Identification and Expression Analysis of Wheat Golden2-like (TaGLK) Gene in Response to Biotic and Abiotic Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the GLK Gene Family in Wheat
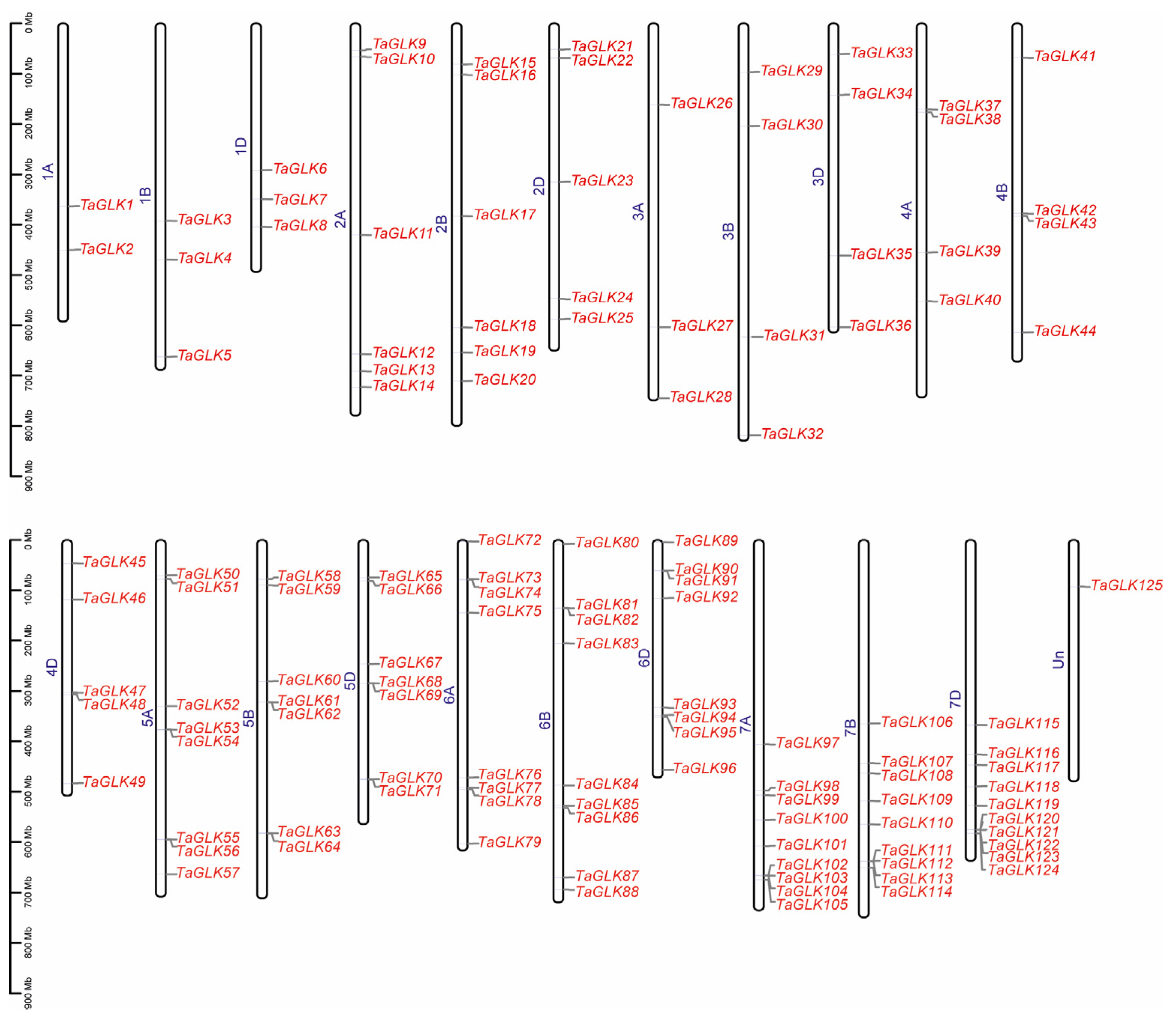
2.2. Physicochemical Properties and Chromosome Mapping Analyses of TaGLKs
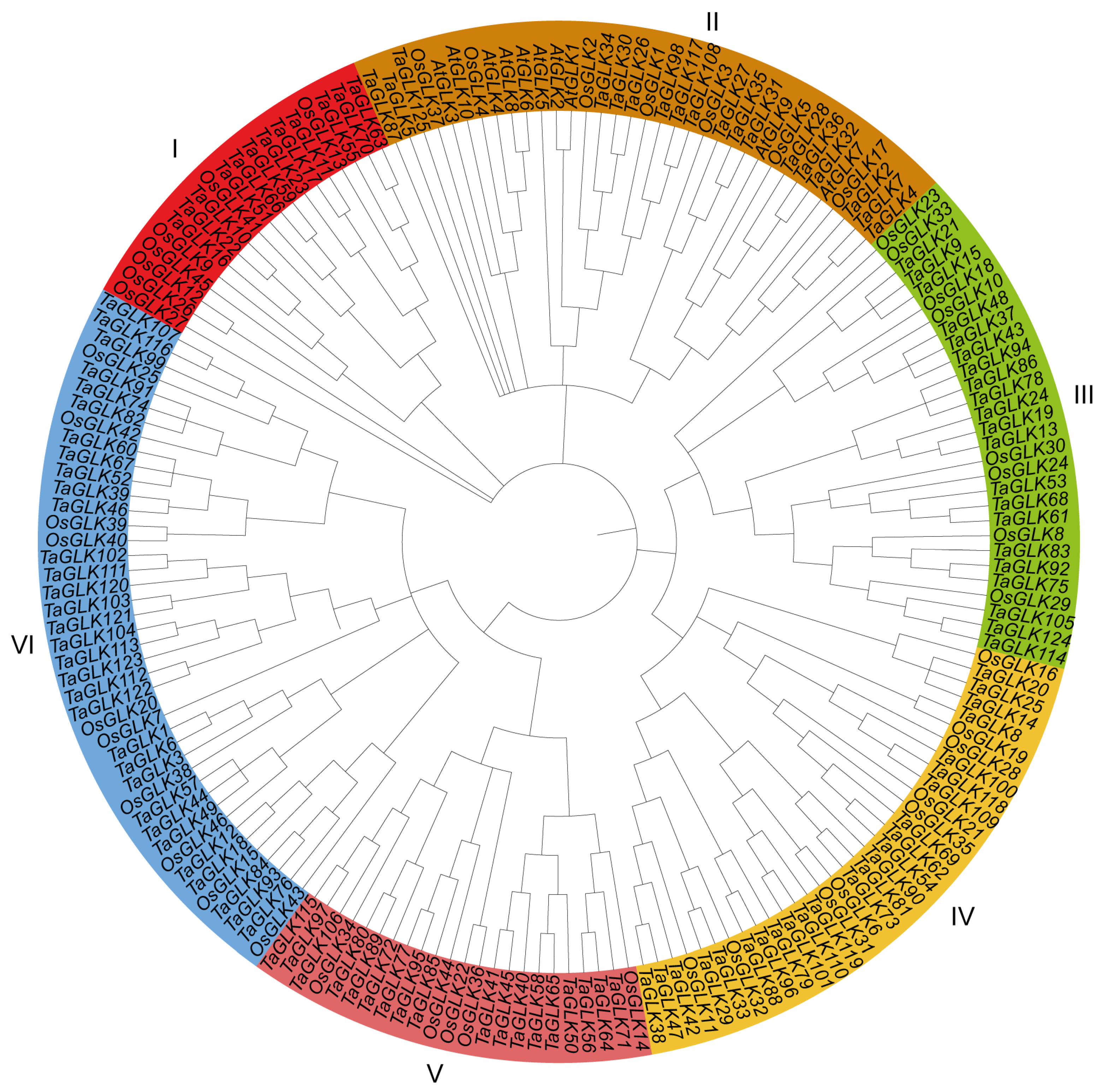
2.3. Phylogenetic Analysis of TaGLK Proteins
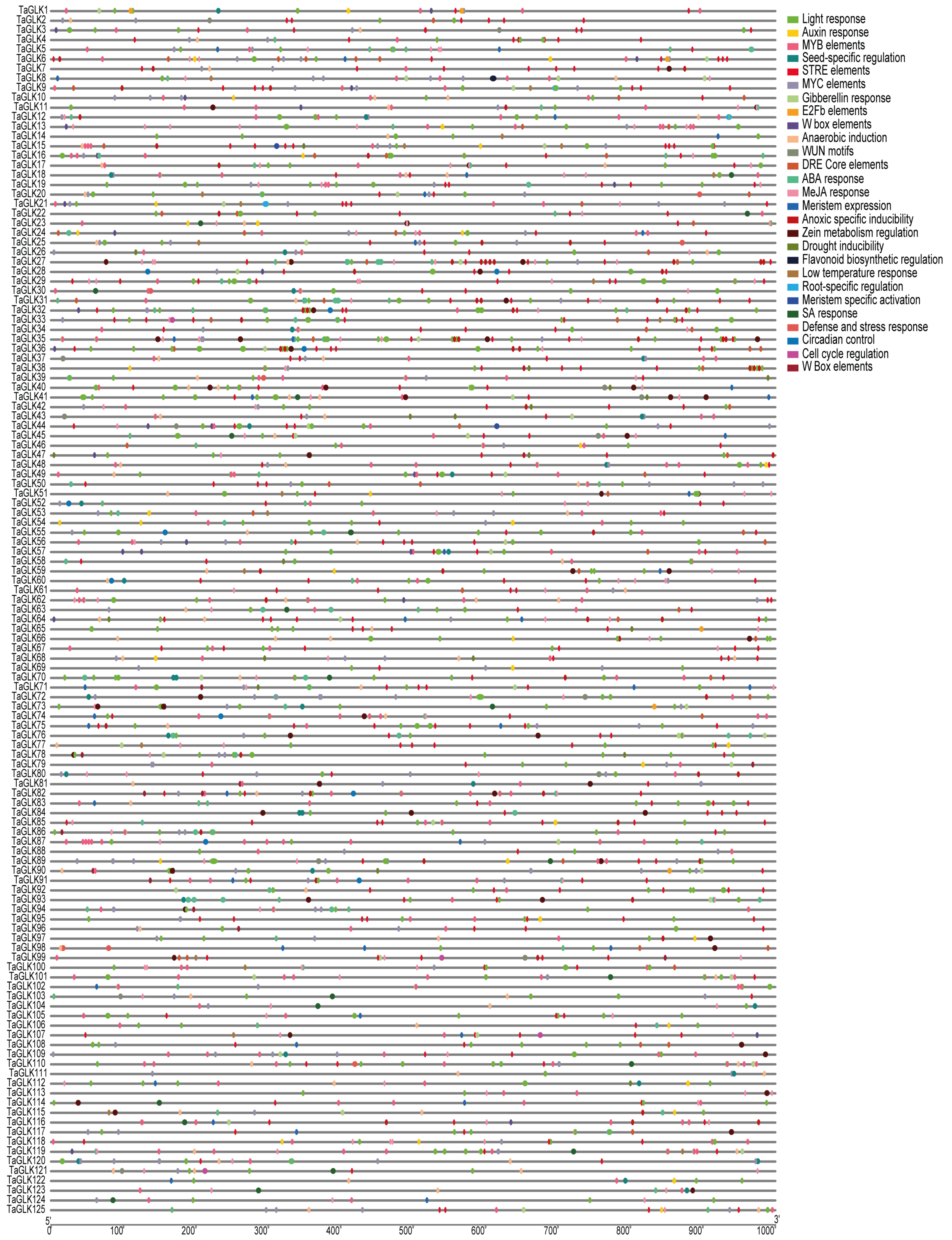
2.4. Gene Structure, Cis-Acting Elements, and Conserved Motif Analyses of TaGLKs
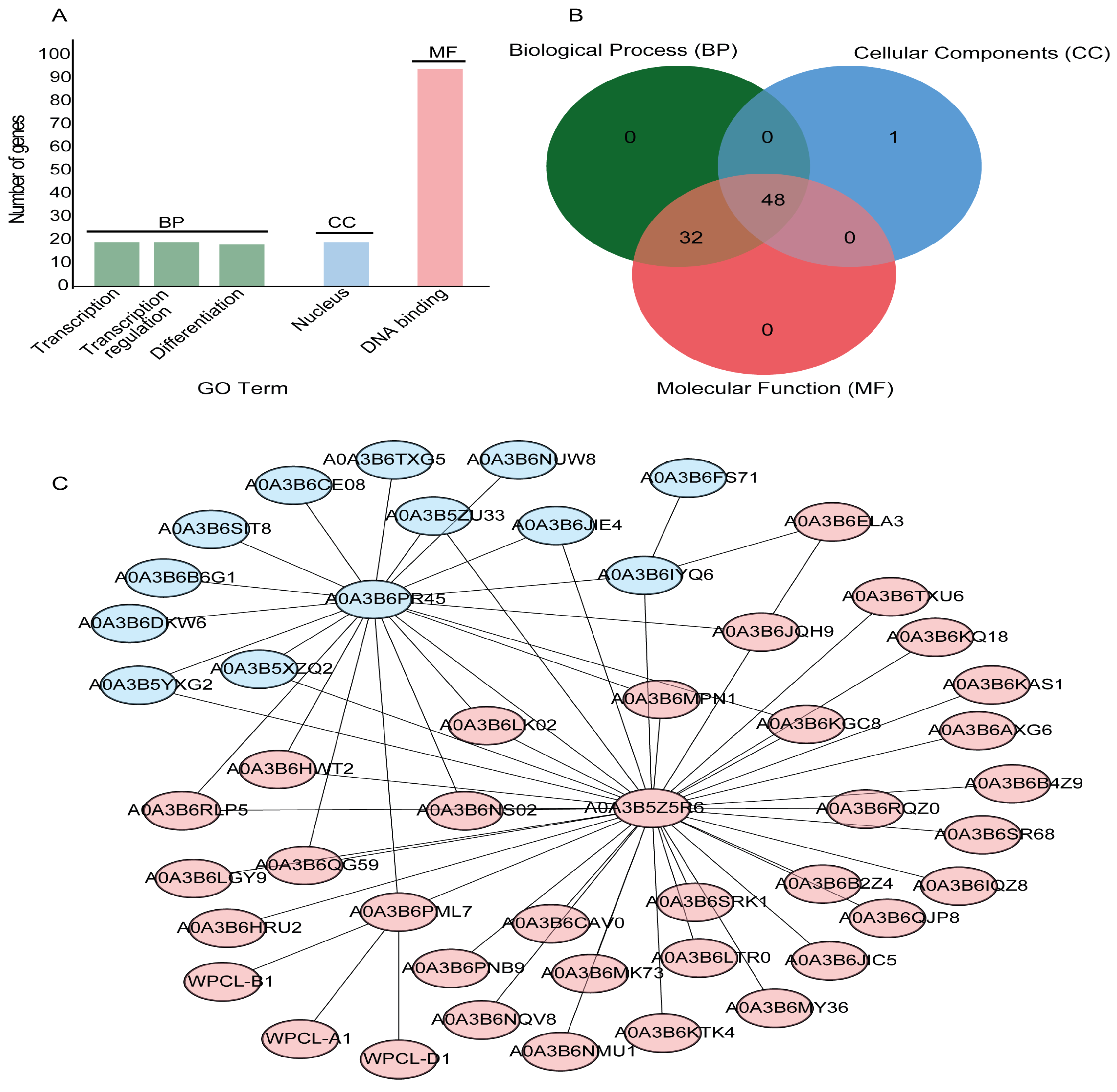
2.5. Gene Ontology (GO) Annotation and Protein–Protein Interaction of TaGLKs
2.6. Expression Data of Wheat TaGLKs Under Abiotic, Biotic, and Developmental Conditions
2.7. Growth and Stress Treatments of Wheat Seedlings
2.8. RT-qPCR Expression Analysis of TaGLKs
3. Results
3.1. Identification and Physicochemical Properties of TaGLKs
3.2. Evolutionary Relationships, Chromosome Mapping, and Domains of TaGLK Proteins
3.3. Conserved Motifs, Gene Structures, and Cis-Acting Elements of TaGLKs
3.4. GO Functional Annotation and Protein–Protein Interaction of TaGLKs
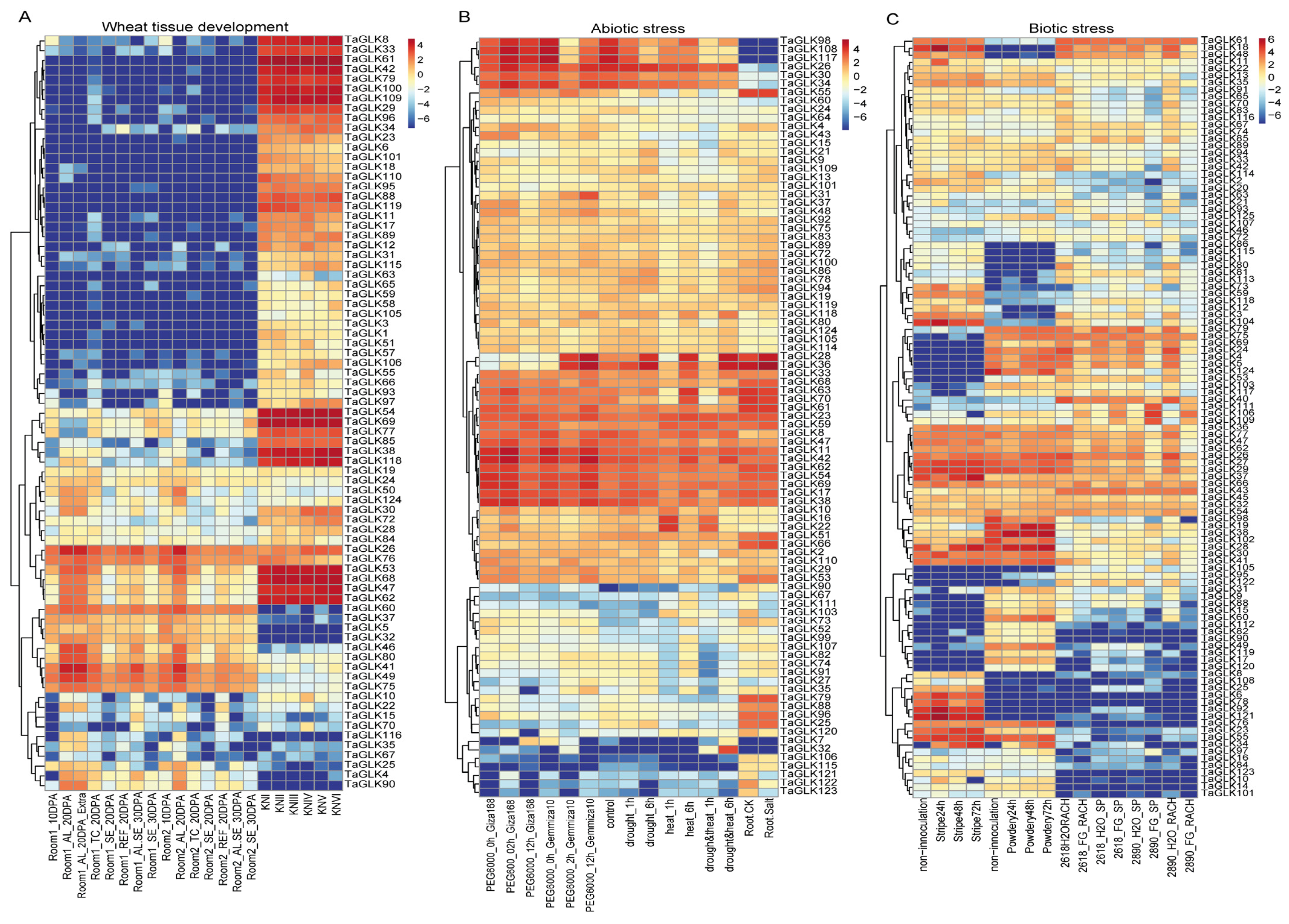
3.5. Analysis of TaGLKs’ Expression Under Wheat Abiotic, Biotic, and Developmental Conditions
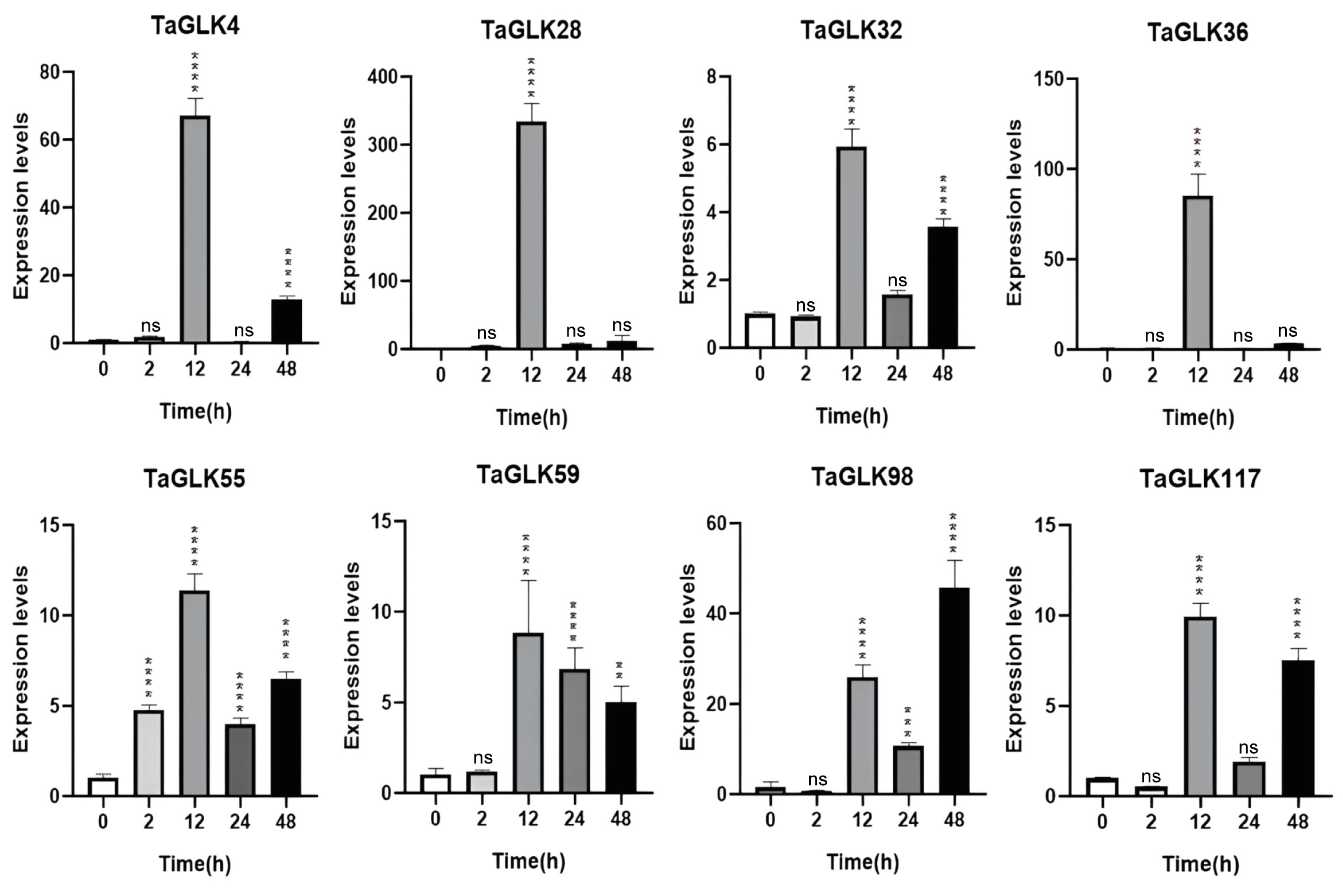
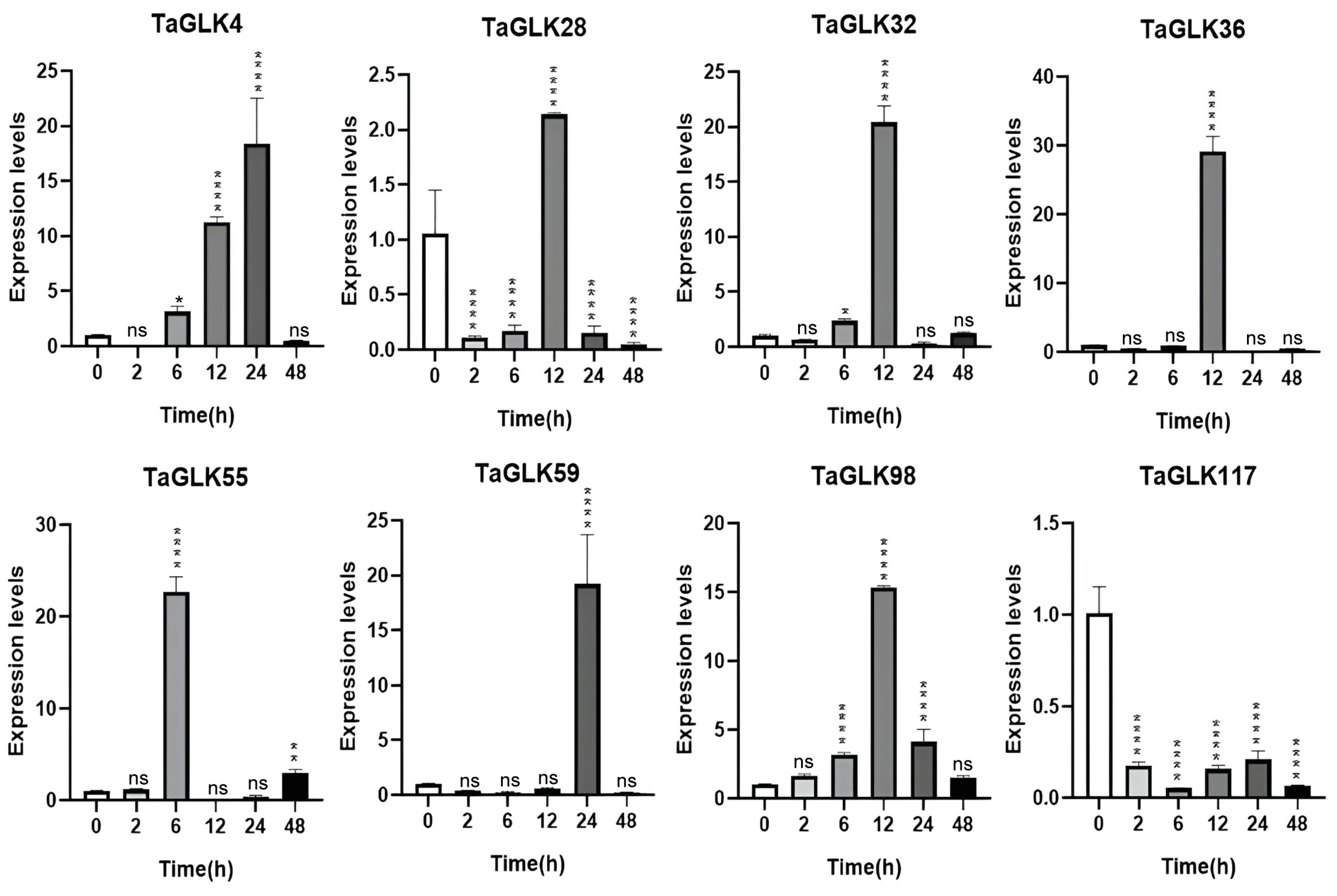
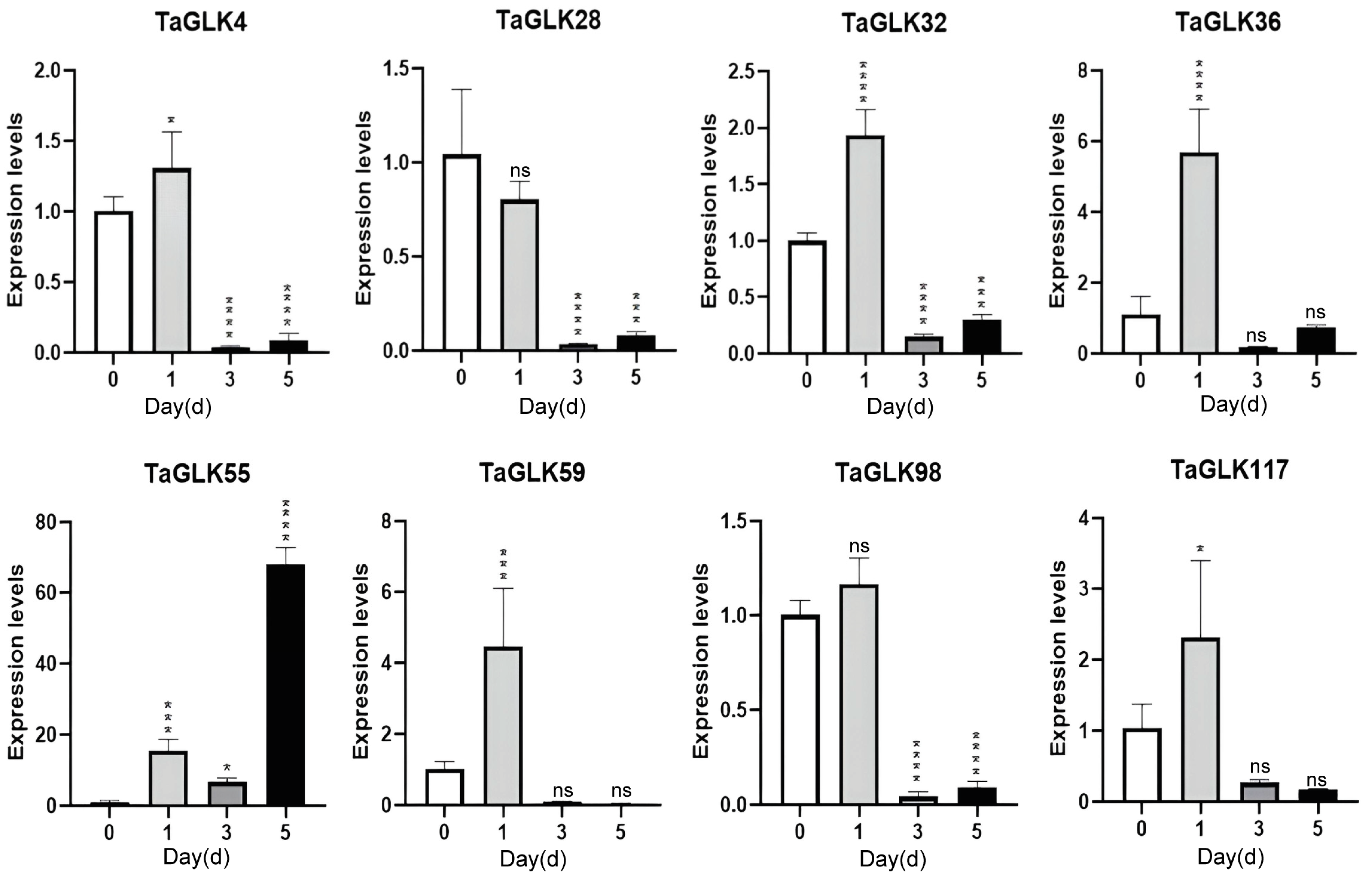
3.6. RT-qPCR Analysis of Wheat Biotic and Abiotic Stress Responses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bohnert, H.J.; Gong, Q.; Li, P.; Ma, S. Unraveling abiotic stress tolerance mechanisms–getting genomics going. Curr. Opin. Plant Biol. 2006, 9, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Zaikina, E.A.; Rumyantsev, S.D.; Sarvarova, E.; Kuluev, B. Transcription factor genes involved in plant response to abiotic stress factors. Ecol. Genet. 2019, 17, 47–58. [Google Scholar] [CrossRef]
- Song, A.; Wu, D.; Fan, Q.; Tian, C.; Chen, S.; Guan, Z.; Xin, J.; Zhao, K.; Chen, F. Transcriptome-wide identification and expression profiling analysis of chrysanthemum trihelix transcription factors. Int. J. Mol. Sci. 2016, 17, 198. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Lin, K.H.; Ho, S.L.; Chiang, C.M.; Chi, M.Y. Enhancing the abiotic stress tolerance of plants: From chemical treatment to biotechnological approaches. Physiol. Plant. 2018, 164, 452–466. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, C.; Shao, X.; Hu, Z.; Li, J.; Wang, P.; Wang, A.; Yu, J.; Shi, K. Transcriptomic and genetic approaches reveal an essential role of the NAC transcription factor SlNAP1 in the growth and defense response of tomato. Hortic. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- Kou, X.; Wang, S.; Wu, M.; Guo, R.; Xue, Z.; Meng, N.; Tao, X.; Chen, M.; Zhang, Y. Molecular Characterization and Expression Analysis of NAC Family Transcription Factors in Tomato. Plant Mol. Biol. Rep. 2014, 32, 501–516. [Google Scholar] [CrossRef]
- Li, M.; Lee, K.P.; Liu, T.; Dogra, V.; Duan, J.; Li, M.; Xing, W.; Kim, C. Antagonistic modules regulate photosynthesis-associated nuclear genes via GOLDEN2-LIKE transcription factors. Plant Physiol. 2021, 188, 2308–2324. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Rossini, L.; Cribb, L.; Martin, D.J.; Langdale, J.A. The maize golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 2001, 13, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Han, X.Y.; Li, P.X.; Zou, L.J.; Tan, W.R.; Zheng, T.; Zhang, D.W.; Lin, H.H. GOLDEN2-LIKE transcription factors coordinate the tolerance to Cucumber mosaic virus in Arabidopsis. Biochem. Biophys. Res. Commun. 2016, 477, 626–632. [Google Scholar] [CrossRef]
- Wu, R.; Guo, L.; Wang, R.; Zhang, Q.; Yao, H. Genome-Wide Identification and Characterization of G2-Like Transcription Factor Genes in Moso Bamboo (Phyllostachys edulis). Molecules 2022, 27, 5491. [Google Scholar] [CrossRef] [PubMed]
- Yasumura, Y.; Moylan, E.C.; Langdale, J.A. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell 2005, 17, 1894–1907. [Google Scholar] [CrossRef]
- Murmu, J.; Wilton, M.; Allard, G.; Pandeya, R.; Desveaux, D.; Singh, J.; Subramaniam, R. Arabidopsis GOLDEN2-LIKE (GLK) transcription factors activate jasmonic acid (JA)-dependent disease susceptibility to the biotrophic pathogen Hyaloperonospora arabidopsidis, as well as JA-independent plant immunity against the necrotrophic pathogen Botrytis cinerea. Mol. Plant Pathol. 2014, 15, 174–184. [Google Scholar]
- Chen, M.; Ji, M.; Wen, B.; Liu, L.; Li, S.; Chen, X.; Gao, D.; Li, L. GOLDEN 2-LIKE transcription factors of plants. Front. Plant Sci. 2016, 7, 1509. [Google Scholar] [CrossRef]
- Wang, H.; Seo, J.K.; Gao, S.; Cui, X.; Jin, H. Silencing of AtRAP, a target gene of a bacteria-induced small RNA, triggers antibacterial defense responses through activation of LSU2 and down-regulation of GLK1. New Phytol. 2017, 215, 1144–1155. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Choi, H.; Ti, T.; Gwak, D.; Ha, S.H. Pleiotropic properties of GOLDEN2-LIKE transcription factors for crop improvement. Appl. Biol. Chem. 2023, 66, 81. [Google Scholar] [CrossRef]
- Fan, P.; Nie, L.; Jiang, P.; Feng, J.; Lv, S.; Chen, X.; Bao, H.; Guo, J.; Tai, F.; Wang, J.; et al. Transcriptome analysis of Salicornia europaea under saline conditions revealed the adaptive primary metabolic pathways as early events to facilitate salt adaptation. PLoS ONE 2013, 8, e80595-18. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Choi, B.; Yun, A.; Son, N.; Ahn, G.; Cha, J.Y.; Kim, W.Y.; Hwang, I. Long-term ABA promotes GLK1 degradation through COP1 in a light intensity-dependent manner to suppress chloroplast development. Plant Cell Environ. 2021, 44, 3034–3048. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, L.; Zhang, B.; Zeng, L.; Li, L. AhHDA1-mediated AhGLK1 promoted chlorophyll synthesis and photosynthesis regulates recovery growth of peanut leaves after water stress. Plant Sci. 2020, 294, 110461. [Google Scholar] [CrossRef]
- Liu, J.; Mehari, T.G.; Xu, Y.; Umer, M.J.; Hou, Y.; Wang, Y.; Peng, R.; Wang, K.; Cai, X.; Zhou, Z.; et al. GhGLK1 a key candidate gene from GARP family enhances cold and drought stress tolerance in cotton. Front. Plant Sci. 2021, 12, 759312. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Liu, Y.; Wang, T.J.; Meng, Q.; Yin, H.; Wang, X.; Wu, Y.; Nan, N.; Liu, B.; Xu, Z.Y. GOLDEN 2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiol. 2019, 179, 1844–1860. [Google Scholar] [CrossRef] [PubMed]
- Savitch, L.V.; Allard, G.; Seki, M.; Robert, L.S.; Tinker, N.A.; Huner, N.P.; Shinozaki, K.; Singh, J. The effect of overexpression of two Brassica CBF/DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus. Plant Cell Physiol. 2005, 46, 1525–1539. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lee, J.; Choi, B.; Park, Y.; Sim, H.J.; Kim, H.; Hwang, I. Physiological and molecular processes associated with long duration of ABA treatment. Front. Plant Sci. 2018, 9, 176. [Google Scholar] [CrossRef]
- Khalid, A.; Hameed, A.; Tahir, M.F. Wheat quality: A review on chemical composition, nutritional attributes, grain anatomy, types, classification, and function of seed storage proteins in bread making quality. Front. Nutr. 2023, 10, 1053196. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Hameed, A.; Shamim, S.; Ahmad, J. Divergence in single kernel characteristics and grain nutritional profiles of wheat genetic resource and association among traits. Front. Nutr. 2022, 8, 805446. [Google Scholar] [CrossRef]
- Wattoo, F.M.; Rana, R.M.; Fiaz, S. Transcriptional Factors’ Response Under Biotic Stress in Wheat. In Transcription Factors for Biotic Stress Tolerance in Plants; Springer International Publishing: Cham, Switzerland, 2022; pp. 129–141. [Google Scholar]
- Liu, H.; Yang, Y.; Zhang, L. Zinc Finger-Homeodomain Transcriptional Factors (ZF-HDs) in Wheat (Triticum aestivum L.): Identification, Evolution, Expression Analysis and Response to Abiotic Stresses. Plants 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.; Wei, S.; Gao, Y.; Pei, H.; Geng, R.; Lu, Z.; Wang, P.; Zhou, W. Maize GOLDEN2-LIKE proteins enhance drought tolerance in rice by promoting stomatal closure. Plant Physiol. 2024, 194, 774–786. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 60 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 8, 1296–1297. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ma, S.; Wang, M.; Wu, J.; Guo, W.; Chen, Y.; Li, G.; Wang, Y.; Shi, W.; Xia, G.; Fu, D.; et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 2021, 14, 1965–1968. [Google Scholar] [CrossRef]
- Lyu, F.; Han, F.; Ge, C.; Mao, W.; Chen, L.; Hu, H.; Chen, G.; Lang, Q.; Fang, C. OmicStudio: A composable bioinformatics cloud platform with real-time feedback that can generate high-quality graphs for publication. Imeta 2023, 2, e85. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Geng, Y.; Liu, Y.; Chen, S.; Cao, S.; Li, W.; Chen, H.; Ma, D.; Yin, J. Genome-wide identification and characterization of SRO gene family in wheat: Molecular evolution and expression profiles during different stresses. Plant Physiol. Biochem. PPB 2020, 154, 590–611. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Su, C.; Wang, Y.; Xu, X.; Li, Y.; Ma, D. Genome-wide identification of Glutathione peroxidase (GPX) family genes and silencing TaGPX3.2A reduced disease resistance in wheat. Plant Physiol. Biochem. PPB 2023, 204, 108139. [Google Scholar] [CrossRef]
- Jiang, C.; Cao, S.; Wang, Z.; Xu, H.; Liang, J.; Liu, H.; Wang, G.; Ding, M.; Wang, Q.; Gong, C.; et al. An expanded subfamily of G-protein-coupled receptor genes in Fusarium graminearum required for wheat infection. Nat. Microbiol. 2019, 4, 1582–1591. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Verdeja, T.; Lundgren, M.R. GOLDEN2-LIKE transcription factors: A golden ticket to improve crops? Plants People Planet 2023, 6, 79–93. [Google Scholar] [CrossRef]
- Xiong, B.; Gong, Y.; Li, Q.; Li, L.; Mao, H.; Liao, L.; Wang, X.; Deng, H.; Zhang, M.; Wang, Z. Genome-Wide Analysis of the GLK Gene Family and the Expression under Different Growth Stages and Dark Stress in Sweet Orange (Citrus sinensis). Horticulturae 2022, 8, 1076. [Google Scholar] [CrossRef]
- Chen, H.; Qin, L.; Wang, X. Identification and Evolutionary Analysis of the GOLDEN 2-LIKE Gene Family in Foxtail Millet. Trop. Plant Biol. 2022, 15, 301–318. [Google Scholar] [CrossRef]
- Qin, M.; Zhang, B.; Gu, G.; Yuan, J.; Yang, X.; Yang, J.; Xie, X. Genome-Wide Analysis of the G2-Like Transcription Factor Genes and Their Expression in Different Senescence Stages of Tobacco (Nicotiana tabacum L.). Front. Genet. 2021, 12, 787. [Google Scholar] [CrossRef]
- Zhao, Z.; Shuang, J.; Li, Z.; Xiao, H.; Liu, Y.; Wang, T.; Wei, Y.; Hu, S.; Wan, S.; Peng, R. Identification of the Golden-2-like transcription factors gene family in Gossypium hirsutum. PeerJ 2021, 9, e12484. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, Y.; Han, G.; Zhou, L.; Ali, A.; Zhu, S.; Li, X. Molecular Evolution and Genetic Variation of G2-Like Transcription Factor Genes in Maize. PLoS ONE 2016, 11, e0161763. [Google Scholar] [CrossRef]
- Ni, J.; Bai, S.; Gao, L.; Qian, M.; Zhong, L.; Teng, Y. Identification, classification, and transcription profiles of the B-type response regulator family in pear. PLoS ONE 2017, 12, e0171523. [Google Scholar] [CrossRef]
- Liu, J.F. Bioinformatics Analysis of Tomato G2-Like Transcription Factor Family and Identification of Resistance-Related Genes. Doctoral Thesis, College of Horticulture & Landscape Architecture, West Lafayette, IN, USA, 2018. [Google Scholar]
- Alam, I.; Manghwar, H.; Zhang, H.; Yu, Q.; Ge, L. Identification of GOLDEN2-like transcription factor genes in soybeans and their role in regulating plant development and metal ion stresses. Front. Plant Sci. 2022, 13, 1052659. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytologist. 2020, 227, 698–713. [Google Scholar] [CrossRef]
- Li, X.; Lin, F.; Li, C.; Du, L.; Liu, Z.; Shi, W.; Lv, J.; Cao, X.; Lan, Y.; Fan, Y. Golden 2-like transcription factor contributes to the major QTL against rice black-streaked dwarf virus disease. Theor. Appl. Genet. 2022, 135, 4233–4243. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, J.; Liu, P.; Bimpong, D.; Shen, J.; Liu, X.; Wang, S.; Li, Y.; Wang, Y.; Ma, D. Identification and Expression Analysis of Wheat Golden2-like (TaGLK) Gene in Response to Biotic and Abiotic Stress. Agronomy 2024, 14, 3070. https://doi.org/10.3390/agronomy14123070
Xiang J, Liu P, Bimpong D, Shen J, Liu X, Wang S, Li Y, Wang Y, Ma D. Identification and Expression Analysis of Wheat Golden2-like (TaGLK) Gene in Response to Biotic and Abiotic Stress. Agronomy. 2024; 14(12):3070. https://doi.org/10.3390/agronomy14123070
Chicago/Turabian StyleXiang, Junhui, Pingu Liu, Daniel Bimpong, Jiayi Shen, Xusi Liu, Siting Wang, Yan Li, Youning Wang, and Dongfang Ma. 2024. "Identification and Expression Analysis of Wheat Golden2-like (TaGLK) Gene in Response to Biotic and Abiotic Stress" Agronomy 14, no. 12: 3070. https://doi.org/10.3390/agronomy14123070
APA StyleXiang, J., Liu, P., Bimpong, D., Shen, J., Liu, X., Wang, S., Li, Y., Wang, Y., & Ma, D. (2024). Identification and Expression Analysis of Wheat Golden2-like (TaGLK) Gene in Response to Biotic and Abiotic Stress. Agronomy, 14(12), 3070. https://doi.org/10.3390/agronomy14123070






