YOLO_SSP: An Auto-Algorithm to Detect Mature Soybean Stem Nodes Based on Keypoint Detection
Abstract
1. Introduction
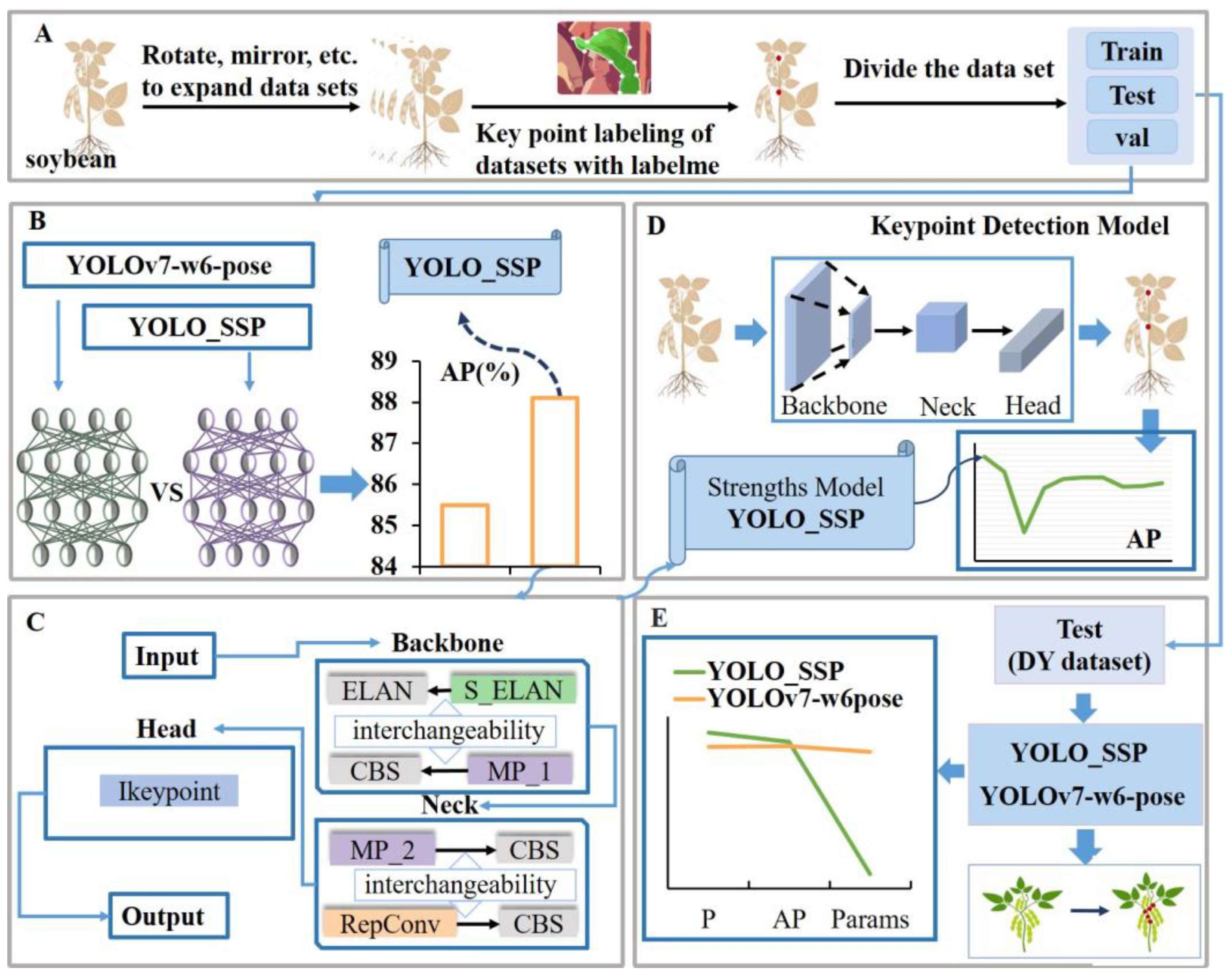
2. Materials and Methods
2.1. Materials Acquisition
2.2. Experimental Procedure
2.2.1. Data Preprocessing
2.2.2. YOLOv7-W6-Pose
2.2.3. YOLO_Soybean Stalk Pose (YOLO_SSP)
2.2.4. Advanced Feature Extraction
2.2.5. Low-Level Feature Extraction
2.2.6. Feature Optimization and Enhancement
2.3. Experimental Environment
2.4. Evaluation Metrics
3. Results
3.1. Model Improvement Experiment
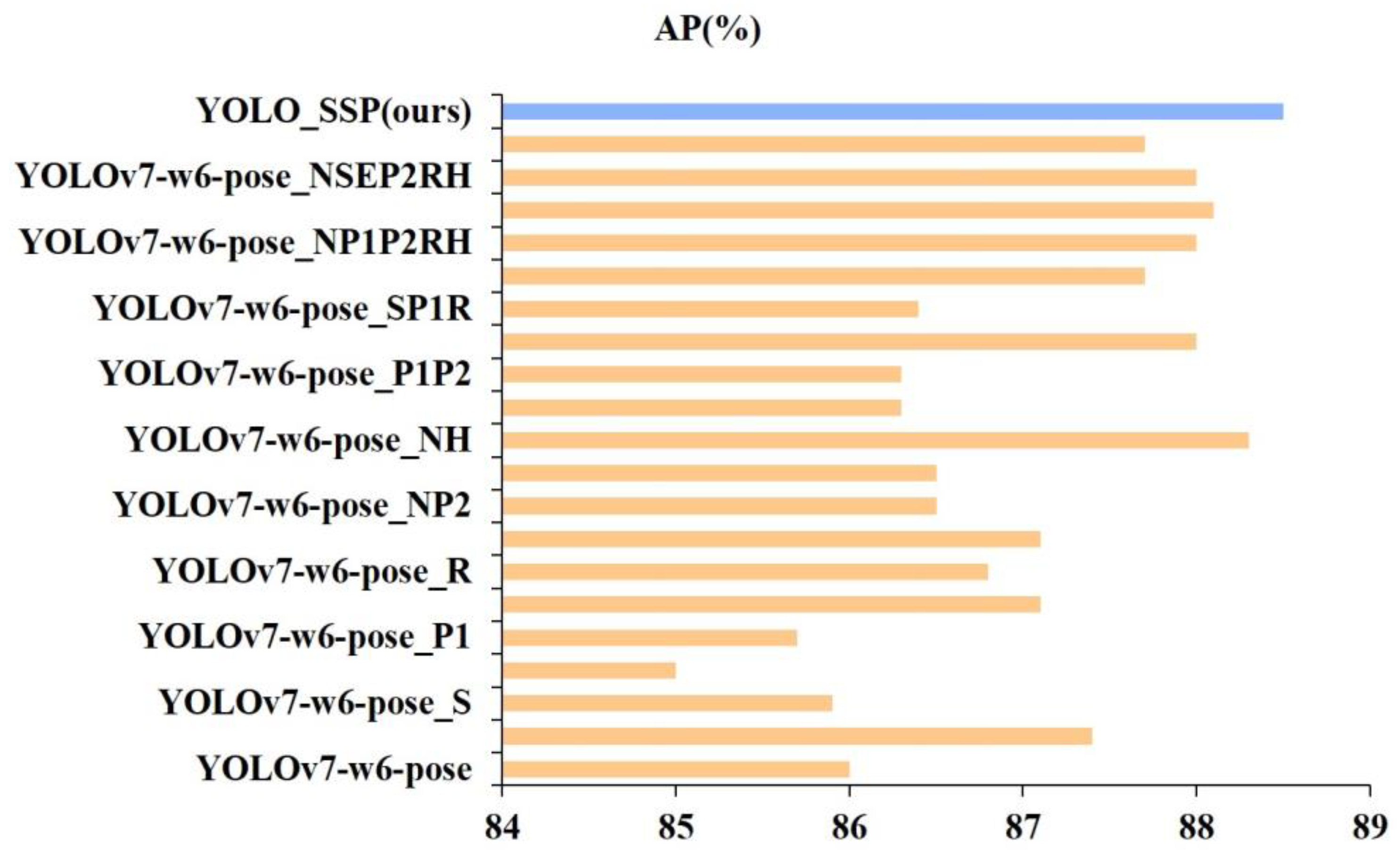
3.2. Ablation Experiment
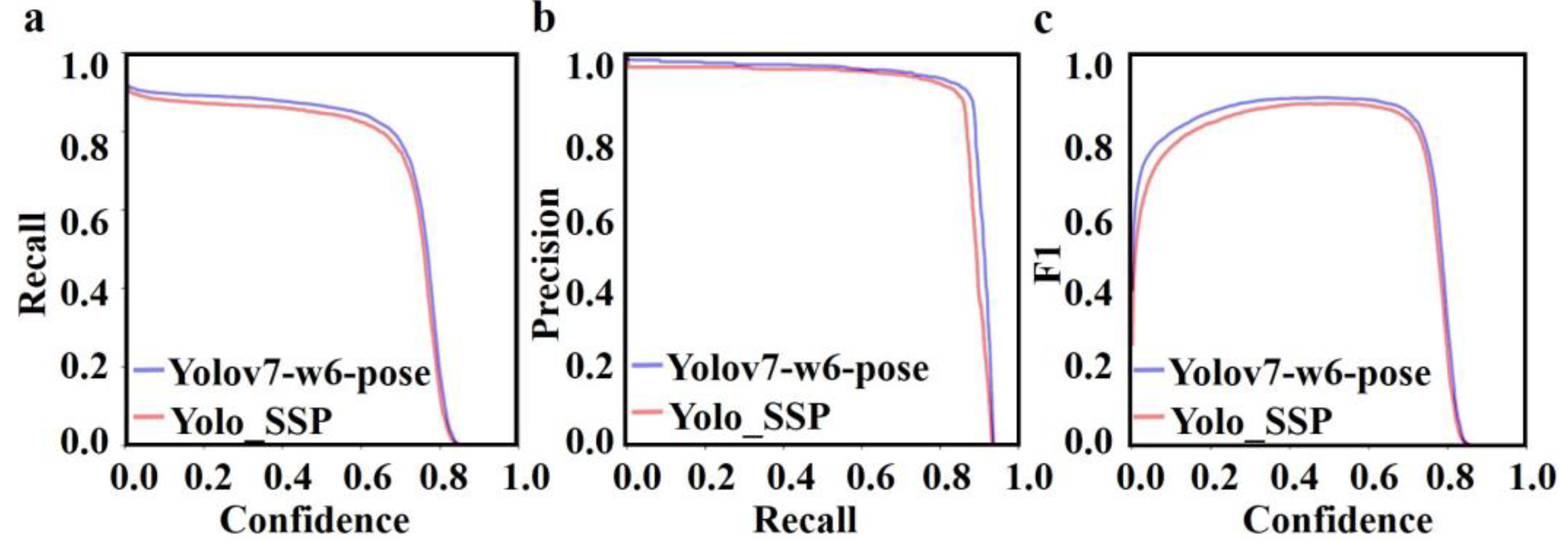
3.3. Comparative Experiment
3.4. DY Dataset Testing Experiment
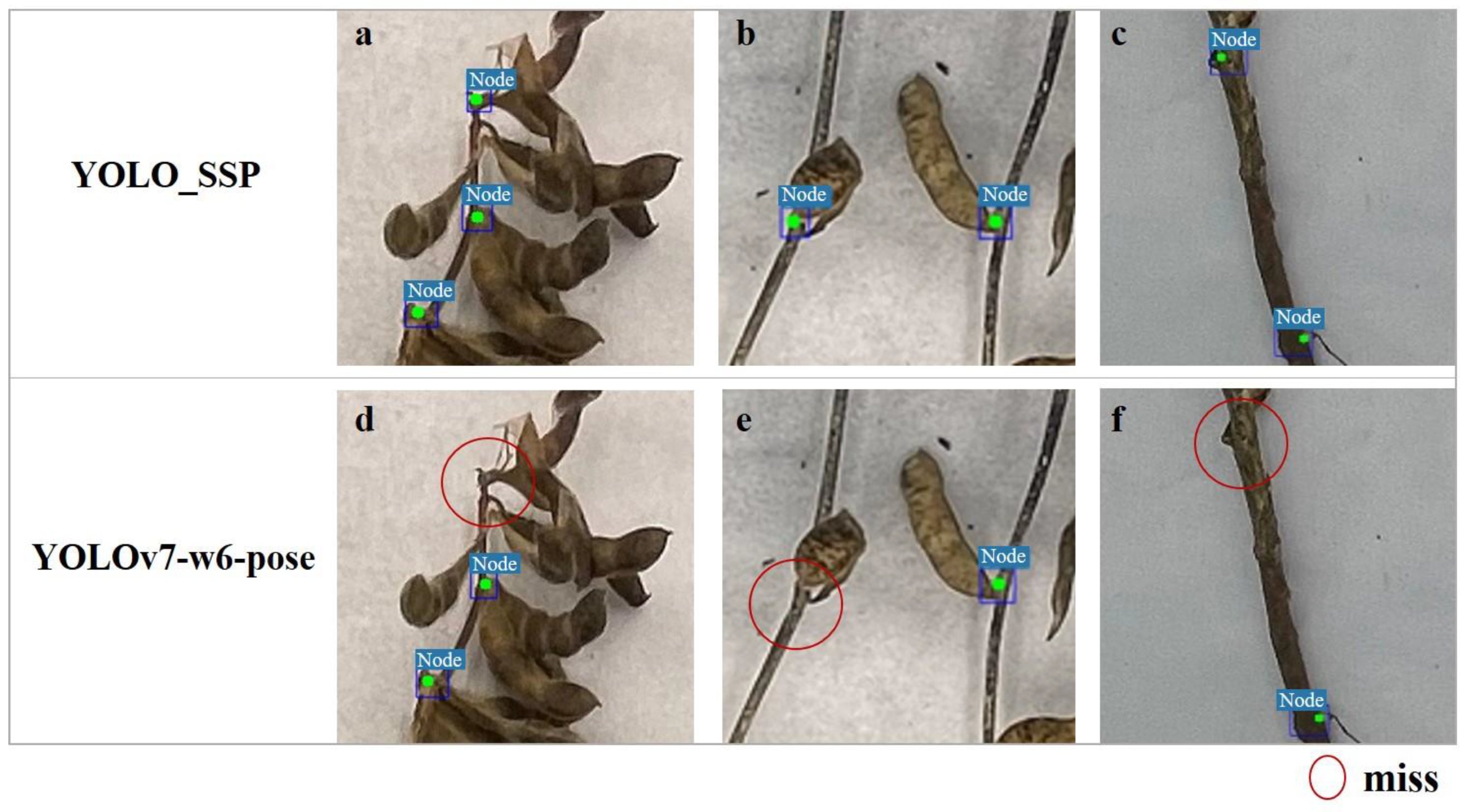
3.5. Visualization Experiment
4. Discussion
4.1. Advantages of Using Yolov7 as a Baseline Model
4.2. Advantages of YOLO_SSP in Mature Soybean Stem Node Detection
4.3. Specificity of the S_ELAN or ELAN-H Module
4.4. YOLO Comparative Advantages of the Models in the Pose Series
4.5. A Non-Critical Exploration of Speed
4.6. Limitations of This Study
4.7. Future Work
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| YOLO_SSP | You Only Look Once _Soybean Stalk Pose |
| AHTIDZ | Agricultural High-Tech Industrial Demonstration Zone |
| S_ELAN | Small_Effective Low-Level Aggregation Network |
| ELAN | Effective Low-Level Aggregation Network |
| ELAN-H | Effective Low-Level Aggregation Network for High-resolution feature aggregation |
| CBS | Conv-BatchNorm-ReLU |
| BN | BatchNorm |
| SPP | Spatial Pyramid Pooling |
| CSCP | Contextual Spatial Pyramid Pooling |
| CSP | Cross Stage Partial |
References
- Jarecki, W.; Migut, D. Comparison of Yield and Important Seed Quality Traits of Selected Legume Species. Agronomy 2022, 12, 2667. [Google Scholar] [CrossRef]
- Nehbandani, A.; Soltani, A.; Hajjarpoor, A.; Dadrasi, A.; Nourbakhsh, F. Comprehensive yield gap analysis and optimizing agronomy practices of soybean in Iran. J. Agric. Sci. 2020, 158, 739–747. [Google Scholar] [CrossRef]
- Świątkiewicz, M.; Witaszek, K.; Sosin, E.; Pilarski, K.; Szymczyk, B.; Durczak, K. The Nutritional Value and Safety of Genetically Unmodified Soybeans and Soybean Feed Products in the Nutrition of Farm Animals. Agronomy 2021, 11, 1105. [Google Scholar] [CrossRef]
- Dorman, S.J.; Hopperstad, K.A.; Reich, B.J.; Kennedy, G.; Huseth, A.S. Soybeans as a non-Bt refuge for Helicoverpa zea in maize-cotton agroecosystems. Agric. Ecosyst. Environ. 2021, 322, 107642. [Google Scholar] [CrossRef]
- Li, Y.; Yu, H.; Liu, L.; Liu, Y.; Huang, L.; Tan, H. Transcriptomic and physiological analyses unravel the effect and mechanism of halosulfuron-methyl on the symbiosis between rhizobium and soybean. Ecotoxicol. Environ. Saf. 2022, 247, 114248. [Google Scholar] [CrossRef]
- Adler, P.R.; Hums, M.E.; McNeal, F.M.; Spatari, S. Evaluation of environmental and cost tradeoffs of producing energy from soybeans for on-farm use. J. Clean. Prod. 2019, 210, 1635–1649. [Google Scholar] [CrossRef]
- Kebede, E. Contribution, utilization, and improvement of legumes-driven biological nitrogen fixation in agricultural systems. Front. Sustain. Food Syst. 2021, 5, 767998. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, J.; Ye, H.; Ali, M.L.; Chen, P.; Nguyen, H.T. Yield estimation of soybean breeding lines under drought stress using unmanned aerial vehicle-based imagery and convolutional neural network. Biosyst. Eng. 2021, 204, 90–103. [Google Scholar] [CrossRef]
- Yang, Q.; Lin, G.; Lv, H.; Wang, C.; Yang, Y.; Liao, H. Environmental and genetic regulation of plant height in soybean. BMC Plant Biol. 2021, 21, 63. [Google Scholar] [CrossRef]
- Vogel, J.T.; Liu, W.; Olhoft, P.; Crafts-Brandner, S.J.; Pennycooke, J.C.; Christiansen, N. Soybean yield formation physiology—A foundation for precision breeding based improvement. Front. Plant Sci. 2021, 12, 719706. [Google Scholar] [CrossRef]
- Xu, C.; Li, R.; Song, W.; Wu, T.; Sun, S.; Hu, S.; Han, T.; Wu, C. Responses of branch number and yield component of soybean cultivars tested in different planting densities. Agriculture 2021, 11, 69. [Google Scholar] [CrossRef]
- Staniak, M.; Czopek, K.; Stępień-Warda, A.; Kocira, A.; Przybyś, M. Cold stress during flowering alters plant structure, yield and seed quality of different soybean genotypes. Agronomy 2021, 11, 2059. [Google Scholar] [CrossRef]
- Staniak, M.; Szpunar-Krok, E.; Kocira, A. Responses of soybean to selected abiotic stresses—Photoperiod, temperature and water. Agriculture 2023, 13, 146. [Google Scholar] [CrossRef]
- Li, W.; Wang, P.; Zhao, H.; Sun, X.; Yang, T.; Li, H.; Hou, Y.; Liu, C.; Siyal, M.; Raja Veesar, R. QTL for main stem node number and its response to plant densities in 144 soybean FW-RILs. Front. Plant Sci. 2021, 12, 666796. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Shan, F.; Wang, C.; Yan, C.; Dong, S.; Xu, Y.; Gong, Z.; Ma, C. Internode elongation pattern, internode diameter and hormone changes in soybean (Glycine max) under different shading conditions. Crop Pasture Sci. 2020, 71, 679–688. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, C.; Zhang, R.; Ma, C.; Dong, S.; Gong, Z. The relationship between internode elongation of soybean stems and spectral distribution of light in the canopy under different plant densities. Plant Prod. Sci. 2021, 24, 326–338. [Google Scholar] [CrossRef]
- Fu, M.; Wang, Y.; Ren, H.; Du, W.; Yang, X.; Wang, D.; Cheng, Y.; Zhao, J.; Gai, J. Exploring the QTL–allele constitution of main stem node number and its differentiation among maturity groups in a Northeast China soybean population. Crop Sci. 2020, 60, 1223–1238. [Google Scholar] [CrossRef]
- Burroughs, C.H.; Montes, C.M.; Moller, C.A.; Mitchell, N.G.; Michael, A.M.; Peng, B.; Kimm, H.; Pederson, T.L.; Lipka, A.E.; Bernacchi, C.J.; et al. Reductions in leaf area index, pod production, seed size, and harvest index drive yield loss to high temperatures in soybean. J. Exp. Bot. Exp. Bot. 2022, 74, 1629–1641. [Google Scholar] [CrossRef]
- Takpah, D.; Asghar, M.A.; Raza, A.; Javed, H.H.; Ullah, A.; Huang, X.; Saleem, K.; Xie, C.; Xiao, X.; Clement, K.S.; et al. Metabolomics Analysis Reveals Soybean Node Position Influence on Metabolic Profile of Soybean Seed at Various Developmental Stages. J. Plant Growth Regul. 2023, 42, 6788–6800. [Google Scholar] [CrossRef]
- Lahiri, S.; Reisig, D.D.; Dean, L.L.; Reay-Jones, F.P.F.; Greene, J.K.; Carter, T.E.J.; Mian, R.; Fallen, B.D. Mechanisms of Soybean Host-Plant Resistance Against Megacopta cribraria (Hemiptera: Plataspidae). Environ. Entomol. 2020, 49, 876–885. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, H.; Chen, Y. An automated phenotyping method for Chinese Cymbidium seedlings based on 3D point cloud. Plant Methods 2024, 20, 151. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Liu, S.; Chen, H.; Chang, Y.; Yu, Y.; Ma, W.; Wang, Y.; Xie, H. A Precise Method for Identifying 3D Circles in Freeform Surface Point Clouds. IEEE Trans. Instrum. Meas. 2025, 74, 5023713. [Google Scholar] [CrossRef]
- Wang, B.; Yang, M.; Cao, P.; Liu, Y. A novel embedded cross framework for high-resolution salient object detection. Appl. Intell. 2025, 55, 277. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, Y.; Wen, W.; Gu, S.; Lu, X.; Guo, X. The future of Internet of Things in agriculture: Plant high-throughput phenotypic platform. J. Clean. Prod. 2021, 280, 123651. [Google Scholar] [CrossRef]
- Tholkapiyan, M.; Aruna Devi, B.; Bhatt, D.; Saravana Kumar, E.; Kirubakaran, S.; Kumar, R. Performance Analysis of Rice Plant Diseases Identification and Classification Methodology. Wirel. Pers. Commun. 2023, 130, 1317–1341. [Google Scholar] [CrossRef]
- Ghanem, M.E.; Marrou, H.; Sinclair, T.R. Physiological phenotyping of plants for crop improvement. Trends Plant Sci. 2015, 20, 139–144. [Google Scholar] [CrossRef]
- Guo, Y.; Gao, Z.; Zhang, Z.; Li, Y.; Hu, Z.; Xin, D.; Chen, Q.; Zhu, R. Automatic and accurate acquisition of stem-related phenotypes of mature soybean based on deep learning and directed search algorithms. Front. Plant Sci. 2022, 13, 906751. [Google Scholar] [CrossRef]
- Li, Z.; Guo, R.; Li, M.; Chen, Y.; Li, G. A review of computer vision technologies for plant phenotyping. Comput. Electron. Agric. 2020, 176, 105672. [Google Scholar] [CrossRef]
- Li, S.; Yan, Z.; Guo, Y.; Su, X.; Cao, Y.; Jiang, B.; Yang, F.; Zhang, Z.; Xin, D.; Chen, Q.; et al. SPM-IS: An auto-algorithm to acquire a mature soybean phenotype based on instance segmentation. Crop J. 2022, 10, 1412–1423. [Google Scholar] [CrossRef]
- Guo, X.; Li, J.; Zheng, L.; Zhang, M.; Wang, M. Acquiring soybean phenotypic parameters using Re-YOLOv5 and area search algorithm. Trans. Chin. Soc. Agric. Eng. 2022, 38, 186–194. [Google Scholar]
- Zhou, W.; Chen, Y.; Li, W.; Zhang, C.; Xiong, Y.; Zhan, W.; Huang, L.; Wang, J.; Qiu, L. SPP-extractor: Automatic phenotype extraction for densely grown soybean plants. Crop J. 2023, 11, 1569–1578. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zheng, L.; Yang, H.; Zhang, M.; Wu, T.; Sun, S.; Tomasetto, F.; Wang, M. A synthetic datasets based instance segmentation network for High-throughput soybean pods phenotype investigation. Expert Syst. Appl. 2022, 192, 116403. [Google Scholar] [CrossRef]
- Zhang, C.; Lu, X.; Ma, H.; Hu, Y.; Zhang, S.; Ning, X.; Hu, J.; Jiao, J. High-Throughput Classification and Counting of Vegetable Soybean Pods Based on Deep Learning. Agronomy 2023, 13, 1154. [Google Scholar] [CrossRef]
- Lu, W.; Du, R.; Niu, P.; Xing, G.; Luo, H.; Deng, Y.; Shu, L. Soybean yield preharvest prediction based on bean pods and leaves image recognition using deep learning neural network combined with GRNN. Front. Plant Sci. 2022, 12, 791256. [Google Scholar] [CrossRef]
- He, H.; Ma, X.; Guan, H.; Wang, F.; Shen, P. Recognition of soybean pods and yield prediction based on improved deep learning model. Front. Plant Sci. 2023, 13, 1096619. [Google Scholar] [CrossRef]
- Yang, S.; Zheng, L.; Chen, X.; Zabawa, L.; Zhang, M.; Wang, M. Transfer learning from synthetic in-vitro soybean pods dataset for in-situ segmentation of on-branch soybean pods. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 1666–1675. [Google Scholar]
- Ning, S.; Chen, H.; Zhao, Q.; Wang, Y. Detection of pods and stems in soybean based on IM-SSD+ ACO algorithm. Trans. Chin. Soc. Agric. Mach. 2021, 52, 182–190. [Google Scholar]
- Li, M.; Jia, T.; Wang, H.; Ma, B.; Lu, H.; Lin, S.; Cai, D.; Chen, D. Ao-detr: Anti-overlapping detr for x-ray prohibited items detection. IEEE Trans. Neural Netw. Learn. Syst. 2024, 1–15. [Google Scholar] [CrossRef]
- Li, Y.; Xu, S.; Zhu, Z.; Wang, P.; Li, K.; He, Q.; Zheng, Q. EFC-YOLO: An efficient surface-defect-detection algorithm for steel strips. Sensors 2023, 23, 7619. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, W.; Zhuang, J.; Zhang, R.; Du, X. Detection Technique Tailored for Small Targets on Water Surfaces in Unmanned Vessel Scenarios. J. Mar. Sci. Eng. 2024, 12, 379. [Google Scholar] [CrossRef]
- Wang, C.; Jiang, J.; Daneva, M.; Van Sinderen, M. CoolTeD: A web-based collaborative labeling tool for the textual dataset. In Proceedings of the 2022 IEEE International Conference on Software Analysis, Evolution and Reengineering (SANER), Honolulu, HI, USA, 15–18 March 2022; IEEE: New Orleans, LA, USA, 2022; pp. 613–617. [Google Scholar]
- Belissent, N.; Peña, J.M.; Mesías-Ruiz, G.A.; Shawe-Taylor, J.; Pérez-Ortiz, M. Transfer and zero-shot learning for scalable weed detection and classification in UAV images. Knowl.-Based Syst. 2024, 292, 111586. [Google Scholar] [CrossRef]
- Sykas, D.; Sdraka, M.; Zografakis, D.; Papoutsis, I. A sentinel-2 multiyear, multicountry benchmark dataset for crop classification and segmentation with deep learning. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 15, 3323–3339. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, Q.; Hu, W. Human action capture method based on binocular camera. In Proceedings of the 2024 IEEE International Conference on Smart Internet of Things (SmartIoT), Shenzhen, China, 14–16 November 2024; IEEE: New Orleans, LA, USA, 2024; pp. 234–239. [Google Scholar]
- Tîrziu, E.; Vasilevschi, A.; Alexandru, A.; Tudora, E. Enhanced Fall Detection Using YOLOv7-W6-Pose for Real-Time Elderly Monitoring. Future Internet 2024, 16, 472. [Google Scholar] [CrossRef]
- Nardi, V. State of the Art Analysis and Optimization of Human Pose Estimation Algorithms. Master’s Thesis, Politecnico di Milano, Milano, Italy, 2022. [Google Scholar]
- Humeau-Heurtier, A. Texture feature extraction methods: A survey. IEEE Access 2019, 7, 8975–9000. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Villena-Rodriguez, A.; Martín-Vega, F.J.; Gómez, G.; Aguayo-Torres, M.C.; Kaddoum, G. Aging-Resistant Wideband Precoding in 5G and Beyond Using 3D Convolutional Neural Networks. arXiv 2024, arXiv:2407.07434. [Google Scholar]
- Zhang, Y.; Zhang, H.; Huang, Q.; Han, Y.; Zhao, M. DsP-YOLO: An anchor-free network with DsPAN for small object detection of multiscale defects. Expert. Syst. Appl. 2024, 241, 122669. [Google Scholar] [CrossRef]
- Wang, C.; Bochkovskiy, A.; Liao, H.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7464–7475. [Google Scholar]
- Wang, L.; Li, X.; Chen, X.; Zhou, B. CenterNet-Elite: A Small Object Detection Model for Driving Scenario. Ieee Access 2025. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE T Pattern Anal. 2015, 37, 1904–1916. [Google Scholar] [CrossRef]
- Karmouch, A.; Galis, A.; Giaffreda, R.; Kanter, T.; Jonsson, A.; Karlsson, A.M.; Glitho, R.; Smirnov, M.; Kleis, M.; Reichert, C. Contextware research challenges in ambient networks. In Proceedings of the Mobility Aware Technologies and Applications: First International Workshop, MATA 2004, Florianópolis, Brazil, 20–22 October 2004; Proceedings 1. Springer: Berlin/Heidelberg, Germany, 2004; pp. 62–77. [Google Scholar]
- Zhao, X.Y.; He, Y.X.; Zhang, H.T.; Ding, Z.T.; Zhou, C.A.; Zhang, K.X. A quality grade classification method for fresh tea leaves based on an improved YOLOv8x-SPPCSPC-CBAM model. Sci. Rep. 2024, 14, 4166. [Google Scholar] [CrossRef]
- Zafar, A.; Aamir, M.; Mohd Nawi, N.; Arshad, A.; Riaz, S.; Alruban, A.; Dutta, A.K.; Almotairi, S. A comparison of pooling methods for convolutional neural networks. Appl. Sci. 2022, 12, 8643. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J. Recent advances in convolutional neural networks. Pattern Recogn. 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, G.; Zhou, B. An efficient feature aggregation network for small object detection in UAV aerial images. J. Supercomput. 2025, 81, 1–26. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, T.; Fang, C.; Zheng, H.; Ma, C.; Wu, Z. A detection method for dead caged hens based on improved YOLOv7. Comput. Electron. Agric. 2024, 226, 109388. [Google Scholar] [CrossRef]
- Chen, J.; Shen, Y.; Liang, Y.; Wang, Z.; Zhang, Q. YOLO-SAD: An Efficient SAR Aircraft Detection Network. Appl. Sci. 2024, 14, 3025. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, Z.; Zhao, H.; Hou, Y.; Liu, Y.; Zuo, X.; Dang, L. A multi-scale object detector based on coordinate and global information aggregation for UAV aerial images. Remote Sens. 2023, 15, 3468. [Google Scholar] [CrossRef]
- Liu, K.; Sun, Q.; Sun, D.; Peng, L.; Yang, M.; Wang, N. Underwater target detection based on improved YOLOv7. J. Mar. Sci. Eng. 2023, 11, 677. [Google Scholar] [CrossRef]
- Gallo, I.; Rehman, A.U.; Dehkordi, R.H.; Landro, N.; La Grassa, R.; Boschetti, M. Deep object detection of crop weeds: Performance of YOLOv7 on a real case dataset from UAV images. Remote Sens. 2023, 15, 539. [Google Scholar] [CrossRef]
- Guarnido-Lopez, P.; Ramirez-Agudelo, J.; Denimal, E.; Benaouda, M. Programming and Setting Up the Object Detection Algorithm YOLO to Determine Feeding Activities of Beef Cattle: A Comparison between YOLOv8m and YOLOv10m. Animals 2024, 14, 2821. [Google Scholar] [CrossRef]
- Du, X.; Meng, Z.; Ma, Z.; Lu, W.; Cheng, H. Tomato 3D pose detection algorithm based on keypoint detection and point cloud processing. Comput. Electron. Agric. 2023, 212, 108056. [Google Scholar] [CrossRef]
- Du, X.; Meng, Z.; Ma, Z.; Zhao, L.; Lu, W.; Cheng, H.; Wang, Y. Comprehensive visual information acquisition for tomato picking robot based on multitask convolutional neural network. Biosyst. Eng. 2024, 238, 51–61. [Google Scholar] [CrossRef]
- Chen, J.; Ma, A.; Huang, L.; Li, H.; Zhang, H.; Huang, Y.; Zhu, T. Efficient and lightweight grape and picking point synchronous detection model based on key point detection. Comput. Electron. Agric. 2024, 217, 108612. [Google Scholar] [CrossRef]
- Shuai, L.; Mu, J.; Jiang, X.; Chen, P.; Zhang, B.; Li, H.; Wang, Y.; Li, Z. An improved YOLOv5-based method for multi-species tea shoot detection and picking point location in complex backgrounds. Biosyst. Eng. 2023, 231, 117–132. [Google Scholar] [CrossRef]
- Wang, J.; Tan, D.; Sui, L.; Guo, J.; Wang, R. Wolfberry recognition and picking-point localization technology in natural environments based on improved Yolov8n-Pose-LBD. Comput. Electron. Agric. 2024, 227, 109551. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, X.; Shi, Y.; Zhang, X.; Wu, Y.; Wang, D.; Huang, X.; Wang, Y.; Wang, J.; Zhang, J. Keypoint detection and diameter estimation of cabbage (Brassica oleracea L.) heads under varying occlusion degrees via YOLOv8n-CK network. Comput. Electron. Agric. 2024, 226, 109428. [Google Scholar] [CrossRef]
- Yang, G.; Jing, H. Multiple convolutional neural network for feature extraction. In Advanced Intelligent Computing Theories and Methodologies, Proceedings of the 11th International Conference, ICIC 2015, Fuzhou, China, 20–23 August 2015; Proceedings, Part II 11; Springer: Berlin/Heidelberg, Germany, 2015; pp. 104–114. [Google Scholar]
- Zhao, S.; Cai, T.; Peng, B.; Zhang, T.; Zhou, X. GAM-YOLOv8n: Enhanced feature extraction and difficult example learning for site distribution box door status detection. Wirel. Netw. 2023, 30, 6939–6950. [Google Scholar] [CrossRef]
- Han, K.; Wang, Y.; Tian, Q.; Guo, J.; Xu, C.; Xu, C. Ghostnet: More features from cheap operations. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 1580–1589. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Acikgoz, H. An automatic detection model for cracks in photovoltaic cells based on electroluminescence imaging using improved YOLOv7. Signal Image Video Process. 2024, 18, 625–635. [Google Scholar] [CrossRef]
- Wu, T.; Dong, Y. YOLO-SE: Improved YOLOv8 for Remote Sensing Object Detection and Recognition. Appl. Sci. 2023, 13, 12977. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, LA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Lin, T.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Zhang, Z.; He, T.; Zhang, H.; Zhang, Z.; Xie, J.; Li, M. Bag of freebies for training object detection neural networks. arXiv 2019, arXiv:1902.04103. [Google Scholar]
- Oliveira, F.; Da Silva, D.Q.; Filipe, V.; Pinho, T.M.; Cunha, M.; Cunha, J.B.; Dos Santos, F.N. Enhancing Grapevine Node Detection to Support Pruning Automation: Leveraging State-of-the-Art YOLO Detection Models for 2D Image Analysis. Sensors 2024, 24, 6774. [Google Scholar] [CrossRef]
- Shrestha, B.; Kulkarni, A.; Ahmed, F.; Sadhotra, A.; Maseeh, M.; Baig, S. Designing Efficient Sugarcane Node Cutting Machines: A Novel Approach. Iup J. Mech. Eng. 2024, 17, 41. [Google Scholar]
- Wen, C.; Guo, H.; Li, J.; Hou, B.; Huang, Y.; Li, K.; Nong, H.; Long, X.; Lu, Y. Application of improved YOLOv7-based sugarcane stem node recognition algorithm in complex environments. Front. Plant Sci. 2023, 14, 1230517. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Li, Y.; Xiao, Y.; Diao, Y.; Liao, H.; Zhang, Y.; Chen, X.; Wu, W.; Wen, C.; Li, S. Sugarcane stem node identification algorithm based on improved YOLOv5. PLoS ONE 2023, 18, e0295565. [Google Scholar] [CrossRef]
- Hu, J.; Li, G.; Mo, H.; Lv, Y.; Qian, T.; Chen, M.; Lu, S. Crop Node Detection and Internode Length Estimation Using an Improved YOLOv5 Model. Agriculture 2023, 13, 473. [Google Scholar] [CrossRef]
- Zhou, S.; Duan, X.; Zhou, J. Human pose estimation based on frequency domain and attention module. Neurocomputing 2024, 604, 128318. [Google Scholar] [CrossRef]
- Bustos, J.P.R. Application of DEEP LEARNING to the Processing of TERRESTRIAL LiDAR Data for the Evaluation of ARCHITECTURAL FEATURES and Functioning of Fruit Trees. Ph.D. Thesis, Université de Montpellier, Montpellier, France, 2024. [Google Scholar]
- Cieslak, M.; Govindarajan, U.; Garcia, A.; Chandrashekar, A.; Hadrich, T.; Mendoza-Drosik, A.; Michels, D.L.; Pirk, S.; Fu, C.; Palubicki, W. Generating Diverse Agricultural Data for Vision-Based Farming Applications. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 5422–5431. [Google Scholar]
- Wäldchen, J.; Rzanny, M.; Seeland, M.; Mäder, P. Automated plant species identification—Trends and future directions. PLoS Comput. Biol. 2018, 14, e1005993. [Google Scholar] [CrossRef]
- Ubbens, J.R.; Stavness, I. Deep plant phenomics: A deep learning platform for complex plant phenotyping tasks. Front. Plant Sci. 2017, 8, 1190. [Google Scholar] [CrossRef]
- Dosovitskiy, A. An image is worth 16×16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Wu, Z.; Sun, J.; Zhang, Y.; Wei, Z.; Chanussot, J. Recent developments in parallel and distributed computing for remotely sensed big data processing. Proc. IEEE 2021, 109, 1282–1305. [Google Scholar] [CrossRef]
- Issac, A. AI-Enabled Big Data Pipeline for Plant Phenotyping and Application in Cotton Bloom Detection and Counting. Master’s Thesis, University of Georgia, Athens, GA, USA, 2023. [Google Scholar]
- Koh, E.; Sunil, R.S.; Lam, H.Y.I.; Mutwil, M. Confronting the data deluge: How artificial intelligence can be used in the study of plant stress. Comput. Struct. Biotec 2024, 23, 3454–3466. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Hu, X.; Song, J.; Guo, J.; Lu, B. An Overview of High-Throughput Crop Phenotyping: Platform, Image Analysis, Data Mining, and Data Management. Plant Funct. Genom. Methods Protoc. 2024, 1, 3–38. [Google Scholar]
- Yang, W.; Duan, L.; Chen, G.; Xiong, L.; Liu, Q. Plant phenomics and high-throughput phenotyping: Accelerating rice functional genomics using multidisciplinary technologies. Curr. Opin. Plant Biol. 2013, 16, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Rasley, J.; Rajbhandari, S.; Ruwase, O.; He, Y. Deepspeed: System Optimizations Enable Training Deep Learning Models with over 100 Billion Parameters. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, CA, Virtual Event, 6–10 July 2020; pp. 3505–3506. [Google Scholar]
- Ghimire, D.; Kil, D.; Kim, S. A survey on efficient convolutional neural networks and hardware acceleration. Electronics 2022, 11, 945. [Google Scholar] [CrossRef]
- Shuvo, M.M.H.; Islam, S.K.; Cheng, J.; Morshed, B.I. Efficient acceleration of deep learning inference on resource-constrained edge devices: A review. Proc. IEEE 2022, 111, 42–91. [Google Scholar] [CrossRef]
- Luo, Z.; Xue, W.; Chae, J.; Fu, G. SKP: Semantic 3D Keypoint Detection for Category-Level Robotic Manipulation. IEEE Robot. Autom. Let. 2022, 7, 5437–5444. [Google Scholar] [CrossRef]
- Xu, D.; Zheng, T.; Zhang, Y.; Yang, X.; Fu, W. Multi-person 3D pose estimation from multi-view without extrinsic camera parameters. Expert Syst. Appl. 2025, 266, 126114. [Google Scholar] [CrossRef]





| Experiment | Public Online Dataset | DY Dataset |
|---|---|---|
| Model improvement experiment | √ | × |
| Ablation experiment | √ | × |
| Comparative experiment | √ | × |
| DY dataset testing experiment | × | √ |
| Visualization experiment | × | √ |
| Model | P (%) | R (%) | AP (%) | Params (M) |
|---|---|---|---|---|
| YOLOv7-w6-pose | 89.85 | 84.85 | 85.5 | 79.87 |
| YOLO_SSP | 89.59 | 86.97 | 88.1 | 44.28 |
| Model | NoReOrg (N) | S_ELAN (S) | ELAN-H (E) | MP_1 (P1) | MP_2 (P2) | RepConv (R) | Head_3 (H) | P (%) | R (%) | A P(%) | Params (M) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| YOLOv7-w6-pose | × | × | × | × | × | × | × | 89.9 | 83.9 | 86.0 | 79.81 |
| YOLOv7-w6-pose_N | √ | × | × | × | × | × | × | 89.6 | 85.8 | 87.4 | 79.86 |
| YOLOv7-w6-pose_S | × | √ | × | × | × | × | × | 90.3 | 82.5 | 85.9 | 77.02 |
| YOLOv7-w6-pose_E | × | × | √ | × | × | × | × | 89.3 | 83.5 | 85.0 | 79.81 |
| YOLOv7-w6-pose_P1 | × | × | × | √ | × | × | × | 88.5 | 84.1 | 85.7 | 73.01 |
| YOLOv7-w6-pose_P2 | × | × | × | × | √ | × | × | 89.3 | 85.2 | 87.1 | 79.81 |
| YOLOv7-w6-pose_R | × | × | × | × | × | √ | × | 89.0 | 84.5 | 86.8 | 80.80 |
| YOLOv7-w6-pose_H | × | × | × | × | × | × | √ | 91.0 | 84.3 | 87.1 | 53.54 |
| YOLOv7-w6-pose_NP2 | √ | × | × | × | √ | × | × | 89.3 | 84.5 | 86.5 | 79.86 |
| YOLOv7-w6-pose_NR | √ | × | × | × | × | √ | × | 89.8 | 83.5 | 86.5 | 80.85 |
| YOLOv7-w6-pose_NH | √ | × | × | × | × | × | √ | 90.6 | 85.9 | 88.3 | 53.59 |
| YOLOv7-w6-pose_P2R | × | × | × | × | √ | √ | × | 89.9 | 82.7 | 86.3 | 80.80 |
| YOLOv7-w6-pose_P1P2 | × | × | × | √ | √ | × | × | 88.8 | 83.4 | 86.3 | 73.01 |
| YOLOv7-w6-pose_NP2H | √ | × | × | × | √ | × | √ | 90.0 | 86.0 | 88.0 | 53.01 |
| YOLOv7-w6-pose_SP1R | × | √ | × | √ | × | √ | × | 89.4 | 84.3 | 86.4 | 67.05 |
| YOLOv7-w6-pose_NP2RH | √ | × | × | × | √ | √ | √ | 90.2 | 85.9 | 87.7 | 53.71 |
| YOLOv7-w6-pose_NP1P2RH | √ | × | × | √ | √ | √ | √ | 89.6 | 86.4 | 88.0 | 47.94 |
| YOLOv7-w6-pose_NSP1P2RH | √ | √ | × | √ | √ | √ | √ | 91.1 | 86.5 | 88.1 | 44.19 |
| YOLOv7-w6-pose_NSEP2RH | √ | √ | √ | × | √ | √ | √ | 90.5 | 85.2 | 88.0 | 52.10 |
| YOLOv7-w6-pose_NSP2RH | √ | √ | × | × | √ | √ | √ | 90.9 | 85.1 | 87.7 | 52.10 |
| YOLO_SSP | √ | √ | √ | √ | √ | √ | √ | 90.0 | 86.2 | 88.5 | 44.19 |
| Model | R (%) | AP (%) | GFLOPs | Params (M) |
|---|---|---|---|---|
| YOLO_SSP | 86.6 | 87.7 | 118.3 | 44.2 |
| YOLOv7-w6-pose | 84.6 | 85.2 | 101.4 | 79.9 |
| YOLOv7-tiny-pose | 72.2 | 74.9 | 19.9 | 9.6 |
| YOLOv3s-pose | 71.0 | 82.4 | 7.4 | 2.6 |
| YOLOv5n-pose | 73.2 | 83.9 | 25 | 9.4 |
| YOLOv5s-pose | 73.4 | 84.2 | 66.6 | 25.7 |
| YOLOv5m-pose | 73.9 | 84.2 | 45 | 15.6 |
| YOLOv6n-pose | 70.8 | 82.6 | 11.9 | 4.3 |
| YOLOv8n-pose | 71.2 | 82.7 | 8.4 | 3.1 |
| YOLOv10b-pose | 71.5 | 83.2 | 77.5 | 18.4 |
| Model | P (%) | R (%) | AP (%) | Params (M) |
|---|---|---|---|---|
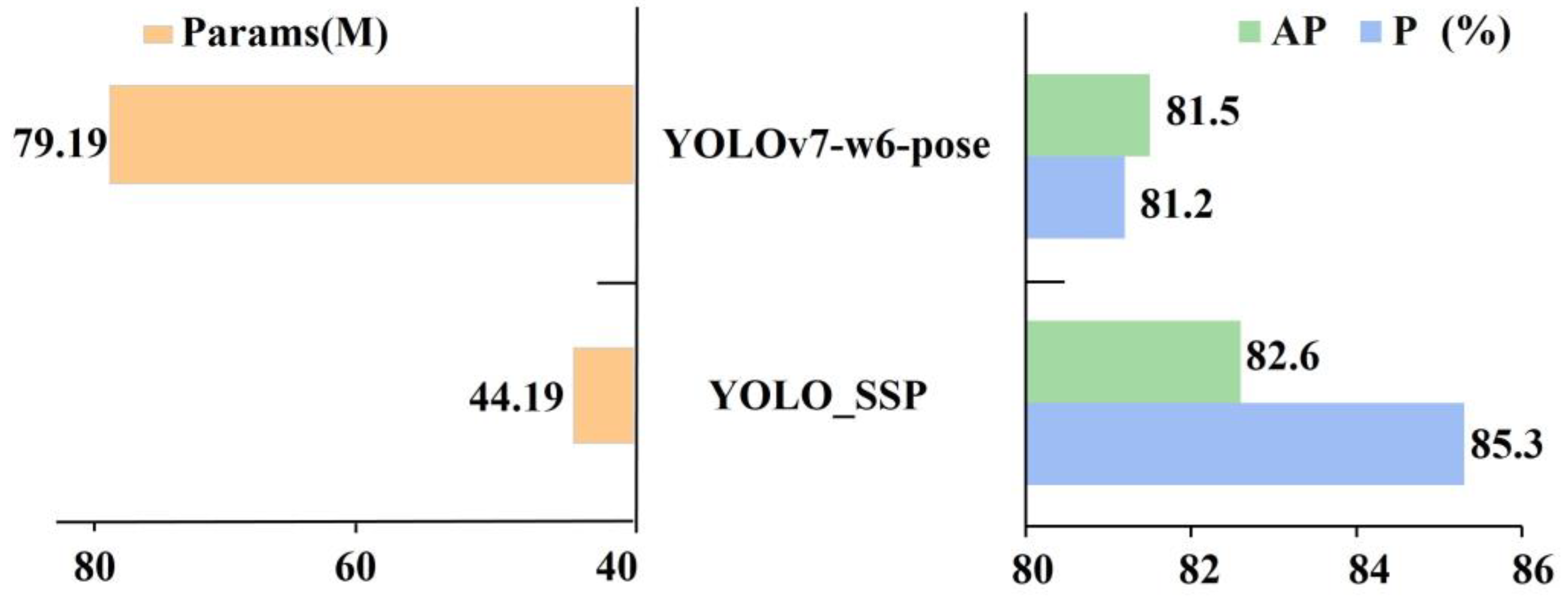
| YOLO_SSP | 85.3 | 76.0 | 82.6 | 44.19 |
| YOLOv7-w6-pose | 81.2 | 79.1 | 81.5 | 79.81 |
| 0 | Detection Point | AP (%) |
|---|---|---|
| YOLOv5s [67] | Tomato Calyx–Fruit Rachis Junction Point | 91.1 |
| Soybean Stem Node Point | 83.9 | |
| YOLOv5s [69] | Tea Buds Keypoints | 71.79 |
| Soybean Stem Node Point | 84.2 | |
| YOLOv8n [68] | Grape Picking Point | 89.7 |
| Soybean Stem Node Point | 82.7 | |
| YOLOv8n [70] | Lycium Barbarum Picking Point | 87.8 |
| Soybean Stem Node Point | 82.7 | |
| YOLOv8n [71] | Cabbage Head Keypoints | 93.5 |
| Soybean Stem Node Point | 82.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Liu, H.; Zhu, H.; Wang, C.; Wang, H.; Han, Z.; Zhao, L.; Liu, F. YOLO_SSP: An Auto-Algorithm to Detect Mature Soybean Stem Nodes Based on Keypoint Detection. Agronomy 2025, 15, 1128. https://doi.org/10.3390/agronomy15051128
Wu Q, Liu H, Zhu H, Wang C, Wang H, Han Z, Zhao L, Liu F. YOLO_SSP: An Auto-Algorithm to Detect Mature Soybean Stem Nodes Based on Keypoint Detection. Agronomy. 2025; 15(5):1128. https://doi.org/10.3390/agronomy15051128
Chicago/Turabian StyleWu, Qiong, Hang Liu, Hongfei Zhu, Cong Wang, Haoyu Wang, Zhongzhi Han, Longgang Zhao, and Fei Liu. 2025. "YOLO_SSP: An Auto-Algorithm to Detect Mature Soybean Stem Nodes Based on Keypoint Detection" Agronomy 15, no. 5: 1128. https://doi.org/10.3390/agronomy15051128
APA StyleWu, Q., Liu, H., Zhu, H., Wang, C., Wang, H., Han, Z., Zhao, L., & Liu, F. (2025). YOLO_SSP: An Auto-Algorithm to Detect Mature Soybean Stem Nodes Based on Keypoint Detection. Agronomy, 15(5), 1128. https://doi.org/10.3390/agronomy15051128






