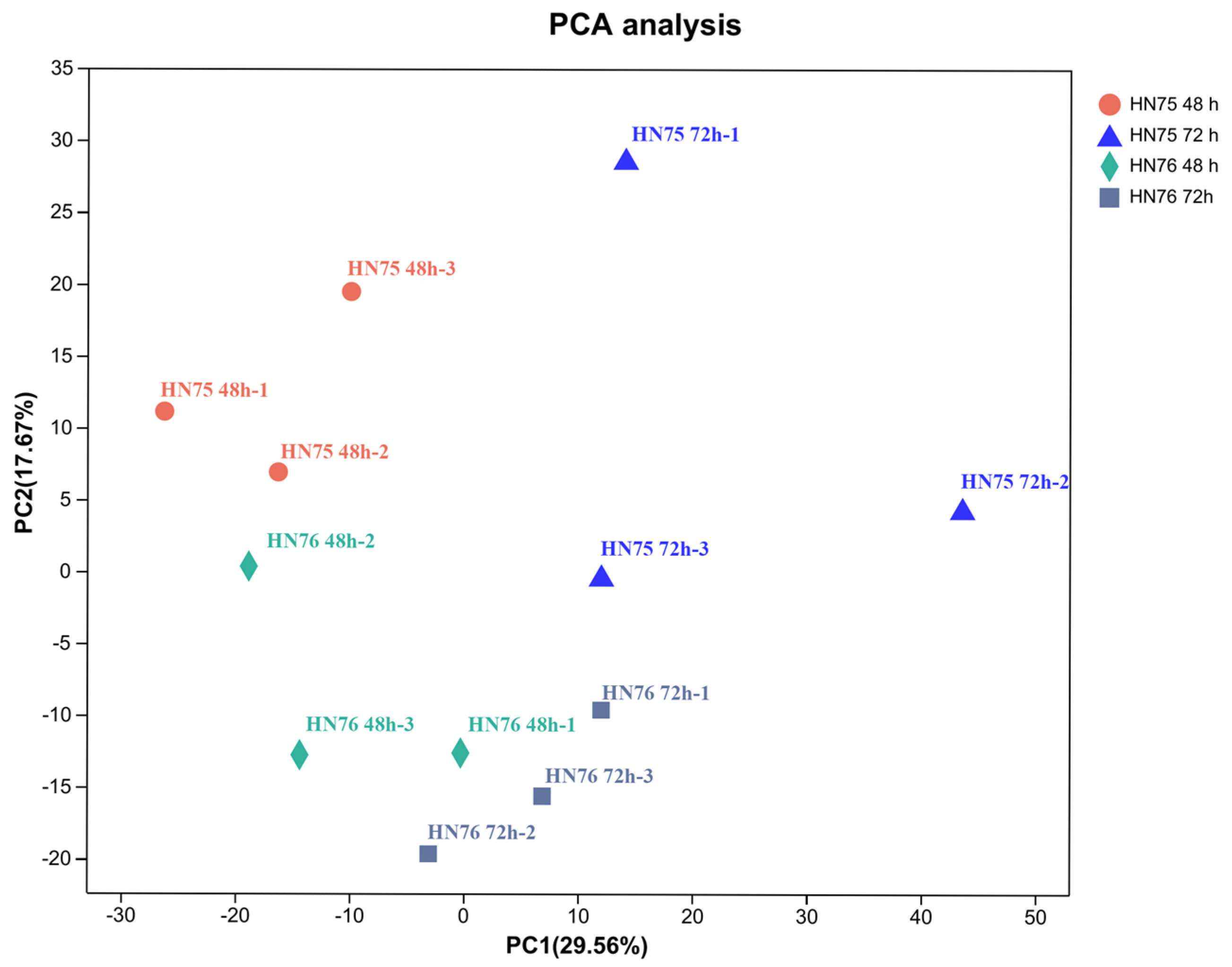
3.2. Principal Component Analysis of the Transcriptome of HN75 and HN76
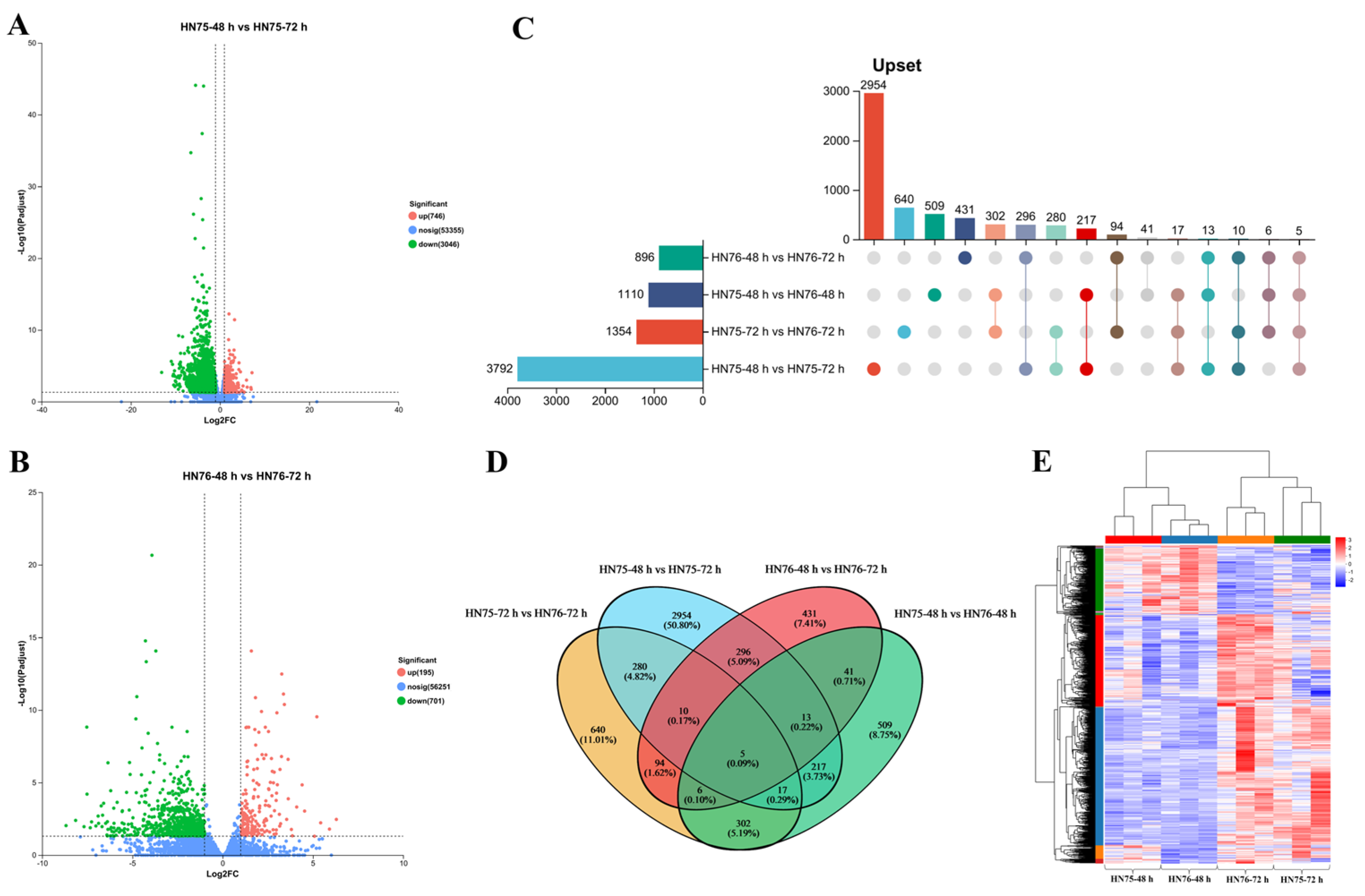
3.3. Identification of DEGs Between HN75 and HN76
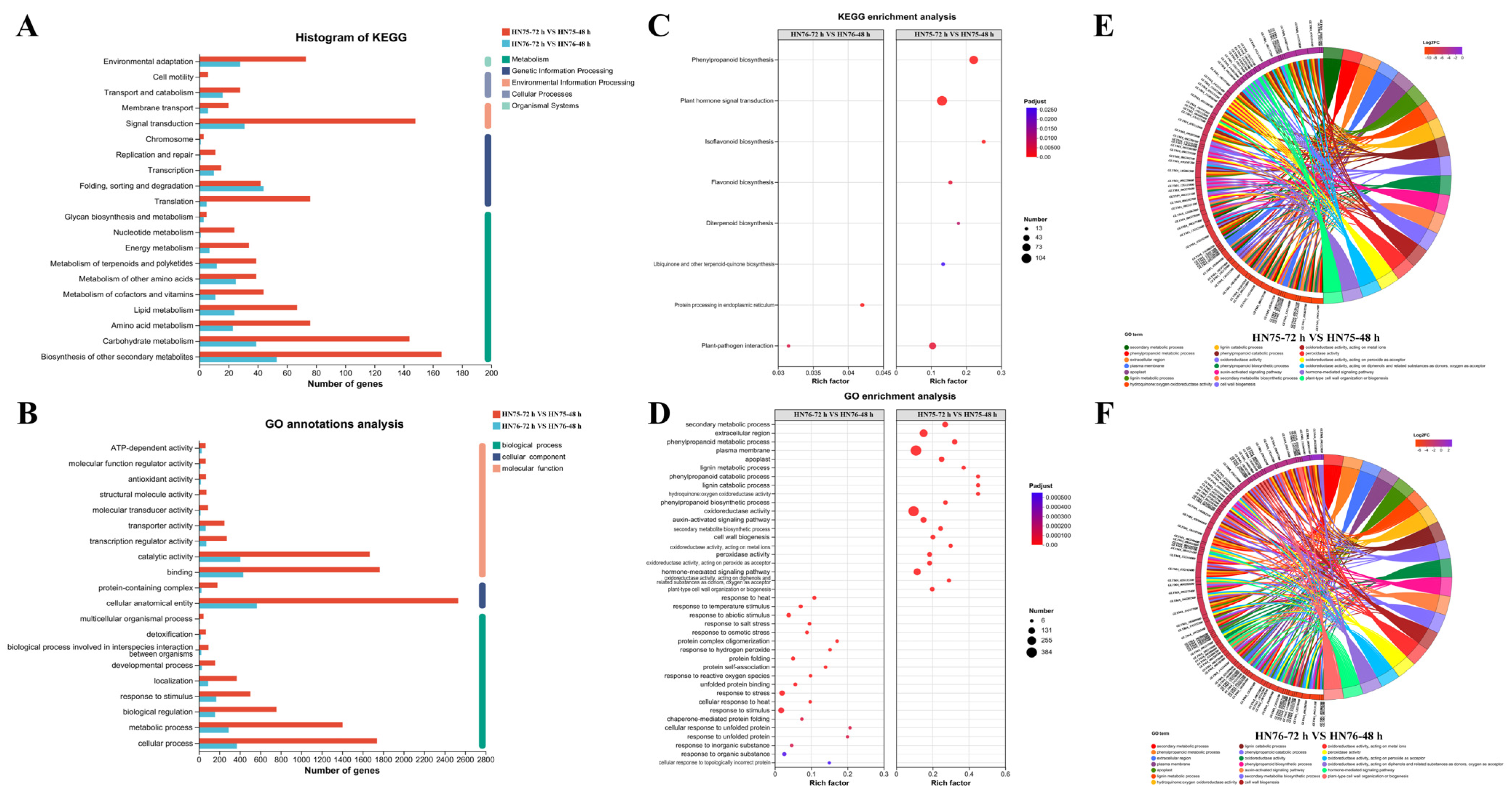
3.4. GO and KEGG Enrichment Analysis of DEGs Between HN75 and HN76
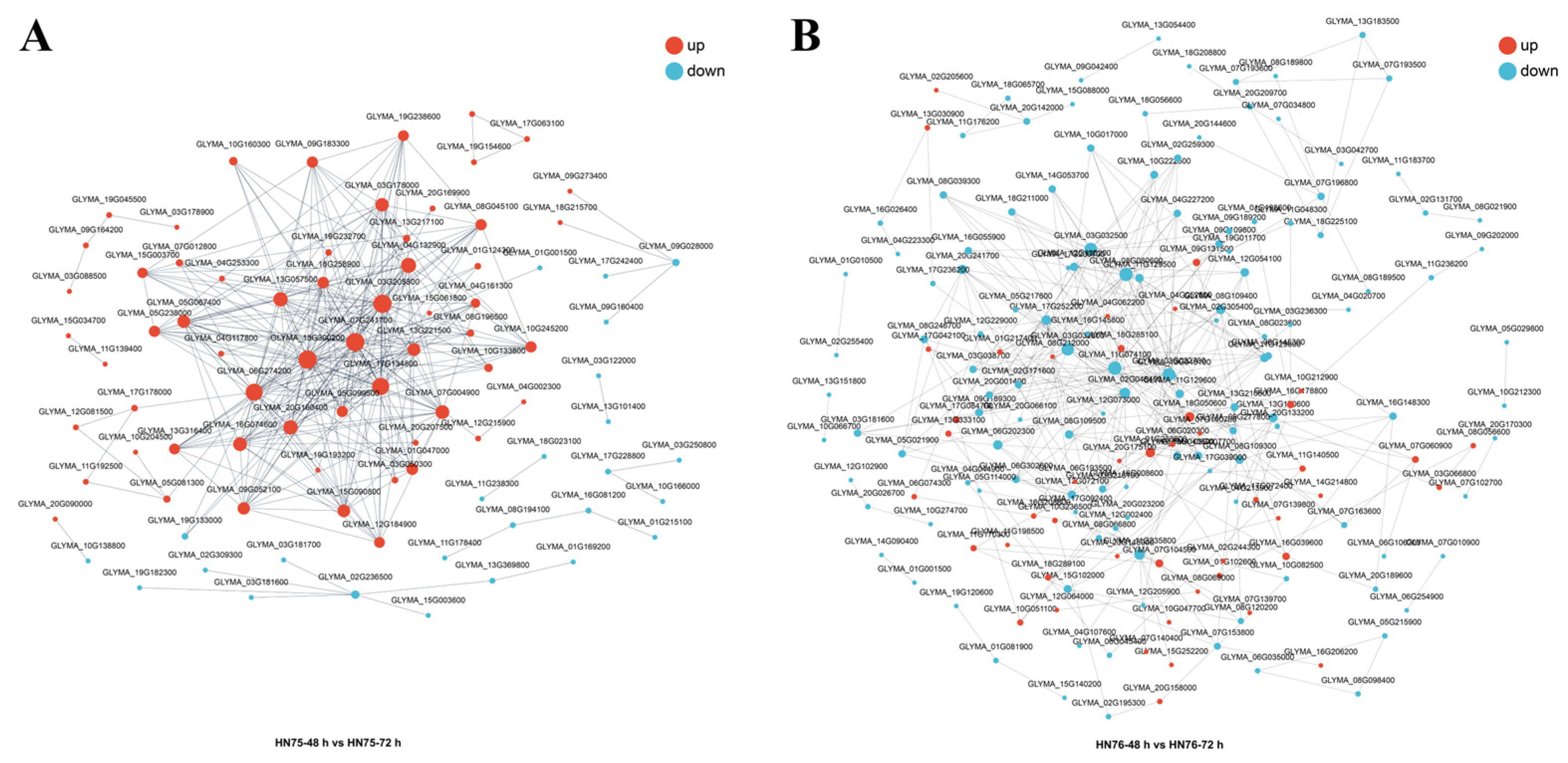
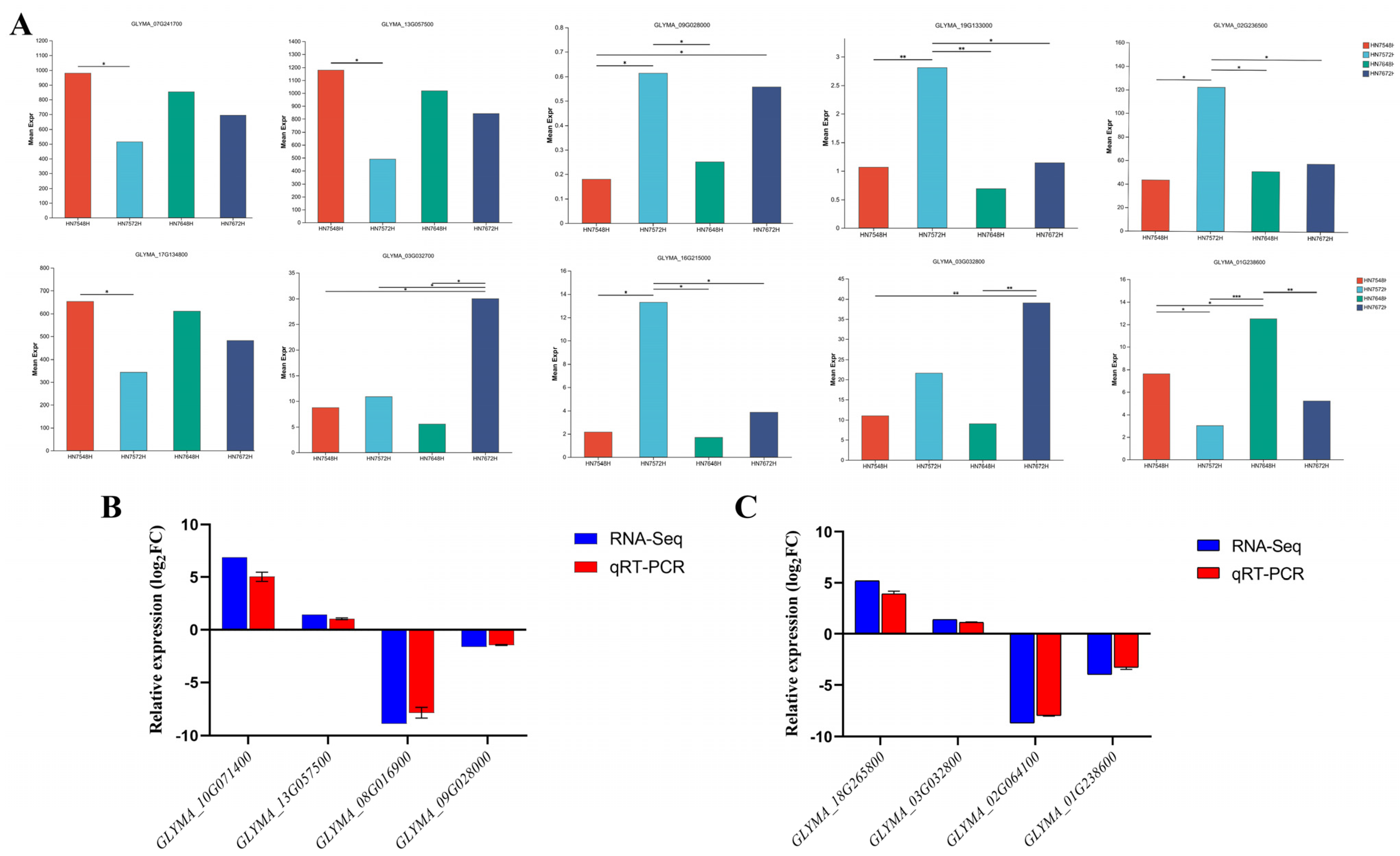
3.5. Screening and Quantitative Analysis of Key Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and Environmental Regulation of Root Development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef]
- Liu, H.; Fu, X. Phytohormonal Networks Facilitate Plant Root Developmental Adaptations to Environmental Changes. Sci. Bull. 2024, 69, 709–713. [Google Scholar] [CrossRef]
- Morffy, N.J.; Strader, L.C. Locally Sourced: Auxin Biosynthesis and Transport in the Root Meristem. Dev. Cell 2018, 47, 262–264. [Google Scholar] [CrossRef]
- Yang, B.; Sun, Y.; Minne, M.; Ge, Y.; Yue, Q.; Goossens, V.; Mor, E.; Callebaut, B.; Bevernaege, K.; Winne, J.M.; et al. SPL13 Controls a Root Apical Meristem Phase Change by Triggering Oriented Cell Divisions. Science 2024, 386, eado4298. [Google Scholar] [CrossRef]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis Root Development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef]
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef]
- Siqueira, J.A.; Otoni, W.C.; Araújo, W.L. The Hidden Half Comes into the Spotlight: Peeking inside the Black Box of Root Developmental Phases. Plant Commun. 2022, 3, 100246. [Google Scholar] [CrossRef]
- Liu, W.; Xu, L. Recruitment of IC-WOX Genes in Root Evolution. Trends Plant Sci. 2018, 23, 490–496. [Google Scholar] [CrossRef]
- Willig, J.-J.; Guarneri, N.; Van Loon, T.; Wahyuni, S.; Astudillo-Estévez, I.E.; Xu, L.; Willemsen, V.; Goverse, A.; Sterken, M.G.; Lozano-Torres, J.L.; et al. Transcription Factor WOX11 Modulates Tolerance to Cyst Nematodes via Adventitious Lateral Root Formation. Plant Physiol. 2024, 195, 799–811. [Google Scholar] [CrossRef]
- Zhang, T.; Ge, Y.; Cai, G.; Pan, X.; Xu, L. WOX-ARF Modules Initiate Different Types of Roots. Cell Rep. 2023, 42, 112966. [Google Scholar] [CrossRef]
- Xiong, H.; He, H.; Chang, Y.; Miao, B.; Liu, Z.; Wang, Q.; Dong, F.; Xiong, L. Multiple Roles of NAC Transcription Factors in Plant Development and Stress Responses. J. Integr. Plant Biol. 2025, 67, 510–538. [Google Scholar] [CrossRef]
- Rodríguez-García, D.R.; Rondón Guerrero, Y.D.C.; Ferrero, L.; Rossi, A.H.; Miglietta, E.A.; Aptekmann, A.A.; Marzol, E.; Martínez Pacheco, J.; Carignani, M.; Berdion Gabarain, V.; et al. Transcription Factor NAC1 Activates Expression of Peptidase-Encoding AtCEPs in Roots to Limit Root Hair Growth. Plant Physiol. 2023, 194, 81–93. [Google Scholar] [CrossRef]
- Chen, D.; Chai, S.; McIntyre, C.L.; Xue, G.-P. Overexpression of a Predominantly Root-Expressed NAC Transcription Factor in Wheat Roots Enhances Root Length, Biomass and Drought Tolerance. Plant Cell Rep. 2018, 37, 225–237. [Google Scholar] [CrossRef]
- Huang, K.-L.; Ma, G.-J.; Zhang, M.-L.; Xiong, H.; Wu, H.; Zhao, C.-Z.; Liu, C.-S.; Jia, H.-X.; Chen, L.; Kjorven, J.O.; et al. The ARF7 and ARF19 Transcription Factors Positively Regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis Roots. Plant Physiol. 2018, 178, 413–427. [Google Scholar] [CrossRef]
- Qi, L.; Kwiatkowski, M.; Chen, H.; Hoermayer, L.; Sinclair, S.; Zou, M.; Del Genio, C.I.; Kubeš, M.F.; Napier, R.; Jaworski, K.; et al. Adenylate Cyclase Activity of TIR1/AFB Auxin Receptors in Plants. Nature 2022, 611, 133–138. [Google Scholar] [CrossRef]
- Roychoudhry, S.; Kepinski, S. Auxin in Root Development. Cold Spring Harb. Perspect. Biol. 2021, 17, a039933. [Google Scholar] [CrossRef]
- Sheldrake, A.R. The Production of Hormones in Higher Plants. Biol. Rev. 1973, 48, 509–559. [Google Scholar] [CrossRef]
- Barker, R.; Fernandez Garcia, M.N.; Powers, S.J.; Vaughan, S.; Bennett, M.J.; Phillips, A.L.; Thomas, S.G.; Hedden, P. Mapping Sites of Gibberellin Biosynthesis in the Arabidopsis Root Tip. New Phytol. 2021, 229, 1521–1534. [Google Scholar] [CrossRef]
- Velandia, K.; Correa-Lozano, A.; McGuiness, P.M.; Reid, J.B.; Foo, E. Cell-layer Specific Roles for Gibberellins in Nodulation and Root Development. New Phytol. 2024, 242, 626–640. [Google Scholar] [CrossRef]
- Geem, K.R.; Lim, Y.; Hong, J.; Bae, W.; Lee, J.; Han, S.; Gil, J.; Cho, H.; Ryu, H. Cytokinin Signaling Promotes Root Secondary Growth and Bud Formation in Panax Ginseng. J. Ginseng Res. 2024, 48, 220–228. [Google Scholar] [CrossRef]
- Artner, C.; Benkova, E. Ethylene and Cytokinin: Partners in Root Growth Regulation. Mol. Plant 2019, 12, 1312–1314. [Google Scholar] [CrossRef]
- Luo, X.; Xu, J.; Zheng, C.; Yang, Y.; Wang, L.; Zhang, R.; Ren, X.; Wei, S.; Aziz, U.; Du, J.; et al. Abscisic Acid Inhibits Primary Root Growth by Impairing ABI4-Mediated Cell Cycle and Auxin Biosynthesis. Plant Physiol. 2023, 191, 265–279. [Google Scholar] [CrossRef]
- Maric, A. The Underground Tango: How Ethylene and Auxin Interact to Regulate Cereal Root Angle. Plant Physiol. 2024, 195, 1757–1758. [Google Scholar] [CrossRef]
- Vukašinović, N.; Wang, Y.; Vanhoutte, I.; Fendrych, M.; Guo, B.; Kvasnica, M.; Jiroutová, P.; Oklestkova, J.; Strnad, M.; Russinova, E. Local Brassinosteroid Biosynthesis Enables Optimal Root Growth. Nat. Plants 2021, 7, 619–632. [Google Scholar] [CrossRef]
- Xuan, W.; Beeckman, T. Plant Signaling: Interplay of Brassinosteroids and Auxin in Root Meristems. Curr. Biol. 2021, 31, R1392–R1395. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukašinović, N.; Hsu, C.-W.; Zhang, J.; Vanhoutte, I.; Shahan, R.; Taylor, I.W.; Greenstreet, L.; Heitz, M.; Afanassiev, A.; et al. Brassinosteroid Gene Regulatory Networks at Cellular Resolution in the Arabidopsis Root. Science 2023, 379, eadf4721. [Google Scholar] [CrossRef]
- Bouwmeester, H.J.; Fonne-Pfister, R.; Screpanti, C.; De Mesmaeker, A. Strigolactones: Plant Hormones with Promising Features. Angew. Chem. Int. Ed. 2019, 58, 12778–12786. [Google Scholar] [CrossRef]
- Pasternak, T.; Groot, E.P.; Kazantsev, F.V.; Teale, W.; Omelyanchuk, N.; Kovrizhnykh, V.; Palme, K.; Mironova, V.V. Salicylic Acid Affects Root Meristem Patterning via Auxin Distribution in a Concentration-Dependent Manner. Plant Physiol. 2019, 180, 1725–1739. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Hayashi, K.; Suzuki, T.; Fukaki, H.; Prusinska, J.; Meester, C.; Quareshy, M.; Egoshi, S.; Matsuura, H.; Takahashi, K.; et al. Jasmonic Acid Inhibits Auxin-Induced Lateral Rooting Independently of the CORONATINE INSENSITIVE1 Receptor. Plant Physiol. 2018, 177, 1704–1716. [Google Scholar] [CrossRef]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A Jasmonate Signaling Network Activates Root Stem Cells and Promotes Regeneration. Cell 2019, 177, 942–956.e14. [Google Scholar] [CrossRef]
- Fang, C.; Kong, F. Soybean. Curr. Biol. 2022, 32, R902–R904. [Google Scholar] [CrossRef]
- Sedivy, E.J.; Wu, F.; Hanzawa, Y. Soybean Domestication: The Origin, Genetic Architecture and Molecular Bases. New Phytol. 2017, 214, 539–553. [Google Scholar] [CrossRef]
- Wang, Z.; Han, Q.; Ji, H. GmRj2/Rfg1 Control of Soybean–Rhizobium–Soil Compatibility. Trends Plant Sci. 2024, 29, 7–9. [Google Scholar] [CrossRef]
- Wang, C.; Li, M.; Zhao, Y.; Liang, N.; Li, H.; Li, P.; Yang, L.; Xu, M.; Bian, X.; Wang, M.; et al. SHORT-ROOT Paralogs Mediate Feedforward Regulation of D-Type Cyclin to Promote Nodule Formation in Soybean. Proc. Natl. Acad. Sci. USA 2022, 119, e2108641119. [Google Scholar] [CrossRef]
- Ma, C.; Liu, C.; Yu, Y.; Ma, S.; Pan, S.; Feng, H.; Chen, Q.; Xin, D.; Wu, X.; Wang, J. GmTNRP1, Associated with Rhizobial type-III Effector NopT, Regulates Nitrogenase Activity in the Nodules of Soybean (Glycine max). Food Energy Secur. 2023, 12, e466. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D. Speak, Friend, and Enter: Signalling Systems That Promote Beneficial Symbiotic Associations in Plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Okazaki, S.; Tittabutr, P.; Teulet, A.; Thouin, J.; Fardoux, J.; Chaintreuil, C.; Gully, D.; Arrighi, J.-F.; Furuta, N.; Miwa, H.; et al. Rhizobium–Legume Symbiosis in the Absence of Nod Factors: Two Possible Scenarios with or without the T3SS. ISME J. 2016, 10, 64–74. [Google Scholar] [CrossRef]
- Giraud, E.; Moulin, L.; Vallenet, D.; Barbe, V.; Cytryn, E.; Avarre, J.-C.; Jaubert, M.; Simon, D.; Cartieaux, F.; Prin, Y.; et al. Legumes Symbioses: Absence of Nod Genes in Photosynthetic Bradyrhizobia. Science 2007, 316, 1307–1312. [Google Scholar] [CrossRef]
- Shi, J.; Wang, X.; Wang, E. Mycorrhizal Symbiosis in Plant Growth and Stress Adaptation: From Genes to Ecosystems. Annu. Rev. Plant Biol. 2023, 74, 569–607. [Google Scholar] [CrossRef]
- Yang, J.; Lan, L.; Jin, Y.; Yu, N.; Wang, D.; Wang, E. Mechanisms Underlying Legume–Rhizobium Symbioses. J. Integr. Plant Biol. 2022, 64, 244–267. [Google Scholar] [CrossRef]
- Xu, P.; Wang, E. Diversity and Regulation of Symbiotic Nitrogen Fixation in Plants. Curr. Biol. 2023, 33, R543–R559. [Google Scholar] [CrossRef]
- Xu, D.; Qu, S.; Li, C.; Zhang, Z.; Wang, C. Breeding and high-yield cultivation techniques of a new soybean variety Henong 75 with high quality. Soybean Sci. Technol. 2016, 6, 23–25. [Google Scholar]
- Guo, M.; Guo, T.; Wang, Z.; Zheng, W.; Li, C.; Zhao, H.; Zhang, Z.; Liu, Z. Soybean Variety ‘Henong75’ for Oil: Parental Pedigree and Analysis and Application of Variety Characteristics. Chin. Agric. Sci. Bull. 2019, 35, 11–15. [Google Scholar]
- Wu, L. A new soybean variety Henong 76 and its supporting cultivation techniques. Mod. Agric. 2016, 4, 16–17. [Google Scholar]
- Zheng, W.; Guo, T.; Wang, Z.; Li, C.; Zhang, Z.; Guo, M.; Zhang, Z.; Wang, Q.; Li, Z. Indirect Utilization of American Dwarf Sources and Innovation of Semi-Dwarf Soybean Henong 76. China Seed Ind. 2016, 7, 57–59. [Google Scholar] [CrossRef]
- Li, C.; Wang, Z.; Zhang, W.; Zhao, H.; Zhang, Z.; Xu, J.; Guo, M.; Guo, T. Genetic Analysis of Oil Content-related Molecular Markers in Soybean Variety Henong 75 with High Oil Content. Seed 2020, 39, 1–5+11. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, C.; Guo, T.; Qu, S.; Wang, C.; Zheng, W.; Li, C.; Zhang, Z.; Li, Y.; Guo, M.; et al. Breeding of a New Soybean Variety Henong 75. China Seed Ind. 2016, 6, 67–69. [Google Scholar] [CrossRef]
- Guo, M.; Guo, T.; Wang, Z.; Zheng, W.; Li, C.; Zhao, H.; Zhang, Z.; Xu, J. Characteristics and Pedigree Analysis of A New Soybean Variety ‘Henong76’. Seed 2020, 39, 110–115. [Google Scholar] [CrossRef]
- Li, C.; Guo, T.; Zheng, W.; Zhang, Z.; Zhao, H.; Gai, Z.; Guo, M.; Wang, Z. Effect of Sowing Density and Fertilizer Levels on the Yield Traits of High Dense Planting Soybean Cultivar Henong 76. Soybean Sci. 2017, 36, 727–732+736. [Google Scholar]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python Library for Gene Ontology Analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [PubMed]
- Miao, B.; Dong, W.; Han, Z.; Luo, X.; Ke, C.; You, W. TOmicsVis: An All-in-one Transcriptomic Analysis and Visualization R Package with Shinyapp Interface. iMeta 2023, 2, e137. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, J.; Gao, Y.; Dong, X.; Feng, H.; Yang, M.; Yu, Y.; Liu, C.; Wu, X.; Qi, Z.; et al. The Type III Effector NopL Interacts with GmREM1a and GmNFR5 to Promote Symbiosis in Soybean. Nat. Commun. 2024, 15, 5852. [Google Scholar] [CrossRef]
- Gai, Z.; Zhao, W.; Du, J.; Liu, J.; Cai, L.; Zhang, W.; Guo, T.; Wang, Z.; Zheng, W.; Li, C.; et al. Row Spacing and Density: Effects on Yield and Quality of Semi-dwarf Soybean Variety ‘Henong76’. J. Agric. 2017, 7, 1–5. [Google Scholar]
- Fernández-Aparicio, M.; Cimmino, A.; Evidente, A.; Rubiales, D. Inhibition of Orobanche Crenata Seed Germination and Radicle Growth by Allelochemicals Identified in Cereals. J. Agric. Food Chem. 2013, 61, 9797–9803. [Google Scholar] [CrossRef]
- Chandran, A.E.J.; Finkler, A.; Hait, T.A.; Kiere, Y.; David, S.; Pasmanik-Chor, M.; Shkolnik, D. Calcium Regulation of the Arabidopsis Na+/K+ Transporter HKT1;1 Improves Seed Germination under Salt Stress. Plant Physiol. 2024, 194, 1834–1852. [Google Scholar] [CrossRef]
- Sánchez-Linares, L.; Gavilanes-Ruíz, M.; Díaz-Pontones, D.; Guzmán-Chávez, F.; Calzada-Alejo, V.; Zurita-Villegas, V.; Luna-Loaiza, V.; Moreno-Sánchez, R.; Bernal-Lugo, I.; Sánchez-Nieto, S. Early Carbon Mobilization and Radicle Protrusion in Maize Germination. J. Exp. Bot. 2012, 63, 4513–4526. [Google Scholar] [CrossRef]
- Doerner, P. Arabidopsis Embryogenesis: Radicle Development(s). Curr. Biol. 1995, 5, 110–112. [Google Scholar] [CrossRef]
- Tsuzuki, K.; Suzuki, T.; Kuruma, M.; Nishiyama, K.; Hayashi, K.-I.; Hagihara, S.; Seto, Y. Radicle Growth Regulation of Root Parasitic Plants by Auxin-Related Compounds. Plant Cell Physiol. 2024, 65, 1377–1387. [Google Scholar] [CrossRef]
- Kang, J.; Yim, S.; Choi, H.; Kim, A.; Lee, K.P.; Lopez-Molina, L.; Martinoia, E.; Lee, Y. Abscisic Acid Transporters Cooperate to Control Seed Germination. Nat. Commun. 2015, 6, 8113. [Google Scholar] [CrossRef]
- Moreira, T.B.; Shaw, R.; Luo, X.; Ganguly, O.; Kim, H.-S.; Coelho, L.G.F.; Cheung, C.Y.M.; Rhys Williams, T.C. A Genome-Scale Metabolic Model of Soybean (Glycine max) Highlights Metabolic Fluxes in Seedlings. Plant Physiol. 2019, 180, 1912–1929. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gong, Z.; Han, G.-Z. On the Origins and Evolution of Phytohormone Signaling and Biosynthesis in Plants. Mol. Plant 2023, 16, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Berman, A.; Shani, E. Plant Hormone Transport and Localization: Signaling Molecules on the Move. Annu. Rev. Plant Biol. 2023, 74, 453–479. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Zhang, T.; Jiang, L. Soy Protein: Molecular Structure Revisited and Recent Advances in Processing Technologies. Annu. Rev. Food Sci. Technol. 2021, 12, 119–147. [Google Scholar] [CrossRef]
- Zhou, X.; Yao, X.; He, D.; Sun, H.; Xie, F. Transcriptomic Investigation Unveils the Role of Energy Metabolism under Low Phosphorus and Salt Combined Stress in Soybean (Glycine max). Physiol. Plant. 2024, 176, e14262. [Google Scholar] [CrossRef]
- Wu, J.; Vallenius, T.; Ovaska, K.; Westermarck, J.; Mäkelä, T.P.; Hautaniemi, S. Integrated Network Analysis Platform for Protein-Protein Interactions. Nat. Methods 2009, 6, 75–77. [Google Scholar] [CrossRef]
- Duan, Z.; Li, Q.; Wang, H.; He, X.; Zhang, M. Genetic Regulatory Networks of Soybean Seed Size, Oil and Protein Contents. Front. Plant Sci. 2023, 14, 1160418. [Google Scholar] [CrossRef]
- Freilich, R.; Arhar, T.; Abrams, J.L.; Gestwicki, J.E. Protein–Protein Interactions in the Molecular Chaperone Network. Acc. Chem. Res. 2018, 51, 940–949. [Google Scholar] [CrossRef]

| Database | Version | Source |
|---|---|---|
| NR | Version 2023.07 | https://www.ncbi.nlm.nih.gov/public/ (accessed on 5 January 2025) |
| GO | Version 2023.07 | http://www.geneontology.org/ (accessed on 6 January 2025) |
| Swiss-prot | Version 2023.11 | https://swissmodel.expasy.org/repository?query=Version+2023.11 (accessed on 7 January 2025) |
| Rfam | Version 14.10 | http://rfam.janelia.org/ (accessed on 7 January 2025) |
| eggNOG | Version 2020.06 | http://eggnogdb.embl.de/#/app/home (accessed on 6 January 2025) |
| KEGG | Version 2023.09 | http://www.genome.jp/kegg/ (accessed on 6 January 2025) |
| Trait (n = 30) | HN75 (Mean ± SE) | HN76 (Mean ± SE) |
|---|---|---|
| Podding habit | Semi-determinacy | Semi-determinacy |
| Plant height (cm) | 82.02 ± 1.82 | 80.08 ± 2.36 |
| Node number of main stem | 18.20 ± 1.56 | 18.03 ± 0.87 |
| Number of pods per node | 2.53 ± 0.50 | 3.00 ± 0.82 |
| Number of grains per plant | 95.23 ± 3.32 | 100.10 ± 3.52 |
| Grain weight per plant (g) | 18.50 ± 0.88 | 19.49 ± 0.88 |
| 100-grain weight (g) | 21.13 ± 0.56 | 19.49 ± 0.26 |
| Average oil content (%) | 23.40 ± 0.88 | 21.50 ± 0.97 |
| Average protein content (%) | 35.71 ± 0.43 | 41.39 ± 0.40 |
| Maturity cycle (d) | 170 | 120 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, F.; Mi, L.; Xin, D. Transcriptomic Analysis for Key Periods of Radicle Development in Contrasting Soybean Varieties HN75 and HN76. Agronomy 2025, 15, 1186. https://doi.org/10.3390/agronomy15051186
Yu F, Mi L, Xin D. Transcriptomic Analysis for Key Periods of Radicle Development in Contrasting Soybean Varieties HN75 and HN76. Agronomy. 2025; 15(5):1186. https://doi.org/10.3390/agronomy15051186
Chicago/Turabian StyleYu, Fengli, Liang Mi, and Dawei Xin. 2025. "Transcriptomic Analysis for Key Periods of Radicle Development in Contrasting Soybean Varieties HN75 and HN76" Agronomy 15, no. 5: 1186. https://doi.org/10.3390/agronomy15051186
APA StyleYu, F., Mi, L., & Xin, D. (2025). Transcriptomic Analysis for Key Periods of Radicle Development in Contrasting Soybean Varieties HN75 and HN76. Agronomy, 15(5), 1186. https://doi.org/10.3390/agronomy15051186







