CbCBF2 Integrates JA and BR Signaling to Enhance Oleanolic Acid Biosynthesis in Conyza blinii H. Lév Under Cold Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Nocturnal Low Temperature and Transcriptome
2.3. Phylogenetic Tree
2.4. Differential Expression and Heatmap Analysis
2.5. Correlation Analysis
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Annotations
2.7. Gene Cloning and Vector Construction
2.8. Transient Transformation of C. blinii Leaves
2.9. Real-Time Quantitative PCR (RT-qPCR)
2.10. Determination of Terpenoid Content
3. Results
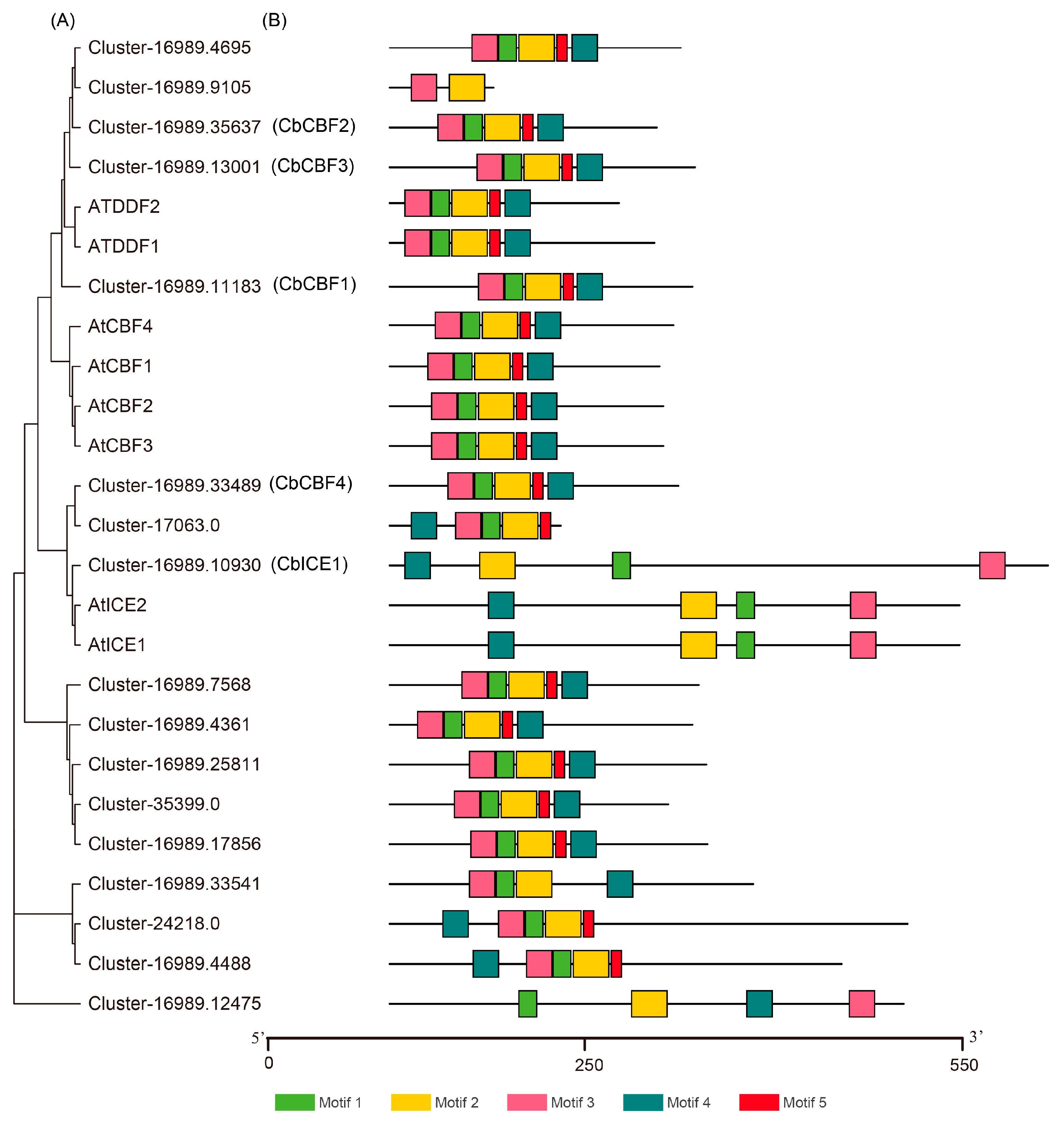
3.1. CbCBF2 Is the Most Active CBF Transcription Factor
3.2. CBF2 Is More Significantly Associated with DEGs Under Low Temperature
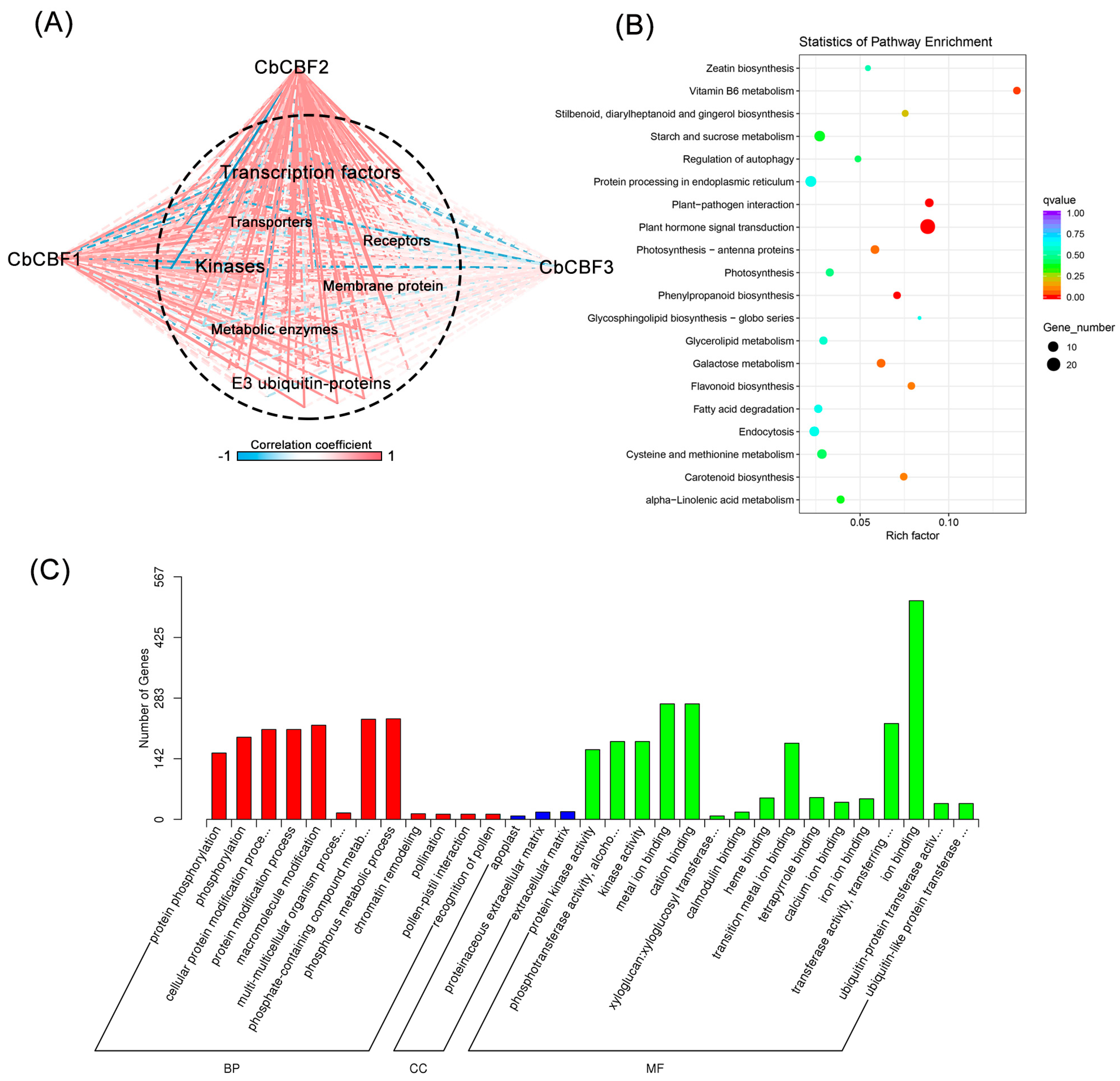
3.3. CBF2 Is the Main Effector Linking Plant Hormones and Terpenoids
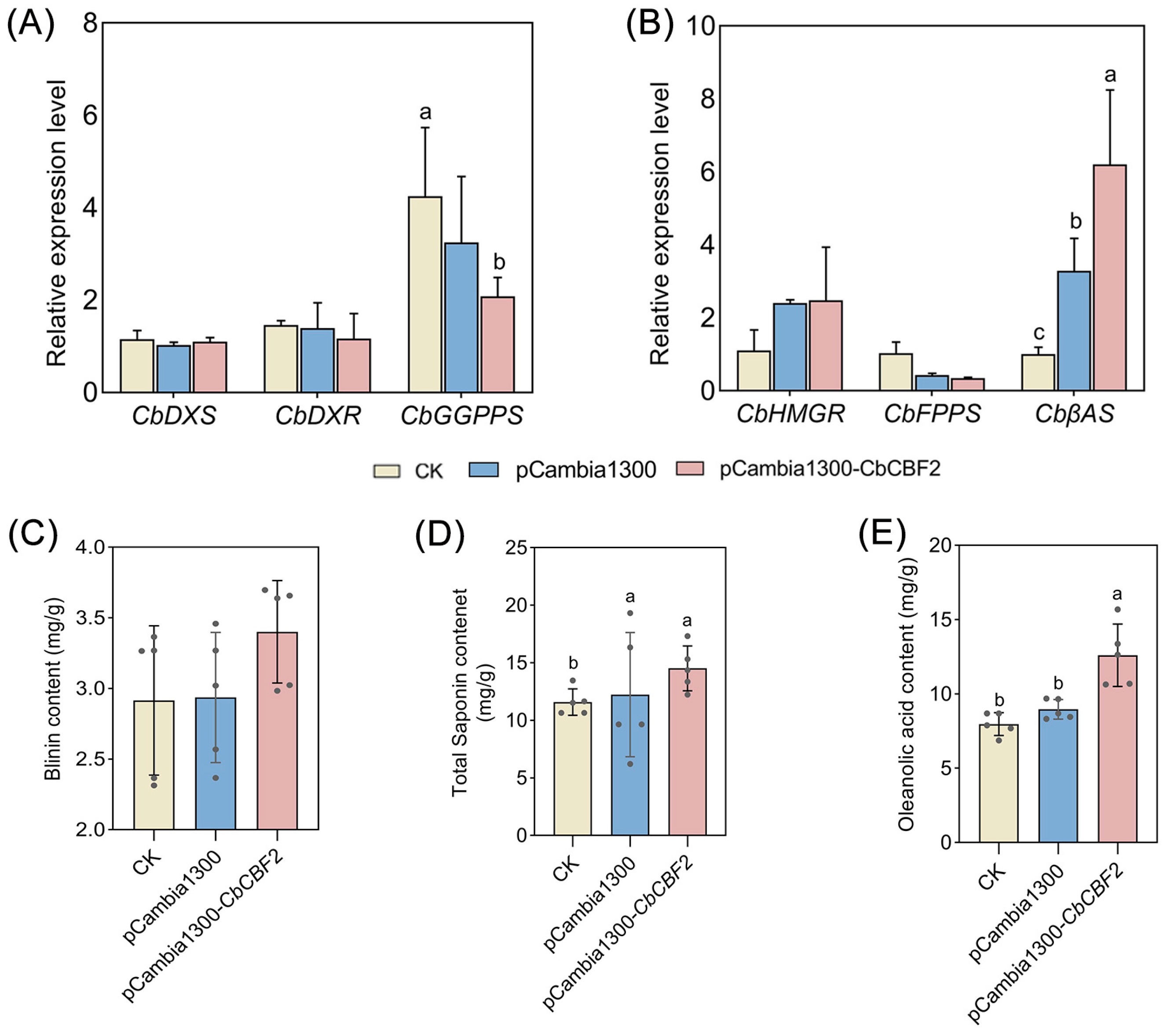
3.4. Overexpression of CbCBF2 Enhances the Activity of MVA Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Theodoridis, S.; Drakou, E.G.; Hickler, T.; Thines, M.; Nogues-Bravo, D. Evaluating natural medicinal resources and their exposure to global change. Lancet Planet. Health 2023, 7, e155–e163. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Fu, X.; Shao, J.; Tang, Y.; Yu, M.; Li, L.; Huang, L.; Tang, K. Transcriptional regulatory network of high-value active ingredients in medicinal plants. Trends Plant Sci. 2023, 28, 429–446. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liu, H.; Qin, P.; Hu, C.; Man, S.; Li, Y.; Liu, Z.; Liu, Z.; Diao, A. Saponin fraction isolated from Conyza blinii H.Lév. demonstrates strong anti-cancer activity that is due to its NF-κB inhibition. Biochem. Biophys. Res. Commun. 2017, 483, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liu, J. The protective activity of Conyza blinii saponin against acute gastric ulcer induced by ethanol. J. Ethnopharmacol. 2014, 158, 358–363. [Google Scholar] [CrossRef]
- Liu, H.; Hu, C.; Sun, N.; Li, Y.; Man, S.; Liu, Z.; Diao, A.; Ma, L. A triterpenoidal saponin fraction of Conyza blinii H.Lév. is a dual-targeting autophagy inhibitor for HeLa cells. RSC Adv. 2017, 7, 24291–24297. [Google Scholar] [CrossRef]
- Yang, M.; Zhou, M.; Shu, M.; Han, Z.; Ma, R.; Chen, Y.; Zheng, T.; Chen, H. The Blinin Accumulation Promoted by CbMYB32 Involved in Conyza blinii Resistance to Nocturnal Low Temperature. Int. J. Mol. Sci. 2023, 24, 7143. [Google Scholar] [CrossRef]
- Yang, M.; Wang, M.; Zhou, M.; Zhang, Y.; Yu, K.; Wang, T.; Bu, T.; Tang, Z.; Zheng, T.; Chen, H. ABA and SA Participate in the Regulation of Terpenoid Metabolic Flux Induced by Low-Temperature within Conyza blinii. Life 2023, 13, 371. [Google Scholar] [CrossRef]
- Zhan, J.; Di, T.; Chen, X.; Zheng, T.; Sun, W.; Yang, M.; Zhou, M.; Shen, Z.; Chen, H.; Su, N. CbMYB108 integrates the regulation of diterpene biosynthesis and trichome development in Conyza blinii against UV-B. Plant Cell Environ. 2024, 47, 1300–1318. [Google Scholar] [CrossRef]
- Zheng, T.; Wang, M.; Zhan, J.; Sun, W.; Yang, Q.; Lin, Z.; Bu, T.; Tang, Z.; Li, C.; Yan, J.; et al. Ferrous iron-induced increases in capitate glandular trichome density and upregulation of CbHO-1 contributes to increases in blinin content in Conyza blinii. Planta 2020, 252, 81. [Google Scholar] [CrossRef]
- Kazemi-Shahandashti, S.S.; Maali-Amiri, R. Global insights of protein responses to cold stress in plants: Signaling, defence, and degradation. J. Plant Physiol. 2018, 226, 123–135. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Jia, Y.; Shi, Y.; Zhang, X.; Song, C.; Gong, Z.; Yang, S. OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 2018, 37, e98228. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef]
- Liu, J.; Shi, Y.; Yang, S. Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 2018, 60, 780–795. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef]
- Li, Q.; Xu, F.; Chen, Z.; Teng, Z.; Sun, K.; Li, X.; Yu, J.; Zhang, G.; Liang, Y.; Huang, X.; et al. Synergistic interplay of ABA and BR signal in regulating plant growth and adaptation. Nat. Plants 2021, 7, 1108–1118. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Dong, X.; Yan, Y.; Jiang, B.; Shi, Y.; Jia, Y.; Cheng, J.; Shi, Y.; Kang, J.; Li, H.; Zhang, D.; et al. The cold response regulator CBF1 promotes Arabidopsis hypocotyl growth at ambient temperatures. EMBO. J. 2020, 39, e103630. [Google Scholar] [CrossRef]
- Guo, Y.; Xiong, L.; Ishitani, M.; Zhu, J.K. An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc. Natl. Acad. Sci. USA 2002, 99, 7786–7791. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.J.; Parkin, I.A. Differential SAGE analysis in Arabidopsis uncovers increased transcriptome complexity in response to low temperature. BMC Genom. 2008, 9, 434. [Google Scholar] [CrossRef]
- Tang, K.; Zhao, L.; Ren, Y.; Yang, S.; Zhu, J.K.; Zhao, C. The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 2020, 62, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, D.; Lu, S.X.; Hu, X.; Huang, R.; Liang, T.; Xu, T.; Tobin, E.M.; Liu, H. Blue Light- and Low Temperature-Regulated COR27 and COR28 Play Roles in the Arabidopsis Circadian Clock. Plant Cell 2016, 28, 2755–2769. [Google Scholar] [CrossRef]
- Wang, P.; Cui, X.; Zhao, C.; Shi, L.; Zhang, G.; Sun, F.; Cao, X.; Yuan, L.; Xie, Q.; Xu, X. COR27 and COR28 encode nighttime repressors integrating Arabidopsis circadian clock and cold response. J. Integr. Plant Biol. 2017, 59, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, X.; Li, M.; Yang, H.; Fu, D.; Lv, J.; Ding, Y.; Gong, Z.; Shi, Y.; Yang, S. The direct targets of CBFs: In cold stress response and beyond. J. Integr. Plant Biol. 2021, 63, 1874–1887. [Google Scholar] [CrossRef]
- Henry, L.K.; Thomas, S.T.; Widhalm, J.R.; Lynch, J.H.; Davis, T.C.; Kessler, S.A.; Bohlmann, J.; Noel, J.P.; Dudareva, N. Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Plants 2018, 4, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Bergman, M.E.; Kortbeek, R.W.J.; Gutensohn, M.; Dudareva, N. Plant terpenoid biosynthetic network and its multiple layers of regulation. Prog. Lipid. Res. 2024, 95, 101287. [Google Scholar] [CrossRef]
- Okada, K. The biosynthesis of isoprenoids and the mechanisms regulating it in plants. Biosci. Biotechnol. Biochem. 2011, 75, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Nadeem, M.; Galagedara, L.; Thomas, R.; Cheema, M. Recent insights into cell responses to cold stress in plants: Signaling, defence, and potential functions of phosphatidic acid. Environ. Exp. Bot. 2022, 203, 105068. [Google Scholar] [CrossRef]
- Castellano, J.M.; Ramos-Romero, S.; Perona, J.S. Oleanolic Acid: Extraction, Characterization and Biological Activity. Nutrients 2022, 14, 623. [Google Scholar] [CrossRef]
- Verma, N.; Raghuvanshi, D.S.; Singh, R.V. Recent advances in the chemistry and biology of oleanolic acid and its derivatives. Eur. J. Med. Chem. 2024, 276, 116619. [Google Scholar] [CrossRef] [PubMed]
- Apel, W.; Schulze, W.X.; Bock, R. Identification of protein stability determinants in chloroplasts. Plant J. 2010, 63, 636–650. [Google Scholar] [CrossRef]
- Duan, S.; Zeng, Y.; Wang, H.-B.; Jin, H.-L. Coordination of genome stability: Novel communication pathways between chloroplasts and other compartments in plant cells. Fundam. Res. 2024. [Google Scholar] [CrossRef]
- Finka, A.; Cuendet, A.F.; Maathuis, F.J.; Saidi, Y.; Goloubinoff, P. Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 2012, 24, 3333–3348. [Google Scholar] [CrossRef]
- He, J.; Yao, L.; Pecoraro, L.; Liu, C.; Wang, J.; Huang, L.; Gao, W. Cold stress regulates accumulation of flavonoids and terpenoids in plants by phytohormone, transcription process, functional enzyme, and epigenetics. Crit. Rev. Biotechnol. 2023, 43, 680–697. [Google Scholar] [CrossRef]
- Bastow, R.; Mylne, J.S.; Lister, C.; Lippman, Z.; Martienssen, R.A.; Dean, C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 2004, 427, 164–167. [Google Scholar] [CrossRef]
- Sung, S.; Amasino, R.M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 2004, 427, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Sun, W.J.; Zhan, J.Y.; Zheng, T.R.; Sun, R.; Wang, T.; Tang, Z.Z.; Bu, T.L.; Li, C.L.; Wu, Q.; Chen, H. The jasmonate-responsive transcription factor CbWRKY24 regulates terpenoid biosynthetic genes to promote saponin biosynthesis in Conyza blinii H. Lév. J. Genet. 2018, 97, 1379–1388. [Google Scholar] [CrossRef]
- Zheng, T.; Zhan, J.; Wang, M.; Sun, W.; Yan, J.; Shan, Z.; Chen, H. Fe induces a dynamic and biased allocation of material flux within terpenoid metabolism controlled by CbNudix in Conyza blinii. Plant Soil 2021, 467, 421–436. [Google Scholar] [CrossRef]
- Haake, V.; Cook, D.; Riechmann, J.L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef]
- Ye, K.; Li, H.; Ding, Y.; Shi, Y.; Song, C.; Gong, Z.; Yang, S. BRASSINOSTEROID-INSENSITIVE2 Negatively Regulates the Stability of Transcription Factor ICE1 in Response to Cold Stress in Arabidopsis. Plant Cell 2019, 31, 2682–2696. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. MAP Kinase Cascades Regulate the Cold Response by Modulating ICE1 Protein Stability. Dev. Cell 2017, 43, 618–629.e615. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Zhang, G.; Deng, J.; Zheng, T.; Liu, M. CbCBF2 Integrates JA and BR Signaling to Enhance Oleanolic Acid Biosynthesis in Conyza blinii H. Lév Under Cold Stress. Agronomy 2025, 15, 1001. https://doi.org/10.3390/agronomy15051001
Yang M, Zhang G, Deng J, Zheng T, Liu M. CbCBF2 Integrates JA and BR Signaling to Enhance Oleanolic Acid Biosynthesis in Conyza blinii H. Lév Under Cold Stress. Agronomy. 2025; 15(5):1001. https://doi.org/10.3390/agronomy15051001
Chicago/Turabian StyleYang, Ming, Guodong Zhang, Junjie Deng, Tianrun Zheng, and Moyang Liu. 2025. "CbCBF2 Integrates JA and BR Signaling to Enhance Oleanolic Acid Biosynthesis in Conyza blinii H. Lév Under Cold Stress" Agronomy 15, no. 5: 1001. https://doi.org/10.3390/agronomy15051001
APA StyleYang, M., Zhang, G., Deng, J., Zheng, T., & Liu, M. (2025). CbCBF2 Integrates JA and BR Signaling to Enhance Oleanolic Acid Biosynthesis in Conyza blinii H. Lév Under Cold Stress. Agronomy, 15(5), 1001. https://doi.org/10.3390/agronomy15051001






