3.1. Non-Cultivar Specific Models
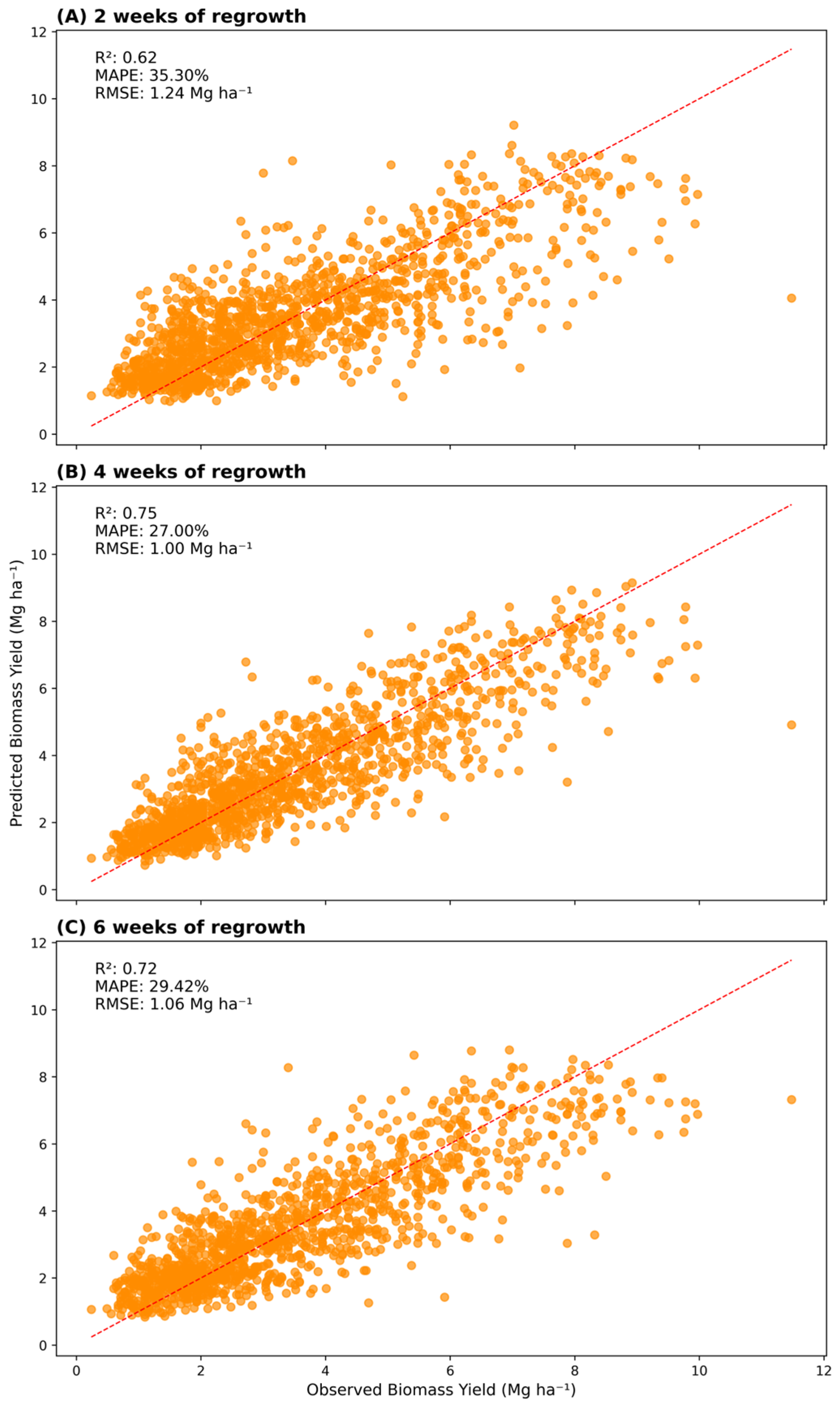
The 2 WOR all-features model (
Figure 1A) had the greatest MAPE and RMSE and the least R
2, meaning it had the greatest error between predicted and observed yield values among all all-features models. Conversely, the 4 WOR all-features model (
Figure 1B) showed the least error between predicted and observed values—the least MAPE, RMSE, and the greatest R
2. Thus, the 6 WOR all-features model (
Figure 1C) presented intermediate MAPE, RMSE, and R
2 (
Figure 1).
Although the 4 WOR all-features model was considered the best-performing model, the other developed all-features models presented good yield predictors compared to existing bermudagrass yield models cited in the current literature. For instance, Starks et al. [
32] used multi-linear regression analysis to develop different empirical prediction models based on six broad wavebands, such as blue (450–520 nm), green (520–600 nm), red (630–690 nm), NIR (760–900 nm), short-wave infrared 1 (SWIR1, 1550–1750 nm), and short-wave infrared 2 (SWIR2, 2080–2300 nm). The authors’ best bermudagrass prediction model uses the first derivative of reflectance value of 895, 1175, 1975, 2025, and 2235 nm wavelengths, achieving an RMSE and R
2 of 1.16 Mg ha
−1 and 0.63, respectively. In another study, Zhao et al. [
33] used multiple regression powered by MAXR (method of maximum R
2 improvement), achieving an RMSE and R
2 of 1.02 Mg ha
−1 and 0.59, respectively, when combining the reflectance value of 565, 725, 1055, 1215, 1265, 1305, 1465, and 1485 nm wavelengths. Both studies showed bermudagrass prediction models with greater RMSE and lesser R
2 than our 4 and 6 WOR all-features models and similar to the 2 WOR all-features model. Moreover, Mosali et al. [
34] achieved a linear regression R
2 of 0.41, predicting yield potential in bermudagrass based on a ratio of NDVI values divided by growing degree days where growing degree days were calculated by subtracting the base temperature from the daily average minimum and maximum temperatures.
Moreover, our yield-predicting all-features model performances (R
2 ranging from 0.62 to 0.75) agreed with existing Random Forest algorithms developed for agricultural purposes. For instance, Zhang et al. [
35] achieved an R
2 of 0.76 by integrating optical, fluorescence, thermal satellite, and environmental data to predict corn yield. Also, the Random Forest algorithm showed improved performance when predicting corn and soybean yield compared to LASSO regression analysis where RF models for corn achieved R
2 equal to 0.7 and 0.69 for soybean compared to LASSO models for corn and soybean achieving R
2 of 0.62 and 0.51, respectively [
36]; moreover, Jeong et al. [
37] found the best RMSE when comparing RF models and multiple linear regression models (MLR) where, when predicting global wheat production, the RMSE for both models was 0.32 and 1.32, respectively; the same trend was observed when predicting corn for grain in the United States (RMSE of 1.13 compared to 1.93), potato in the U.S (RMSE of 2.77 compared to 5.62) and corn for silage (RMSE of 1.9 using RF compared to 4.54 when using MLR).
The all-features models trained with data collected at 2, 4, and 6 WOR showed different feature importance score results (
Figure 2). Normalized difference vegetation index was the most important feature at 2 WOR followed by laser, precipitation, ultrasonic, and N rate. The NDVI has an elevated performance in yield prediction in early plant development stages when soil exposure is present to contrast with vegetation, increasing the range values of ratios between red and NIR wavelengths [
38,
39]. Furthermore, since laser and ultrasonic measurements are related to plant height, which did not vary much at bermudagrasses’ early development, NDVI became crucial by reflecting plants’ overall health and vigor.
At 4 WOR, laser became the most important feature, followed by NDVI, precipitation, ultrasonic, and N rate. At this stage, the plants were mid-vegetative, and the canopy was not completely closed [
40], allowing the laser to play a key role in assessing height and potential vegetation cover gaps. The laser has a 4.8 mm diameter reading spot that precisely distinguishes bare soil, i.e., “zero”, from canopy height [
41].
Conversely, at 6 WOR, the ultrasonic sensor was the most important feature, followed by laser, NDVI, precipitation, and N rate. At this reading event, plant stages varied from late vegetative to anthesis; canopies were denser fully covering the soil (visual inferences only). Ultrasonic sensors capture multiple points within their reading diameter footprint, i.e., 16.61 cm for this experiment (Senix Corporation, 2025), and average them. This average height feature better represents the overall structure and density of the plants when there is less exposed soil due to increased vegetative growth.
After applying the backward feature elimination, most reduced models achieved similar or slightly lower performances than the all-features models.
Table 4 lists all backward feature elimination for all reading events; however, all feature combinations models’ performance is listed in
Table A1,
Table A2 and
Table A3. Backward feature elimination was employed, aiming at a faster processing, simpler, minimalistic algorithm by eliminating less informative features [
30,
31]. Previous authors used the backward feature elimination procedure, which stated model improvements. Elavarasan et al. [
42], when developing prediction models for rice yield using initially 45 features, increased the R
2 from 0.53 to 0.67 and reduced the MAPE from 21.3% to 19% when implementing an aggregation of the correlation-based filter (CFS) and Random Forest recursive feature elimination (RFRFE). When predicting corn yield using an open-source dataset in Eswatini, Fashoto et al. [
43] also observed an increase in the adjusted R
2 from 0.85 to 0.89 when applying a backward features elimination on an 80–20 training/testing proportion when removing the explanatory variable with the highest
p-value.
At 2 WOR, a slight performance decline was observed when eliminating the ultrasonic sensor, followed by an abrupt decline when eliminating the precipitation and laser features.
When the N rate feature was eliminated at 4 WOR, a slight performance decline was observed as well. Then, a minor performance decline was observed with ultrasonic feature elimination, followed by abrupt performance declines when precipitation and NDVI were eliminated.
Likewise, the all-features model performed similarly at the first backward feature elimination at 6 WOR. This model also presented a slight performance decrease when the N rate was eliminated. Then, abrupt performance declines were observed when precipitation, NDVI, and laser were eliminated.
Previous bermudagrass yield prediction models found in the current literature showed R
2 and RMSE of 0.63 and 1.16 Mg ha
−1 [
43] and 0.59 and 1.02 Mg ha
−1 [
33]. Using their performances as a guide, the reduced models containing NDVI, laser, and precipitation features for 2 WOR, laser feature for 4 WOR, and ultrasonic feature for 6 WOR could be considered appropriate.
The previously developed bermudagrass yields predictive models used laser and ultrasonic sensors, which are not commonly adopted in commercial forage enterprises. Thus, their uses might be restricted to research development. Thinking about commercial applications, a model was developed using a combination of NDVI, precipitation, and N rate features. Multispectral sensors that allow NDVI calculation are very popular in agricultural enterprises [
44,
45]. Also, the downside of employing the ultrasonic and laser sensors is that they have 4.3 and 15 m maximum operational distance from the target, respectively, making their use unfeasible on UAVs. This commercially adapted model was developed using readings taken at 4 WOR only because previous models performed best at this reading time.
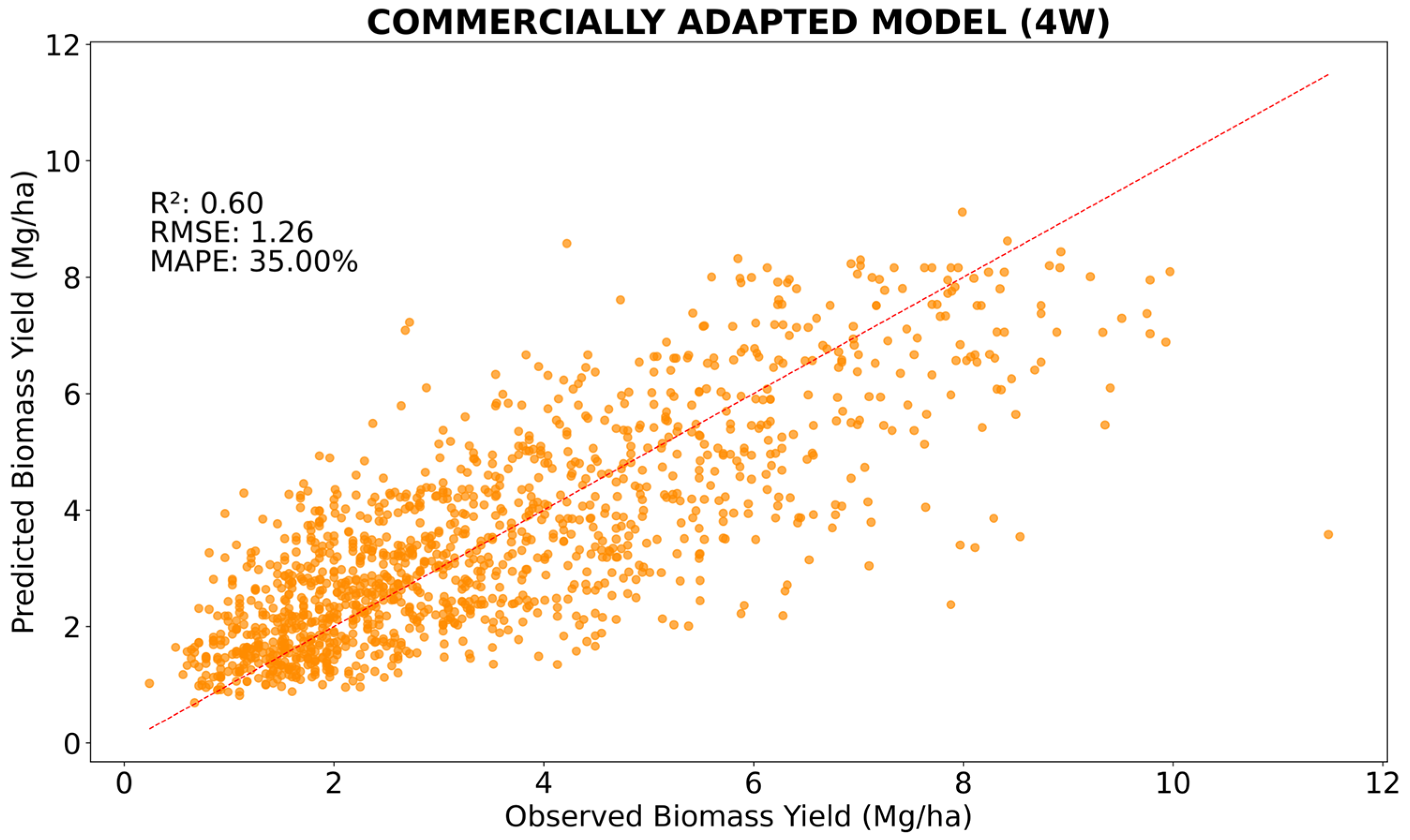
The commercially adapted model (
Figure 3) had an abrupt reduction in its performance (R
2 decreased from 0.75 to 0.60) compared to the all-features model (
Figure 1B). This reduction in performance is related to the removal of the laser feature, which was the feature with the greatest importance (65.5%,
Figure 3) at 4 WOR. However, the commercially adapted model showed similar or superior performance to other bermudagrass yield predictive models in the current literature review already cited above [
34,
35,
44].
3.2. Cultivar-Specific Models
Given that 4 WOR readings produced the highest-performing all-features model, and that this sampling time offers a practical balance between prediction accuracy and timely decision making, cultivar-specific models were subsequently developed. This timepoint allows producers to adjust stocking rates or schedule harvests. A model containing all cultivars’ 4 WOR dataset, except Greenfield, was developed as the hay-type cultivars model. A model containing Greenfield, the only grazing-type cultivar [
25], dataset was developed, followed by other five cultivars-specific models, such as Goodwell, Midland, Midland 99, Ozark, and Tifton 44. All models are listed in
Table 5.
When comparing the all-features model trained with all cultivars (R2, MAPE, and RMSE of 0.75, 26.79, and 1.0, respectively) to all cultivar-specific and hay-type cultivars models, only the cultivar-specific model for Midland99 and Ozark had the same or slightly reduced performance compared to the model trained with all cultivars. Cultivars-specific models for Greenfield, Tifton 44, Goodwell, and hay-type cultivars had reduced performance compared to the model trained with all cultivars. Moreover, the Midland cultivar-specific model had an abrupt performance decline compared to the model trained with all cultivars.
A different scenario was found when comparing the commercially adapted model trained with all cultivars (R2, MAPE, and RMSE of 0.61, 34.1, and 1.24, respectively) to cultivar-specific and hay-type cultivar models. Greenfield, Midland 99, and Ozark had considerable improvement in performance, whereas Goodwell had a slight improvement. The association of hay-type cultivars and Tifton 44 showed a declined performance, and Midland showed an abrupt decline.
All-cultivars, hay-type, and cultivar-specific models built with all features showed higher performance than the commercially adapted ones. This result reinforced the information displayed in
Figure 2, where the laser was the most important feature (i.e., 65.5%) in all-features, all-cultivar models at 4 WOR. Thus, its removal led to a decline in model performance in most commercially adapted models, such as the all-cultivars model, the hay-type cultivar model, Midland 99, Tifton 44, Midland, Ozark, and Goodwell.
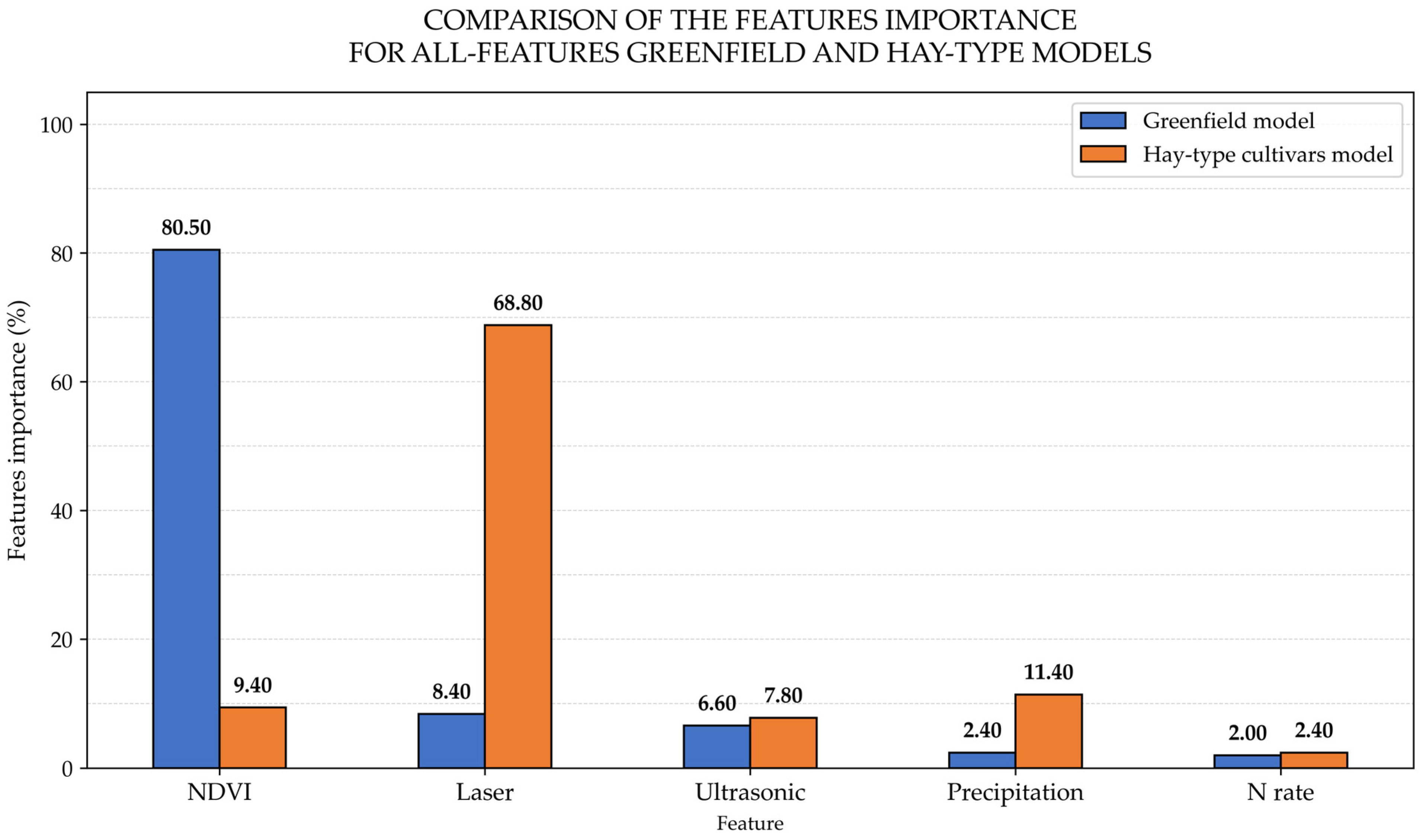
Nevertheless, the Greenfield commercially adapted cultivar model showed a slight increase in its performance when compared to the Greenfield all-features model presenting R
2, MAPE, and RMSE changes of +0.02, −1.12 percent units, and −0.03 Mg ha
−1, respectively. Further features importance examination presented in
Figure 4 revealed that the Greenfield all-features model’s most important input was NDVI (80.50%) followed by laser (68.80%), ultrasonic (7.80%), precipitation (11.40%), and N rate (2.0%). This result differed from the all-features hay-type model, which showed laser as the most important feature (68.80%) followed by precipitation (11.40%), NDVI (9.40%), ultrasonic (7.80%), and N rate (2.40).
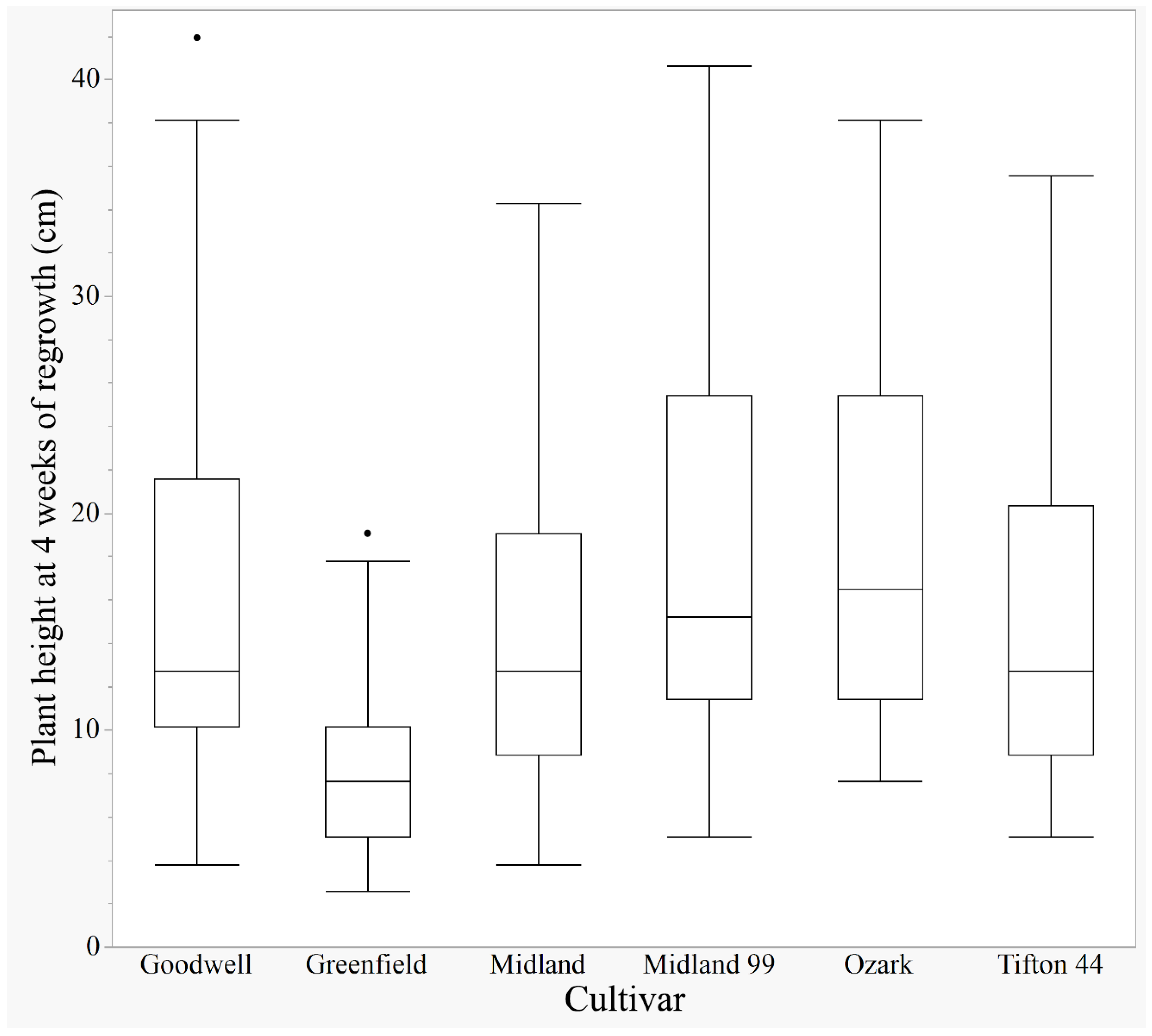
The Greenfield cultivar is short and creeping, and its average height is statistically lower (13.8 cm) than that of all the other hay cultivars (19.5–25 cm) used in this experiment (
Figure A1). Thus, Greenfield did not benefit from the height measurements provided by laser and ultrasonic; otherwise, it mostly benefited from NDVI, which reflects the overall health and vigor of the plants at 4 WOR.