Improvement of Rice Salt Tolerance by Pyramiding Two Genes in Xian and Geng Backgrounds Through CRISPR-Cas9 System
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of hst1-lrrk1, hst1-strk2 and hst1-pc1
2.2. Identification of Mutant Transgenic Plants
2.3. Identification and Evaluation of Salt Tolerance (ST) for Mutant Plants
2.4. Investigation of Yield-Related Traits
2.5. Data Analysis
3. Results
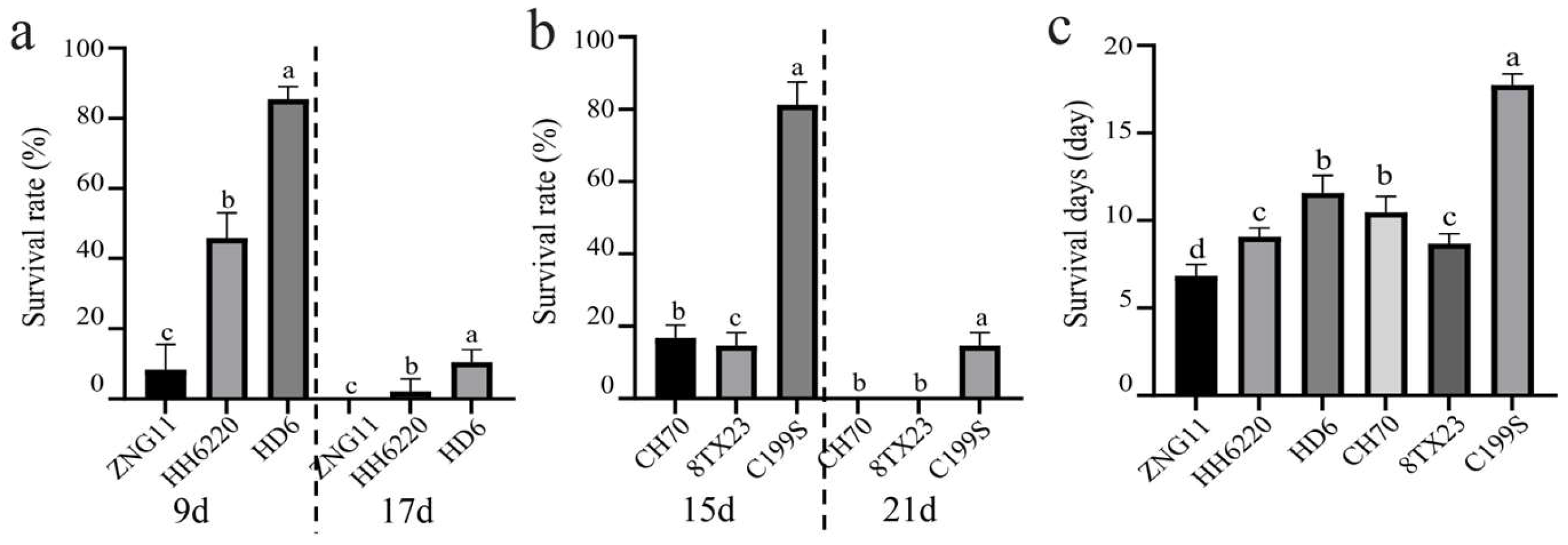
3.1. ST of Wildtype
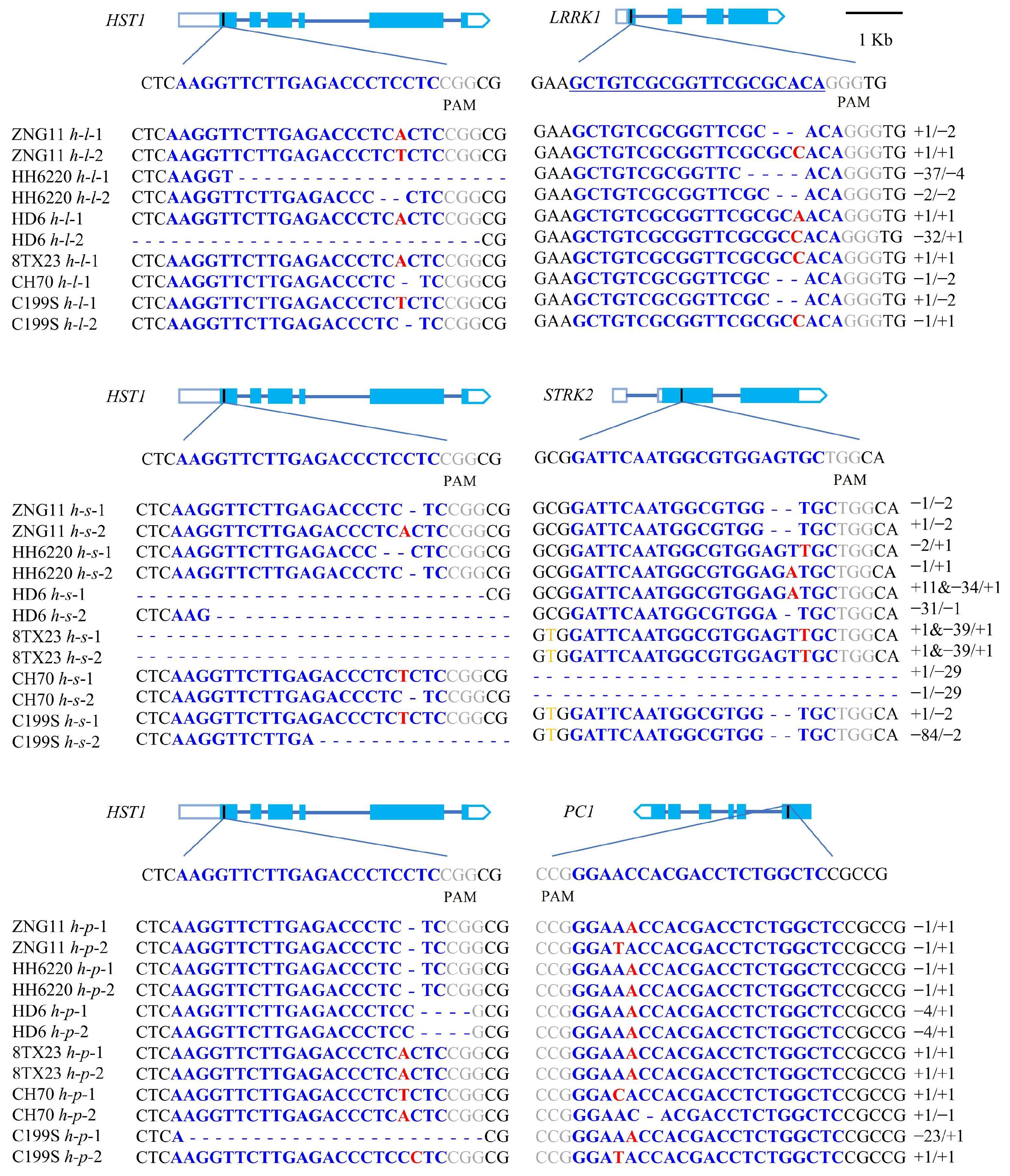
3.2. Acquisition of Transgenic Seedlings and Identification of Homozygotes
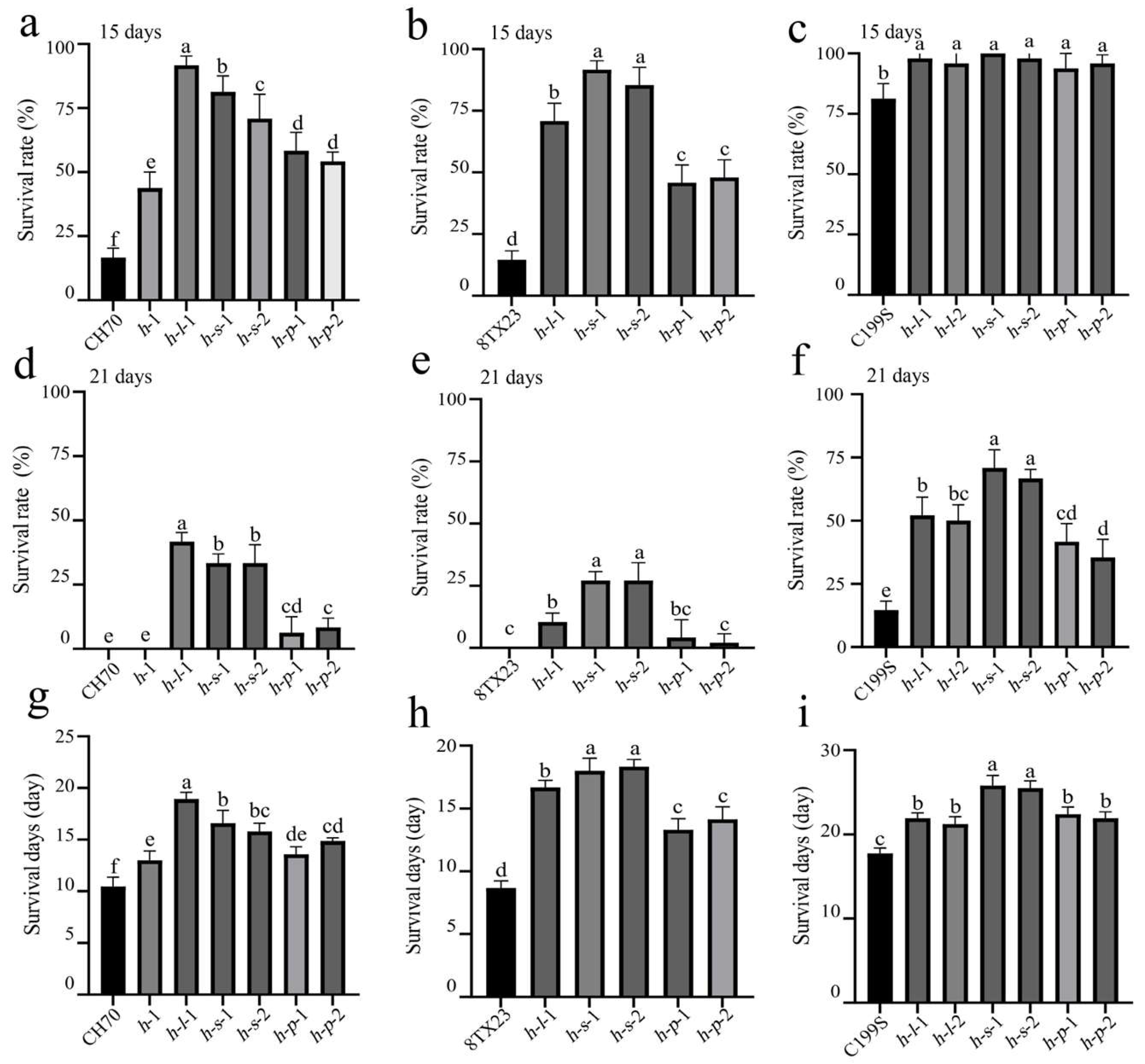
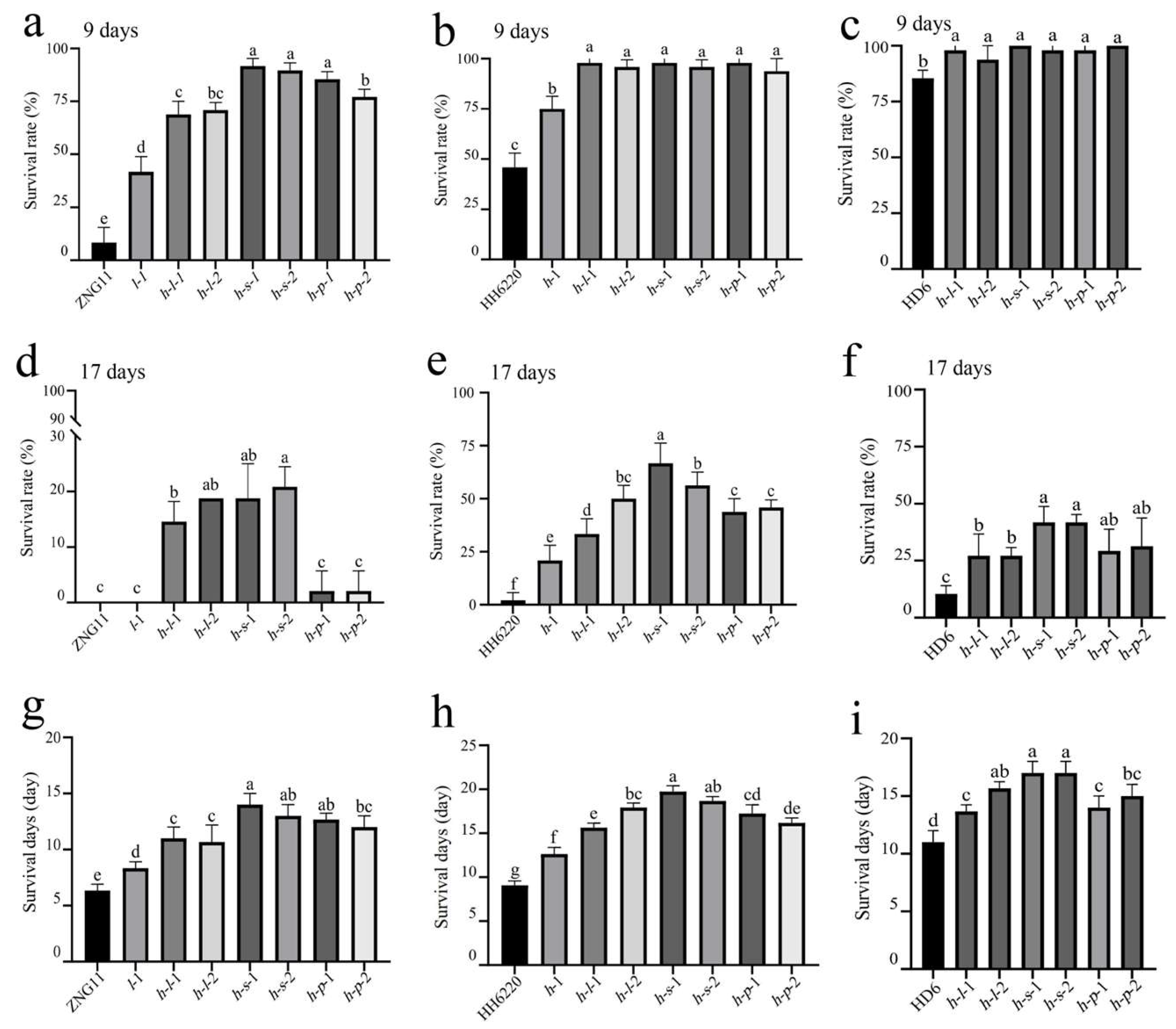
3.3. ST of Gene Mutant Lines
3.4. Identification of Transgenic Vector
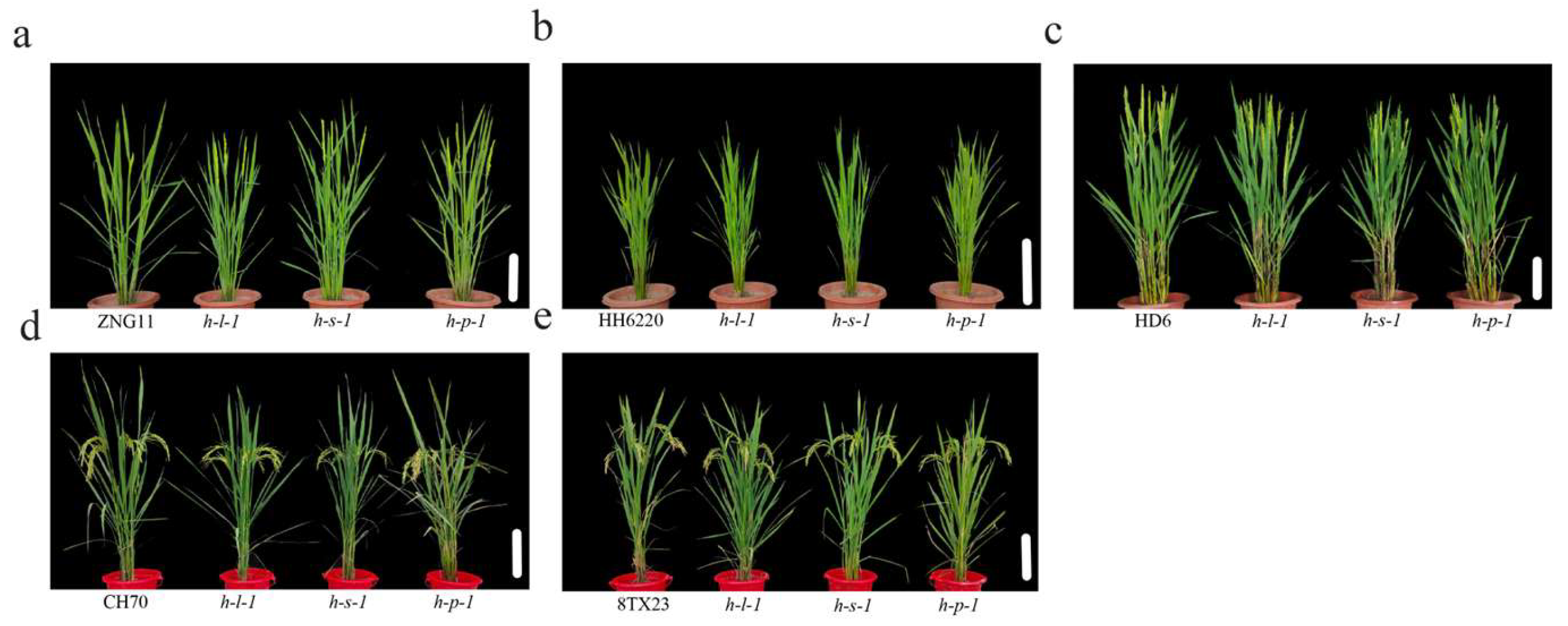
3.5. Morphological Features and Agronomic Traits of the Transgenic Plants
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ZNG11 | Zhongnonggeng 11 |
| HH6220 | Huhan 6220 |
| CH70 | Changhui 70 |
| C199S | Chun199S |
| ST | Salt tolerance |
| SR | Survival rate |
| SD | Survival days |
| GY | Grain yield per plant |
| EPN | Effective panicle number per plant |
| GNP | Grain number per panicle |
| SSR | Seed setting rate |
| TGW | Thousand grain weight |
References
- Qin, H.; Li, Y.; Huang, R. Advances and challenges in the breeding of salt-tolerant rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.A.; Rafii, M.Y.; Yusoff, M.M.; Ali, N.S.; Yusuff, O.; Datta, D.R.; Anisuzzaman, M.; Ikbal, M.F. Advanced breeding strategies and future perspectives of salinity tolerance in rice. Agronomy 2021, 11, 1631. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X. Response mechanisms of plants under Saline-Alkali stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Zhang, C.; Yates, G.; Bailey, M.; Brown, A.; Sadanandom, A. SUMO is a critical regulator of salt stress responses in rice. Plant Physiol. 2016, 170, 2378–2391. [Google Scholar] [CrossRef]
- Nam, M.H.; Bang, E.; Kwon, T.Y.; Kim, Y.; Kim, E.H.; Cho, K.; Park, W.J.; Kim, B.-G.; Yoon, I.S. Metabolite profiling of diverse rice germplasm and identification of conserved metabolic markers of rice roots in response to long-term mild salinity stress. Int. J. Mol. Sci. 2015, 16, 21959–21974. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Choi, W.Y.; Ko, J.C.; Kim, T.S.; Gregorio, G.B. Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta 2003, 216, 1043–1046. [Google Scholar] [CrossRef]
- Hu, T.T.; Liu, C.; Wang, J.k.; Ding, C.W.; Guo, R.L.; Wu, Y.L.; Xu, J.A.; Wang, Y.S. Progress of genetic and breeding on salt tolerance in rice. Mol. Breed. 2009, 7, 110–116. [Google Scholar]
- Huang, X.Y.; Chao, D.Y.; Gao, J.P.; Zhu, M.Z.; Shi, M.; Lin, H.X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef]
- Liu, C.; Lin, J.Z.; Wang, Y.; Tian, Y.; Zheng, H.P.; Zhou, Z.K.; Zhou, Y.B.; Tang, X.D.; Zhao, X.H.; Wu, T.; et al. The protein phosphatase PC1 dephosphorylates and deactivates CatC to negatively regulate H2O2 homeostasis and salt tolerance in rice. Plant Cell 2023, 35, 3604–3625. [Google Scholar] [CrossRef]
- Yan, L. The Biochemical Mechanism of Receptor-like Cytoplasmic Kinase LRRK1 in Salt Stress Regulation in Rice. Ph.D. Thesis, Hunan University, Changsha, China, 2021. [Google Scholar]
- Aycan, M.; Nahar, L.; Baslam, M.; Mitsui, T. B-type response regulator hst1 controls salinity tolerance in rice by regulating transcription factors and antioxidant mechanisms. Plant Physiol. Biochem. 2023, 196, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.F.; Chen, Z.H.; Kong, D.Y.; Bi, J.G.; Zhang, F.Y.; Luo, X.X.; Wang, J.H.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef]
- Han, X.; Chen, Z.; Li, P.; Xu, H.; Liu, K.; Zha, W.; Li, S.; Chen, J.; Yang, G.; Huang, J.; et al. Development of Novel Rice Germplasm for Salt-Tolerance at Seedling Stage Using CRISPR-Cas9. Sustainability 2022, 14, 2621. [Google Scholar] [CrossRef]
- Zhu, B.C.; Su, J.; Chang, M.C.; Verma, D.P.; Fan, Y.L.; Wu, R. Overexpression of aΔ1-pyrroline-5-carboxylate synthetase gene and analysis of tolerance to water and salt stress in transgenic rice. Plant Sci. 1998, 139, 41–48. [Google Scholar] [CrossRef]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, A.; Nakamura, A.; Tagiri, A.; Tanaka, H.; Miyao, A.; Hirochika, H.; Tanaka, Y. Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol. 2004, 45, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, Q.D.; Luo, L.; Yang, T.Y.; Zhang, S.; Hu, Y.B.; Yu, L.; Xu, G.H. Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 2015, 38, 2747–2765. [Google Scholar] [CrossRef]
- Subburaj, S.; Tu, L.; Jin, Y.T.; Bae, S.; Seo, P.J.; Jung, Y.J.; Lee, G.J. Targeted genome editing, an alternative tool for trait improvement in horticultural crops. Hortic. Environ. Biotechnol. 2016, 57, 531–543. [Google Scholar] [CrossRef]
- Tang, X.Y.; Wang, H.M.; Long, Q.Z.; Huang, Y.L.; Lu, M.; Wan, J.L. The application of crispr/cas9 system in rice genome editing. Mol. Plant Breed. 2017, 15, 895–902. [Google Scholar] [CrossRef]
- Sukegawa, S.; Toki, S.; Saika, H. Genome editing technology and its application to metabolic engineering in rice. Rice 2022, 15, 21. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; He, M.; Li, J.; Chen, L.; Huang, Z.F.; Zheng, S.Y.; Zhu, L.Y.; Ni, E.D.; Jiang, D.G.; Zhao, B.R.; et al. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 2016, 6, 37395. [Google Scholar] [CrossRef]
- Xu, R.F.; Yang, Y.C.; Qin, R.Y.; Li, H.; Qiu, C.H.; Li, L.; Wei, P.C.; Yang, J.B. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genom. 2016, 43, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Cai, L.; Wu, H.; Wang, B.; Gu, B.; Cui, S.; Huang, X.; Xu, Z.; Hao, B.; Hou, H.; et al. Fine-tuning rice heading date through multiplex editing of the regulatory regions of key genes by CRISPR-Cas9. Plant Biotechnol. J. 2024, 22, 751–758. [Google Scholar] [CrossRef]
- Senguttuvel, P.; Raveendran, M.; Vijayalakshmi, C.; Thiyagarajan, K.; Bapu, J.R.K.; Viraktamath, B.C. Molecular mechanism of salt tolerance for genetic diversity analysed in association with Na+/K+ ratio through SSR markers in rice (Oryza sativa L.). Int. J. Agric. Res. 2010, 5, 708–719. [Google Scholar] [CrossRef]
- Yu, J.; Zhu, C.; Xuan, W.; An, H.; Tian, Y.; Wang, B.; Chi, W.; Chen, G.; Ge, Y.; Li, J.; et al. Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice. Nat. Commun. 2023, 14, 3550. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Chen, G.; Liu, C.; Gao, Z.; Zhang, Y.; Zhang, A.; Zhu, L.; Hu, J.; Ren, D.; Yu, L.; Xu, G.; et al. Variation in the abundance of OsHAK1 transcript underlies the differential salinity tolerance of an indica and a japonica rice cultivar. Front. Plant Sci. 2018, 8, 2216. [Google Scholar] [CrossRef]
- Kong, W.; Sun, T.; Zhang, C.; Deng, X.; Li, Y. Comparative transcriptome analysis reveals the mechanisms underlying differences in salt tolerance between indica and japonica rice at seedling stage. Front. Plant Sci. 2021, 12, 725436. [Google Scholar] [CrossRef]
- Negrao, S.; Courtois, B.; Ahmadi, N.; Abreu, I.; Saibo, N.; Oliveira, M.M. Recent updates on salinity stress in rice: From physiological to molecular responses. Crit. Rev. Plant Sci. 2011, 30, 329–377. [Google Scholar] [CrossRef]
- Cheng, L.R.; Wang, Y.; Meng, L.J.; Hu, X.; Cui, Y.R.; Sun, Y.; Zhu, L.H.; Ali, J.; Xu, J.L.; Li, Z.K. Identification of salt-tolerant QTLs with strong genetic background effect using two sets of reciprocal introgression lines in rice. Genome 2012, 55, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Yeo, A.R.; Flowers, T.J. Salinity resistance in rice (Oryza sativa L.) and a pyramiding approach to breeding varieties for saline soils. Aust. J. Plant Physiol. 1986, 13, 161–173. [Google Scholar] [CrossRef]
- Chawla, S.; Jain, S.; Jain, V. Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativa L.). Plant Biochem. Biot. 2013, 22, 27–34. [Google Scholar] [CrossRef]
- Gharsallah, C.; Fakhfakh, H.; Grubb, D.; Gorsane, F. Effect of salt stress on ion concentration, proline content, antioxidant enzyme activities and gene expression in tomato cultivars. AoB Plants 2016, 8, plw055. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Crossa, J.; Perez-Rodriguez, P.; Cuevas, J.; Montesinos-Lopez, O.; Jarquin, D.; de Los Campos, G.; Burgueno, J.; Gonzalez-Camacho, J.M.; Perez-Elizalde, S.; Beyene, Y.; et al. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Singh, B.; Dhakate, P.; Rahman, M.; Islam, M.A. Rice, Marker-Assisted Breeding, and Disease Resistance. In Disease Resistance in Crop Plants: Molecular, Genetic and Genomic Perspectives; Wani, S.H., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 83–111. [Google Scholar] [CrossRef]
- Chen, Y.P.; Dan, Z.W.; Li, S.Q. GROWTH REGULATING FACTOR 7-mediated arbutin metabolism enhances rice salt tolerance. Plant Cell 2024, 36, 2834–2850. [Google Scholar] [CrossRef]
- Yin, W.C.; Xiao, Y.H.; Niu, M.; Meng, W.J.; Li, L.L.; Zhang, X.X.; Liu, D.P.; Zhang, G.H.; Qian, Y.W.; Sun, Z.T.; et al. ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell 2020, 32, 2292–2306. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, R.; Meng, Q.S.; Qin, H.; Quan, R.D.; Wei, P.C.; Li, X.Y.; Jiang, L.; Huang, R.F. A natural variation in OsDSK2a modulates plant growth and salt tolerance through phosphorylation by SnRK1A in rice. Plant Biotechnol. J. 2024, 22, 1881–1896. [Google Scholar] [CrossRef]
- Xiang, Y.H.; Yu, J.J.; Liao, B.; Shan, J.X.; Ye, W.W.; Dong, N.Q.; Guo, T.; Kan, Y.; Zhang, H.; Yang, Y.B.; et al. An a/b hydrolase family member negatively regulates salt tolerance but promotes flowering through three distinct functions in rice. Mol. Plant 2022, 15, 1908–1930. [Google Scholar] [CrossRef]
- Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. The Saltol QTL-localized transcription factor OsGATA8 plays an important role in stress tolerance and seed development in Arabidopsis and rice. J. Exp. Bot. 2020, 71, 684–698. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.L.; Yang, J.; Yu, D.Q.; Xu, P. Identification and validation a major QTL from ‘Sea Rice 86’ seedlings conferred salt tolerance. Agronomy 2020, 10, 410. [Google Scholar] [CrossRef]

| Primer Name | Primer Sequence (5′-3′) | Purpose |
|---|---|---|
| HST1-F | AGGTGCTTCAGTTTCAGTTGC | Target site sequencing |
| HST1-R | GTGCACCATAGTAGGCATATT | Target site sequencing |
| LRRK1-F | AAGCTGCGAGGCTGTTCTCCGC | Target site sequencing |
| LRRK1-R | GTTCCAAAACTAAACCAAGATT | Target site sequencing |
| STRK2-F | TGAAGGGGTTCAGGAAGTAGAG | Target site sequencing |
| STRK2-R | CTGGGATTGCTCCAGTTAATTT | Target site sequencing |
| PC1-F | GAGAACATGGACTGGGTGC | Target site sequencing |
| PC1-R | AAGCCGAGACGGACAAGA | Target site sequencing |
| HPT-F | GAGCATATACGCCCGGAGTC | Transgenic analysis |
| HPT-R | CAAGACCTGCCTGAAACCGA | Transgenic analysis |
| Lines | Effective Panicle Number | Grain Number Per Panicle | Seed Setting Rate (%) | 1000-Grain Weight (g) | Grain Yield Per Plant (g) |
|---|---|---|---|---|---|
| ZNG11 | 10.17 ± 1.47 a | 130.92 ± 11.20 a | 91.22 ± 1.05 a | 21.58 ± 0.38 b | 27.21 ± 1.69 a |
| ZNG11_h-l-1 | 9.33 ± 1.67 a | 127.92 ± 7.18 a | 87.83 ± 2.73 b | 21.08 ± 0.4 c | 22.71 ± 2.04 b |
| ZNG11_h-s-1 | 9.25 ± 1.28 a | 140.39 ± 12.87 a | 83.91 ± 2.48c | 21.91 ± 0.45 b | 21.8 ± 2.41 b |
| ZNG11_h-p-1 | 9.67 ± 1.63 a | 129.89 ± 18.42 a | 83.79 ± 2.52 c | 22.91 ± 0.6 a | 23.22 ± 1.89 b |
| HH6220 | 16.88 ± 1.55 ab | 55.34 ± 3.89 a | 89.97 ± 4.88 a | 23.9 ± 0.55 a | 19.06 ± 2.83 a |
| HH6220_h-l-1 | 15.75 ± 1.58 b | 46.31 ± 3.51 b | 77.64 ± 5.56 c | 21.37 ± 1.09 c | 11.54 ± 2.02 c |
| HH6220_h-s-1 | 17.5 ± 2.07 ab | 44.96 ± 8.15 b | 82.32 ± 4.05 b | 22.83 ± 0.61 b | 15.98 ± 2.74 b |
| HH6220_h-p-1 | 17.8 ± 0.84 a | 42.49 ± 4.83 b | 85.73 ± 2.44 b | 23.81 ± 0.87 a | 18.03 ± 2.18 ab |
| HD6 | 10.9 ± 1.79 ab | 143.1 ± 19.84 a | 93.81 ± 1.4 a | 24.64 ± 0.61 a | 36.39 ± 4.13 a |
| HD6_h-l-1 | 9.71 ± 0.76 b | 141.56 ± 12.87 a | 87.8 ± 1.9 b | 24.79 ± 0.66 a | 26.7 ± 5.93 b |
| HD6_h-s-1 | 11.75 ± 1.49 a | 135.41 ± 7.72 a | 76.1 ± 2.58 c | 23.39 ± 0.5 b | 28.08 ± 4.42 b |
| HD6_h-p-1 | 10.86 ± 2.04 ab | 135.04 ± 9.13 a | 86.12 ± 3.95 b | 24.58 ± 0.65 a | 30.27 ± 5.93 b |
| Lines | Effective Panicle Number | Grain Number Per Panicle | Seed Setting Rate (%) | 1000-Grain Weight (g) | Grain Yield Per Plant (g) |
|---|---|---|---|---|---|
| CH70 | 7.25 ± 0.96 a | 194.05 ± 15.33 a | 86.69 ± 6.57 a | 18.04 ± 0.43 b | 20.01 ± 2.28 a |
| CH70_h-l-1 | 7.29 ± 1.8 a | 180.33 ± 18.86 a | 75.94 ± 3.04 c | 17.76 ± 0.37 b | 17.00 ± 4.51 a |
| CH70_h-s-1 | 7.50 ± 1.05 a | 177.13 ± 38.36 a | 77.71 ± 2.69 bc | 17.05 ± 0.39 c | 16.79 ± 2.66 a |
| CH70_h-p-1 | 6.80 ± 0.84 a | 195.19 ± 20.4 a | 81.81 ± 4.91 b | 19.50 ± 0.33 a | 20.60 ± 4.63 a |
| 8TX23 | 8.50 ± 1.29 a | 157.17 ± 13.9 a | 87.87 ± 3.17 a | 15.75 ± 0.45 b | 16.85 ± 3.16 a |
| 8TX23_h-l-1 | 8.0 ± 1.15 a | 128.79 ± 19.13 b | 80.75 ± 5.80 b | 15.40 ± 0.63 bc | 12.05 ± 3.15 b |
| 8TX23_h-s-1 | 8.57 ± 1.40 a | 132.56 ± 16.49 b | 86.28 ± 2.11 ab | 15.04 ± 0.21 c | 13.83 ± 2.26 ab |
| 8TX23_h-p-1 | 8.33 ± 1.21 a | 149.83 ± 16.56 ab | 82.30 ± 6.83 ab | 17.35 ± 0.25 a | 16.23 ± 3.17 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Z.; Zhai, L.; Chen, K.; Zhang, F.; Qiu, X.; Xu, J. Improvement of Rice Salt Tolerance by Pyramiding Two Genes in Xian and Geng Backgrounds Through CRISPR-Cas9 System. Agronomy 2025, 15, 1014. https://doi.org/10.3390/agronomy15051014
Ding Z, Zhai L, Chen K, Zhang F, Qiu X, Xu J. Improvement of Rice Salt Tolerance by Pyramiding Two Genes in Xian and Geng Backgrounds Through CRISPR-Cas9 System. Agronomy. 2025; 15(5):1014. https://doi.org/10.3390/agronomy15051014
Chicago/Turabian StyleDing, Zhihu, Laiyuan Zhai, Kai Chen, Fan Zhang, Xianjin Qiu, and Jianlong Xu. 2025. "Improvement of Rice Salt Tolerance by Pyramiding Two Genes in Xian and Geng Backgrounds Through CRISPR-Cas9 System" Agronomy 15, no. 5: 1014. https://doi.org/10.3390/agronomy15051014
APA StyleDing, Z., Zhai, L., Chen, K., Zhang, F., Qiu, X., & Xu, J. (2025). Improvement of Rice Salt Tolerance by Pyramiding Two Genes in Xian and Geng Backgrounds Through CRISPR-Cas9 System. Agronomy, 15(5), 1014. https://doi.org/10.3390/agronomy15051014







