Genetic Dissection of the Powdery Mildew Resistance in a Cultivated Emmer Wheat Accession
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Pathogens
2.2. Phenotypic Identification of Bgt Isolates
2.3. Genetic Analysis
2.4. RNA Extraction and Sequencing Library Setup for BSR-Seq
2.5. Sequencing and Bioinformatics Analysis
2.6. Candidate Interval Analysis and Gene Mapping
2.7. Differential Expression Gene Analysis, Functional Annotation, and Enrichment Analysis of DEGs
2.8. Microscopic Observation of Powdery Mildew Resistance
2.9. RNA Extraction and qRT-PCR
2.10. Assessment of Linked Markers for MAS
3. Results
3.1. Evaluation and Genetic Analysis of Powdery Mildew Resistance in L709
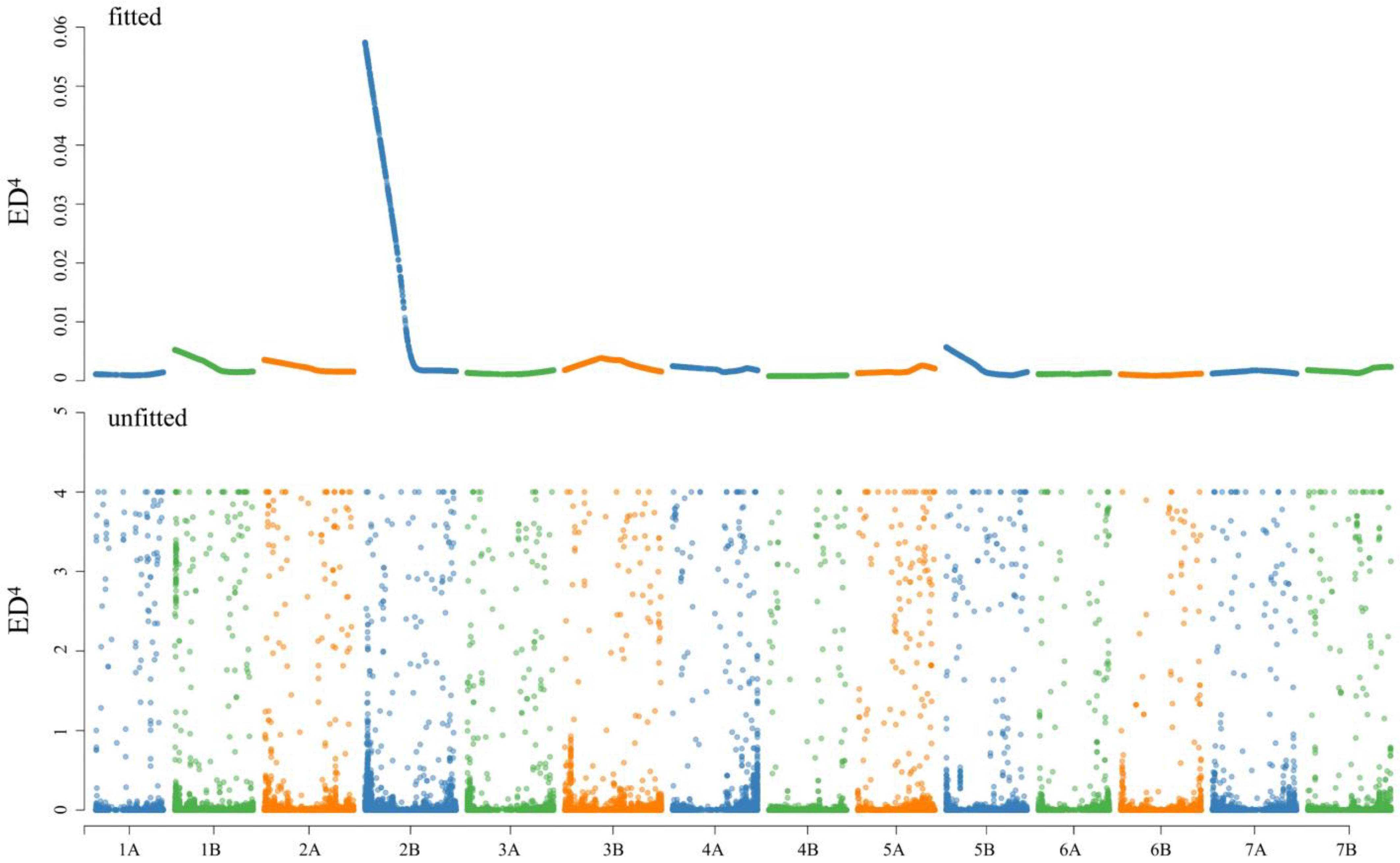
3.2. SNP Calling and Confirmation of Candidate Interval
3.3. Molecular Mapping of PmL709 and Prediction of Candidate Genes
3.4. Comparisons of PmL709 and Known Pm Genes on Chromosome Arm 2BS
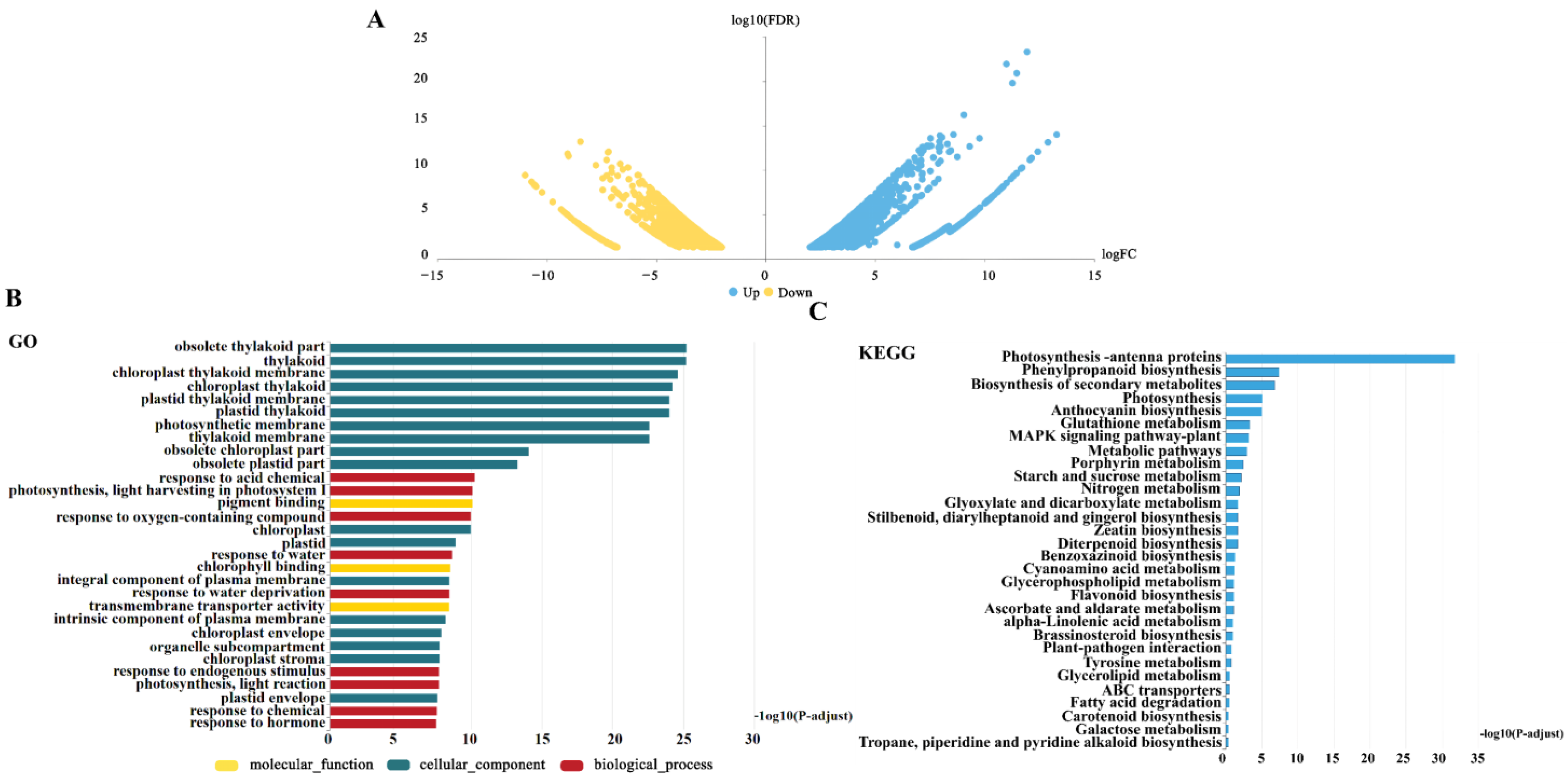
3.5. Discovery and Analysis of DEGs
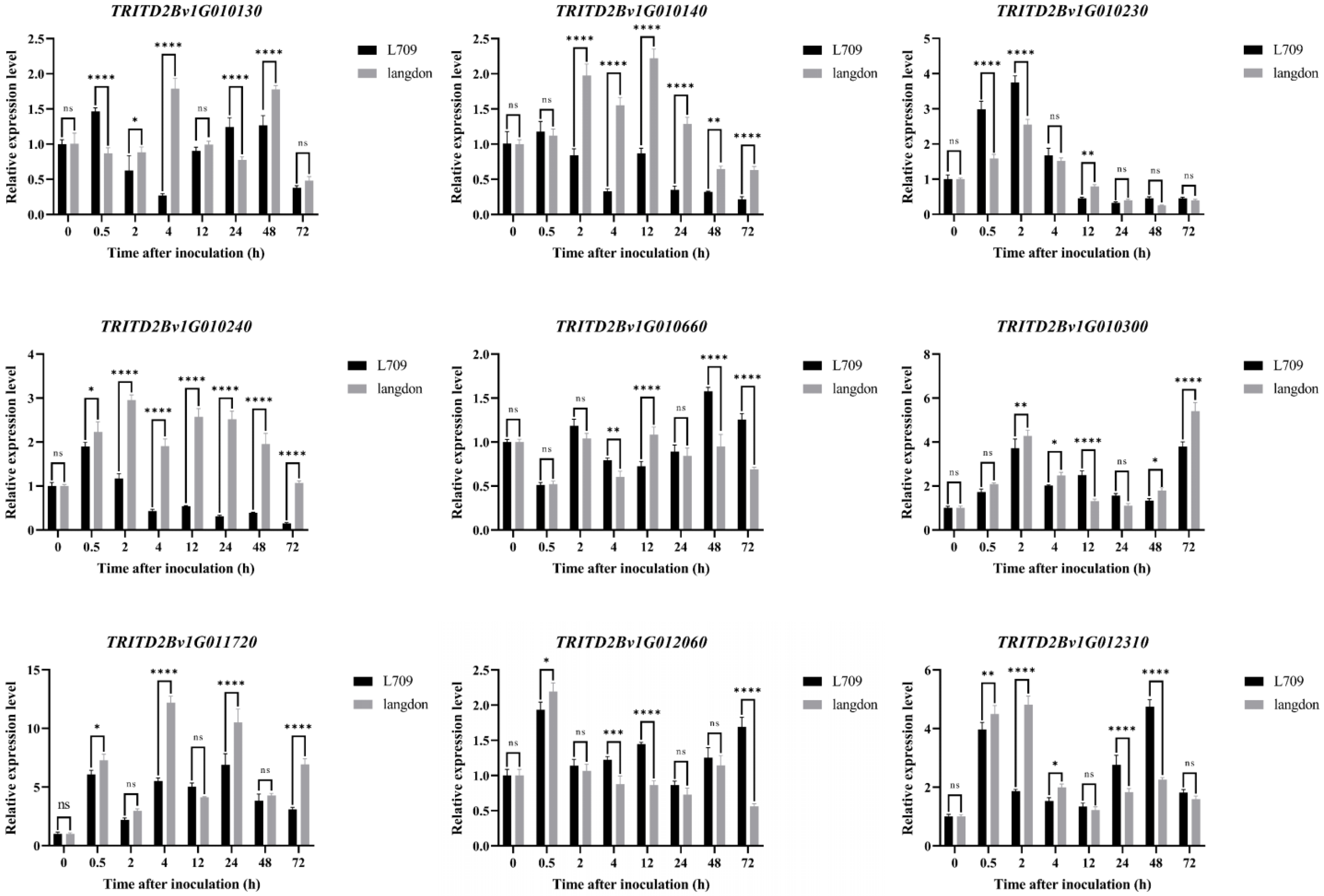
3.6. Validation of Disease Resistance-Related Genes in Cultivated Emmer Wheat L709 via qRT-PCR
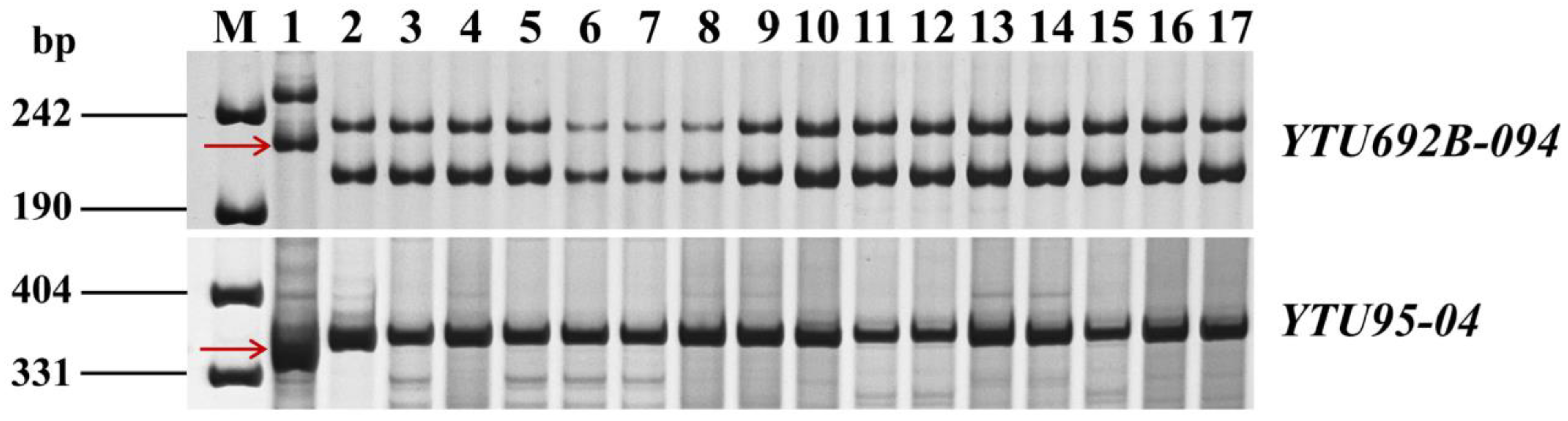
3.7. Molecular Markers for MAS
4. Discussion
5. Conclusions
- (1)
- Cultivated emmer wheat accession L709 is resistant to 27 Bgt isolates and has extremely important breeding value.
- (2)
- The resistance to powdery mildew was conferred by the broad-spectrum resistance gene PmL709 in accession L709. PmL709 was delimited to a 1.7 cM genetic interval on chromosome arm 2BS, corresponding to a 21.82–25.94 Mb physical interval (svevo.v1).
- (3)
- DEGs, including TRITD2Bv1G010130, TRITD2Bv1G010230, TRITD2Bv1G010660, TRITD2Bv1G010300, TRITD2Bv1G012060, TRITD2Bv1G010140, TRITD2Bv1G010240, TRITD2Bv1G011720, and TRITD2Bv1G012310, located in the candidate interval, were induced to be expressed by Bgt invasion and could be candidates during the process of disease resistance response in L709.
- (4)
- The seven flanking markers of PmL709 (including Xdw05, YTU95-04, YTU95-06, YTU95-08, Xdw10, Xdw11, and YTU692B-094) can be used in MAS to track the PmL709 gene when transferring it to susceptible cultivars/lines.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Godfray, H.C.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Sotiropoulos, A.G.; Arango-Isaza, E.; Ban, T.; Barbieri, C.; Bourras, S.; Cowger, C.; Czembor, P.C.; Ben-David, R.; Dinoor, A.; Ellwood, S.R.; et al. Global genomic analyses of wheat powdery mildew reveal association of pathogen spread with historical human migration and trade. Nat. Commun. 2022, 13, 4315. [Google Scholar] [CrossRef]
- Parlange, F.; Roffler, S.; Menardo, F.; Ben-David, R.; Bourras, S.; McNally, K.E.; Oberhaensli, S.; Stirnweis, D.; Buchmann, G.; Wicker, T.; et al. Genetic and molecular characterization of a locus involved in avirulence of Blumeria graminis f. sp. tritici on wheat Pm3 resistance alleles. Fungal. Genet. Biol. 2015, 82, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Mapuranga, J.; Chang, J.; Yang, W. Combating powdery mildew: Advances in molecular interactions between Blumeria graminis f. sp. tritici and wheat. Front. Plant Sci. 2022, 13, 1102908. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Bai, Y.; Wu, G.H.; Zou, S.H.; Chen, Y.F.; Gao, C.X.; Tang, D.Z. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef]
- Wang, B.; Meng, T.; Xiao, B.; Yu, T.Y.; Yue, T.Y.; Jin, Y.L.; Ma, P.T. Fighting wheat powdery mildew: From genes to fields. Theor. Appl. Genet. 2023, 136, 196. [Google Scholar] [CrossRef]
- Zhang, J.D.; Yang, H.; Han, G.H.; Xu, H.X.; Liu, R.S.; Yu, N.N.; Han, R.; Li, Y.X.; Li, J.T.; Dai, Y.T.; et al. Fine mapping of Pm71, a novel powdery mildew resistance gene from emmer wheat. Crop J. 2025, 13, 62–68. [Google Scholar] [CrossRef]
- Li, Y.H.; Wei, Z.Z.; Sela, H.; Govta, L.; Klymiuk, V.; Roychowdhury, R.; Chawla, H.S.; Ens, J.; Wiebe, K.; Bocharova, V.; et al. Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69. Plant Commun. 2024, 5, 100646. [Google Scholar] [CrossRef]
- Wu, X.X.; Bian, Q.; Gao, Y.; Ni, X.Y.; Sun, Y.Q.; Xuan, Y.H.; Cao, Y.Y.; Li, T.Y. Evaluation of resistance to powdery mildew and identification of resistance genes in wheat cultivars. Peer J. 2021, 9, e10425. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Srichumpa, P.; Dudler, R.; Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 2004, 37, 528–538. [Google Scholar] [CrossRef]
- Sánchez-Martín, J.; Keller, B. NLR immune receptors and diverse types of non-NLR proteins control race-specific resistance in Triticeae. Curr. Opin. Plant Biol. 2021, 62, 102053. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Guo, L.; Wang, Z.Z.; Li, B.B.; Li, J.; Li, Y.H.; Qiu, D.; Shi, W.Q.; Yang, L.J.; Wang, N.; et al. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 2020, 11, 680. [Google Scholar] [CrossRef]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.W.; Herrera-Foessel, S.; Lan, C.; Schnippenkoetter, W.; Ayliffe, M.; Huerta-Espino, J.; Lillemo, M.; Viccars, L.; Milne, R.; Periyannan, S.; et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 2015, 47, 1494–1498. [Google Scholar] [CrossRef]
- Jones, J.; Dangl, J. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Zhang, X.; Dodds, P.N.; Bernoux, M. What do we know about NOD-like receptors in plant immunity? Annu. Rev. Phytopathol. 2017, 55, 205–229. [Google Scholar] [CrossRef]
- Wang, G.F.; Ji, J.; El-Kasmi, F.; Dangl, J.L.; Johal, G.; Balint-Kurti, P.J. Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog. 2015, 11, e1004674. [Google Scholar]
- Jin, Y.; Liu, H.; Gu, T.; Xing, L.; Han, G.; Ma, P.; Li, X.; Zhou, Y.; Fan, J.; Li, L.; et al. Pm2b, a CC-NBS-LRR protein, interacts with TaWRKY76-D to regulate powdery mildew resistance in common wheat. Front. Plant Sci. 2022, 13, 973065. [Google Scholar] [CrossRef]
- Kunz, L.; Sotiropoulos, A.G.; Graf, J.; Razavi, M.; Keller, B.; Müller, M.C. The broad use of the Pm8 resistance gene in wheat resulted in hypermutation of the AvrPm8 gene in the powdery mildew pathogen. BMC Biol. 2023, 21, 29. [Google Scholar] [CrossRef]
- Zhu, S.Y.; Liu, C.; Gong, S.J.; Chen, Z.Z.; Chen, R.; Liu, T.L.; Liu, R.K.; Du, H.A.; Guo, R.; Li, G.Y.; et al. Orthologous genes Pm12 and Pm21 from two wild relatives of wheat show evolutionary conservation but divergent powdery mildew resistance. Plant Commun. 2023, 4, 100472. [Google Scholar] [CrossRef] [PubMed]
- He, H.G.; Zhu, S.Y.; Zhao, R.H.; Jiang, Z.N.; Ji, Y.Y.; Ji, J.; Qiu, D.; Li, H.J.; Bie, T.D. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-Spectrum resistance to wheat powdery mildew disease. Mol. Plant 2018, 11, 879–882. [Google Scholar] [CrossRef]
- Shi, X.H.; Wu, P.P.; Hu, J.H.; Qiu, D.; Qu, Y.; Li, Y.H.; Liu, Y.; Gebremariam, T.G.; Xie, J.Z.; Wu, Q.H.; et al. Molecular characterization of all-stage and adult-plant resistance loci against powdery mildew in winter wheat cultivar Liangxing 99 using BSR-Seq technology. Plant Dis. 2021, 105, 3443–3450. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.T.; Wu, L.R.; Xu, Y.F.; Xu, H.X.; Zhang, X.; Wang, W.R.; Liu, C.; Wang, B. Bulked segregant RNA-seq provides distinctive expression profile against powdery mildew in the wheat genotype YD588. Front. Plant Sci. 2021, 12, 764978. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.; Vazquez-Gross, H.; Herin, S.Y.; Hane, D.; Wang, Y.; Gu, Y.Q.; Dubcovsky, J. WheatExp: An RNA-seq expression database for polyploid wheat. BMC Plant Biol. 2015, 15, 299. [Google Scholar] [CrossRef]
- Pankievic, V.C.S.; Camilios-Neto, D.; Bonato, P.; Balsanelli, E.; Tadra-Sfeir, M.Z.; Faoro, H.; Chubatsu, L.S.; Donatti, L.; Wajnberg, G.; Passetti, F.; et al. RNA-Seq transcriptional profiling of herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant Mol. Biol. 2016, 90, 589–603. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, J.Z.; Zhang, H.Z.; Guo, B.M.; Ning, S.Z.; Chen, Y.X.; Lu, P.; Wu, Q.H.; Wu, M.M.; Zhang, D.Y.; et al. Mapping stripe rust resistance gene YrZH22 in Chinese wheat cultivar Zhoumai 22 by bulked segregant RNA-Seq (BSR-Seq) and comparative genomics analyses. Theor. Appl. Genet. 2017, 130, 2191–2201. [Google Scholar] [CrossRef]
- Hao, Z.M.; Geng, M.M.; Hao, Y.R.; Zhang, Y.; Zhang, L.J.; Wen, S.M.; Wang, R.H.; Liu, G.R. Screening for differential expression of genes for resistance to Sitodiplosis mosellana in bread wheat via BSR-Seq analysis. Theor. Appl. Genet. 2019, 132, 3201–3221. [Google Scholar] [CrossRef]
- Chen, G.F.; Zhang, H.; Deng, Z.Y.; Wu, R.G.; Li, D.M.; Wang, M.Y.; Tian, J.C. Genome-wide association study for kernel weight-related traits using SNPs in a Chinese winter wheat population. Euphytica 2016, 212, 173–185. [Google Scholar] [CrossRef]
- Li, Y.H.; Shi, X.H.; Hu, J.H.; Wu, P.P.; Qiu, D.; Qu, Y.F.; Xie, J.Z.; Wu, Q.H.; Zhang, H.J.; Li, Y.; et al. Identification of a recessive gene PmQ conferring resistance to powdery mildew in wheat landrace Qingxinmai using BSR-Seq analysis. Plant Dis. 2020, 104, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.R.; Zhu, T.; He, H.G.; Cao, X.Y.; Li, H.S.; Xu, H.X.; Jia, M.S.; Zhang, L.P.; Song, J.C.; Mirzaghaderi, G.; et al. Genetic dissection of the powdery mildew resistance in wheat breeding line LS5082 using BSR-Seq. Crop J. 2022, 10, 1120–1130. [Google Scholar] [CrossRef]
- Dai, Y.T.; Yu, N.N.; Xu, H.X.; Liu, S.Q.; Zhang, J.D.; Liu, R.S.; Li, J.T.; Li, Y.X.; Xiao, B.; Pan, G.T.; et al. A unique expression profile responding to powdery mildew in wild emmer wheat D430. Int. J. Mol. Sci. 2024, 26, 242. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wu, L.R.; He, H.G.; Song, J.C.; Jia, M.S.; Liu, L.C.; Wang, X.L.; Han, R.; Niu, L.P.; Du, W.X.; et al. Bulked segregant RNA-Seq reveals distinct expression profiling in Chinese wheat cultivar Jimai 23 responding to powdery mildew. Front. Genet. 2020, 11, 474. [Google Scholar] [CrossRef]
- Rahman, S.; Islam, S.; Yu, Z.T.; She, M.Y.; Nevo, E.; Ma, W.J. Current progress in understanding and recovering the wheat genes lost in evolution and domestication. Int. J. Mol. Sci. 2020, 21, 5836. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Amosova, A.V.; Goncharov, N.P.; Macas, J.; Ruban, A.S.; Grechishnikova, I.V.; Zoshchuk, S.A.; Houben, A. A set of cytogenetic markers allows the precise identification of all A-genome chromosomes in diploid and polyploid wheat. Cytogenet. Genome Res. 2015, 146, 71–79. [Google Scholar] [CrossRef]
- Sharma, J.S.; Running, K.L.D.; Xu, S.S.; Zhang, Q.; Peters Haugrud, A.R.; Sharma, S.; McClean, P.E.; Faris, J.D. Genetic analysis of threshability and other spike traits in the evolution of cultivated emmer to fully domesticated durum wheat. Mol. Genet. Genom. 2019, 294, 757–771. [Google Scholar] [CrossRef]
- Zaharieva, M.; Ayana, N.G.; Hakimi, A.A.; Misra, S.C.; Monneveux, P. Cultivated emmer wheat (Triticum dicoccon Schrank), an old crop with promising future: A review. Genet. Resour. Crop Evol. 2010, 57, 937–962. [Google Scholar] [CrossRef]
- Rong, J.K.; Millet, E.; ManisterskiI, J. A new powdery mildew resistance gene: Introgression from wild emmer into common wheat and RFLP-based mapping. Euphytica 2000, 115, 121–126. [Google Scholar] [CrossRef]
- Qian, Z.J.; Han, G.H.; Yu, N.N.; Liu, C.; Han, R.; Jameson, P.E.; Wang, J.J.; Zhao, Y.; Xiao, B.; Liu, R.S.; et al. Fine mapping of the powdery mildew resistance gene PmXQ-0508 in bread wheat. Crop J. 2024, 12, 1176–1184. [Google Scholar] [CrossRef]
- Hua, W.; Liu, Z.J.; Zhu, J.; Xie, C.J.; Yang, T.; Zhou, Y.L.; Duan, X.Y.; Sun, Q.X.; Liu, Z.Y. Identification and genetic mapping of pm42, a new recessive wheat powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2009, 119, 223–230. [Google Scholar] [CrossRef] [PubMed]
- He, H.G.; Liu, R.K.; Ma, P.T.; Du, H.N.; Zhang, H.H.; Wu, Q.H.; Yang, L.J.; Gong, S.J.; Liu, T.L.; Huo, N.X.; et al. Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 2021, 134, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, D.Y.; Ouyang, S.H.; Xie, J.Z.; Wu, Q.H.; Wang, Z.Z.; Cui, Y.; Lu, P.; Zhang, D.; Liu, Z.J.; et al. Dynamic evolution of resistance gene analogs in the orthologous genomic regions of powdery mildew resistance gene MlIW170 in Triticum dicoccoides and Aegilops tauschii. Theor. Appl. Genet. 2015, 128, 1617–1629. [Google Scholar] [CrossRef]
- Ma, P.T.; Xu, H.X.; Han, G.H.; Luo, Q.L.; Xu, Y.F.; Zhang, X.T.; An, D.G.; Li, L.H.; Sun, Y. Characterization of a segregation distortion locus with powdery mildew resistance in a wheat-Thinopyrum intermedium introgression line WE99. Plant Dis. 2016, 100, 1541–1547. [Google Scholar] [CrossRef]
- Cowger, C.; Parks, R.; Kosman, E. Structure and migration in US Blumeria graminis f. sp tritici populations. Phytopathology 2016, 106, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Basandrai, A.K.; Mehta, A.; Basandrai, D. Virulence structure of wheat powdery mildew pathogen, Blumeria graminis tritici: A review. Indian Phytopathol. 2023, 76, 21–45. [Google Scholar] [CrossRef]
- An, D.G.; Zheng, Q.; Zhou, Y.L.; Ma, P.T.; Lv, Z.L.; Li, L.H.; Li, B.; Luo, Q.L.; Xu, H.X.; Xu, Y.F. Molecular cytogenetic characterization of a new wheat-rye 4R chromosome translocation line resistant to powdery mildew. Chromosome Res. 2013, 21, 419–432. [Google Scholar] [CrossRef]
- Vendramin, V.; Ormanbekova, D.; Scalabrin, S.; Scaglione, D.; Maccaferri, M.; Martelli, P.; Salvi, S.; Jurman, I.; Casadio, R.; Cattonaro, F.; et al. Genomic tools for durum wheat breeding: De novo assembly of Svevo transcriptome and SNP discovery in elite germplasm. BMC Genom. 2019, 20, 278. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- McCormick, R.F.; Truong, S.K.; Mullet, J.E. RIG: Recalibration and interrelation of genomic sequence data with the GATK. G3 2015, 5, 655–665. [Google Scholar] [CrossRef]
- Dong, C.H.; Zhang, L.C.; Chen, Z.X.; Xia, C.; Gu, Y.Q.; Wang, J.R.; Li, D.P.; Xie, Z.C.; Zhang, Q.; Zhang, X.Y.; et al. Combining a new exome capture panel with an effective varBScore algorithm accelerates BSA-Based gene cloning in wheat. Front. Plant Sci. 2020, 13, 1249. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Gorsi, B.; Su, Y.C.; Yost, H.J. MMAPPR: Mutation mapping analysis pipeline for pooled RNA seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, S.E.; Daly, M.J.; Lander, E.S.; Lincoln, S.; Daly, M.; Lander, E. Constructing Genetic Maps with Mapmaker/EXP3.0; Technical Report; Whitehead Institute: Cambridge, MA, USA, 1992. [Google Scholar]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.L.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W.Z. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Bourras, S.; McNally, K.E.; Ben-David, R.; Parlange, F.; Roffler, S.; Praz, C.R.; Oberhaensli, S.; Menardo, F.; Stirnweis, D.; Frenkel, Z.; et al. Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 2015, 27, 2991–3012. [Google Scholar]
- Wang, Y.; Huang, L.; Luo, W.; Jin, Y.; Gong, F.; He, J.; Liu, D.; Zheng, Y.; Wu, B. Transcriptome analysis provides insights into the mechanisms underlying wheat cultivar Shumai 126 responding to stripe rust. Gene 2021, 768, 145290. [Google Scholar] [CrossRef]
- Jin, Y.; Han, G.H.; Zhang, W.J.; Bu, B.; Zhao, Y.; Wang, J.J.; Liu, R.S.; Yang, H.; Xu, H.X.; Ma, P.T. Evaluation and genetic dissection of the powdery mildew resistance in 558 wheat accessions. New Crops 2024, 1, 100018. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef]
- Wei, X.B.; Liu, X.Y.; Zhang, X.; Guo, S.Y.; Shi, J.Q. Structural insights into ligand recognition and receptor activation of plant leucine-rich repeat (LRR) transmembrane receptors. New Crops 2024, 1, 100022. [Google Scholar] [CrossRef]
- An, Y.Y.; Zhang, M.X. Advances in understanding the plant-ralstonia solanacearum interactions: Unraveling the dynamics, mechanisms, and implications for crop disease resistance. New Crops 2024, 1, 100014. [Google Scholar] [CrossRef]
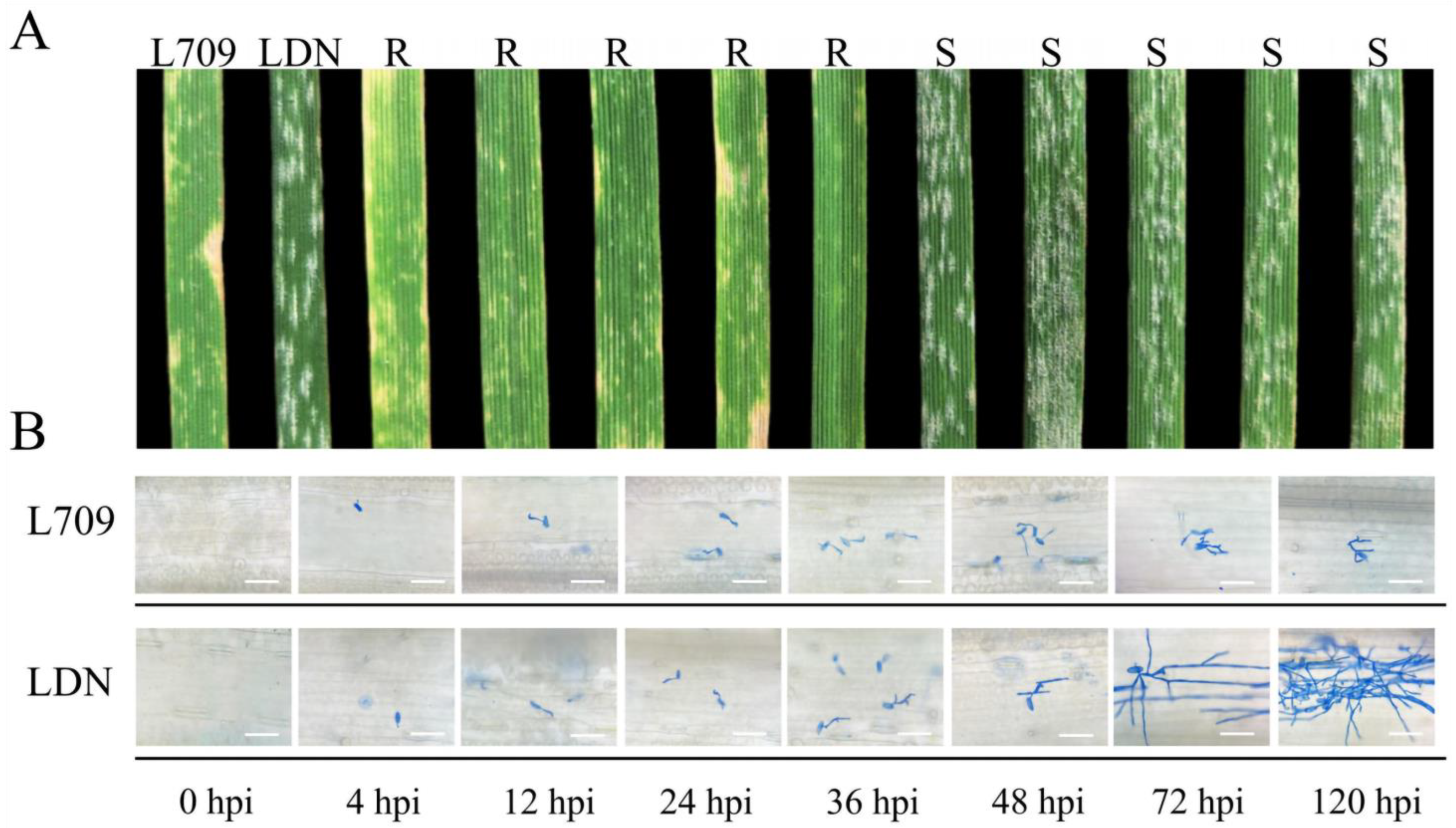

| Parents and Cross | Generation | Observed Ratio | Expected Ratio | χ2 | p |
|---|---|---|---|---|---|
| L709 | RP | R:S = 10:0 | |||
| LDN | SP | R:S = 0:10 | |||
| L709 × LDN F1 | F1 | R:S = 10: 0 | |||
| L709 × LDN F2 | F2 | R:S = 183:63 | 3:1 | 0.05 | 0.83 |
| L709 × LDN F2:3 | F2:3 | HR:Seg:HS = 60:123:63 | 1:2:1 | 0.07 | 0.96 |
| No. | Genes | Physical Genomic Location | Functional Annotation |
|---|---|---|---|
| 1 | TRITD2Bv1G010130 | chr2B:21983992..21987186 | Disease resistance protein |
| 2 | TRITD2Bv1G010140 | chr2B:21994145..21997744 | Disease resistance protein (NBS-LRR class) family |
| 3 | TRITD2Bv1G010230 | chr2B:22398122..22400896 | NBS-LRR-like resistance protein |
| 4 | TRITD2Bv1G010240 | chr2B:22456742..22460374 | NBS-LRR-like resistance protein |
| 5 | TRITD2Bv1G010300 | chr2B:22629909..22631735 | Disease resistance protein (TIR-NBS-LRR class) |
| 6 | TRITD2Bv1G010660 | chr2B:23210823..23214726 | Receptor protein kinase |
| 7 | TRITD2Bv1G011720 | chr2B:24749117..24754598 | Disease resistance protein (TIR-NBS-LRR class) |
| 8 | TRITD2Bv1G012060 | chr2B:25098753..25102541 | Receptor-like protein kinase |
| 9 | TRITD2Bv1G012310 | chr2B:25474820..25483319 | NBS-LRR class disease resistance protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Jin, Y.; Yu, N.; Xu, H.; Sun, X.; Wang, J.; Liu, X.; Zhang, J.; Li, J.; Li, Y.; et al. Genetic Dissection of the Powdery Mildew Resistance in a Cultivated Emmer Wheat Accession. Agronomy 2025, 15, 980. https://doi.org/10.3390/agronomy15040980
Liu R, Jin Y, Yu N, Xu H, Sun X, Wang J, Liu X, Zhang J, Li J, Li Y, et al. Genetic Dissection of the Powdery Mildew Resistance in a Cultivated Emmer Wheat Accession. Agronomy. 2025; 15(4):980. https://doi.org/10.3390/agronomy15040980
Chicago/Turabian StyleLiu, Ruishan, Yuli Jin, Ningning Yu, Hongxing Xu, Xusheng Sun, Jiangchun Wang, Xueqing Liu, Jiadong Zhang, Jiatong Li, Yaoxue Li, and et al. 2025. "Genetic Dissection of the Powdery Mildew Resistance in a Cultivated Emmer Wheat Accession" Agronomy 15, no. 4: 980. https://doi.org/10.3390/agronomy15040980
APA StyleLiu, R., Jin, Y., Yu, N., Xu, H., Sun, X., Wang, J., Liu, X., Zhang, J., Li, J., Li, Y., & Ma, P. (2025). Genetic Dissection of the Powdery Mildew Resistance in a Cultivated Emmer Wheat Accession. Agronomy, 15(4), 980. https://doi.org/10.3390/agronomy15040980






