Abstract
Transcription factors play a key role in plant growth and development. As the largest family of plant-specific transcription factors, the NAC family plays a central role in coordinating plant growth and development and environmental adaptation through its unique molecular design paradigm of “fixed N-terminal structural domain + variable C-terminal regulatory domain”. This review systematically analyses the multidimensional regulatory mechanisms of NAC transcription factors in developmental processes such as cell wall remodelling, root system architecture, leaf senescence and fruit ripening, and reveals their molecular basis for responding to biotic/abiotic stresses through strategies such as hormone signalling integration (ABA, SA, JA, etc.), antioxidant defence activation and metabolic reprogramming. The study found that NAC proteins precisely control plant growth through multiple regulatory mechanisms and have evolved to form both conservative and diverse functional modules, which are of great value for crop improvement. However, research still faces three major challenges: the NAC regulatory network in different crops is still unclear, the coordinated response to multiple stresses has not been solved, and the ecological risks of gene editing have not been assessed. To this end, this paper proposes to build an ‘NAC regulatory map database’ and use synthetic biology and artificial intelligence technology to design smarter, stress-tolerant and high-yielding crops, overcoming the limitations of traditional research.
1. Introduction
In the 1960s and 1970s, scientists discovered that transcription factors can specifically bind to DNA and regulate gene expression. Subsequently, their regulatory mechanisms were gradually revealed, and genome sequencing has promoted research on plant transcription factor families such as MYB, bZIP, WRKY, and NAC. Transcription factors play a key role in growth and development, hormone signalling, and environmental responses. Since the 21st century, the development of genomics, single-cell omics, and gene editing technology has expanded research from single-gene analysis to complex regulatory networks, deepened the understanding of crop trait regulation, and promoted genetic improvement and precision breeding.
The regulation of gene expression is a fundamental process in plant growth, development, and stress responses, with transcription factors (TFs) playing a crucial role in orchestrating these processes. Among the diverse families of TFs, the NAC (NAM, ATAF, and CUC) family stands out as one of the most plant-specific and functionally diverse groups. First identified in P. hybrida in 1996, NAC transcription factors have since been characterized across numerous plant species, highlighting their evolutionary conservation and functional significance [1,2]. These TFs are characterized by a highly conserved N-terminal domain and a highly variable C-terminal region, which together confer their functional diversity [3].
Over the past three decades, extensive research has elucidated the multifaceted roles of NAC transcription factors in plant biology. These factors are involved in regulating secondary metabolite synthesis, plant growth and development, and responses to both abiotic and biotic stresses. For instance, NAC TFs have been shown to modulate the synthesis of secondary metabolites such as anthocyanins, allicin, and ginsenosides, which are critical for plant adaptation and have potential therapeutic benefits [4,5,6]. Additionally, NAC TFs play key roles in developmental processes, including secondary cell wall formation, root development, leaf senescence, and fruit ripening [7,8]. Their involvement in stress responses is equally significant, with studies demonstrating their roles in enhancing tolerance to drought, salt, heavy metals, and pathogens [9].
Despite these advances, our understanding of NAC transcription factors is still incomplete. Current research has focused mainly on the gene cloning and functional analysis of model plants such as Arabidopsis and rice (Oryza sativa L.) [1,2]. In particular, the mechanisms by which NAC TFs regulate downstream target genes and integrate hormonal and environmental signals have been the focus of research in model plants, while the exploration of their role in other species is limited [10,11,12]. Consequently, there is an urgent need for in-depth studies on a broader range of plant species to elucidate these mechanisms. This review aims to provide a comprehensive overview of the structural features, functions, and regulatory mechanisms of NAC transcription factors, integrating recent findings to highlight their significance in plant growth, development, and stress responses. By summarizing the current knowledge and identifying gaps, we hope to lay the foundation for future research that will further unravel the complex roles of NAC TFs in plant biology.
In comparison with previous reviews, this paper summarizes and analyses the overview of recent research on new NAC transcription factors through various aspects of plant growth and development and stress.
NAC transcription factors, which are composed of a conserved N-terminal structural domain and a differentiated C-terminal regulatory domain, are involved in different growth periods in different species, providing us with a good basis for theoretical studies. In addition, related studies such as single-cell transcriptome and protein interactions have revealed that NAC factors play important roles in developmental events such as cell wall remodelling and root architecture. In addition to the major model plants, we also summarize the number of NAC transcription factors in other plants based on related studies, totalling 56 species, and sort them by number to help readers more intuitively feel the differences among different species.
Notably, we identified the limitations of existing studies focusing on model plants [1,2], and mined and summarized the key regulators of agronomic traits such as wheat TaNAC48 and apple MdNAC1. Related theoretical studies provide new ideas and directions for obtaining high-trait transgenic plants [13,14].
2. NAC Transcription Factor
Following the initial identification of these transcription factors, NAC factors have been utilised to enhance the gene level of numerous crops, including plants such as rice (Oryza sativa L.) and tobacco (Nicotiana tabacum L.). Furthermore, the presence of NAC transcription factors has been detected in numerous other plant species, including corn (Zea mays), cotton (G. hirsutum L.) and other plants (Table 1).

Table 1.
Total number of NACTFs in different plants.
The NAC domain, highly conserved and specific to the N-terminal end of NAC proteins, is the most important structural feature. It is capable of recognizing and binding to specific cis-binding elements. The NAC domain can be subdivided into five sub-domains, namely A, B, C, D and E. Among them, the A, C and D sub-domains are highly conserved across different species and carry a positive charge. Notably, the C and D subdomains facilitate protein binding to DNA, and carry a positive charge. The B and E sub-domains exhibit species-dependent variations, which may correlate with the functional diversity of the NAC family. In contrast, the C-terminal is a multifunctional regulatory domain with relatively simple sequences, often consisting of repetitive amino acid sequences [47,48,49] (Figure 1).
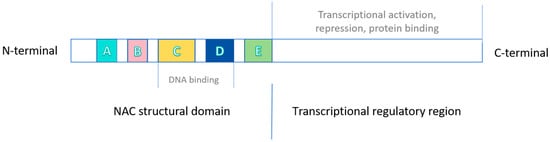
Figure 1.
Schematic structure of NAC transcription factors. Note: A–E represent the sub-structural domains further subdivided within the NAC domain.
3. Studies on the Involvement of NAC Transcription Factors in Plant Growth and Development
NAC transcription factors have been identified as key regulators of plant developmental processes, regulating the development of nutrient and reproductive organs, promoting the formation of secondary cell walls and lateral roots, and participating in leaf senescence and fruit ripening during plant growth and development [7,8].
3.1. Regulation of Plant Cell Secondary Wall Growth
The plant cell wall consists of three distinct layers: the intercellular layer, the primary cell wall, and the secondary cell wall (SCW). The secondary cell wall is formed by the continuous accumulation of materials on the inner surface of the primary wall, outside the cell membrane. It plays a crucial role in plant growth and development by maintaining plant morphology, transporting nutrients and water, and providing structural support. Rich in cellulose, hemicellulose and lignin, the secondary cell wall maintains the integrity of the plant cell [50,51,52].
NAC transcription factors play a crucial role in regulating the synthesis of the plant cell secondary wall. For instance, AtSND1, a member of the NAC family in the Arabidopsis genome, regulates the expression of genes involved in secondary wall synthesis, leading to significant accumulation of secondary walls. Similarly, AtSND2, another NAC family member, is pivotal in regulating genes associated with cellulose, hemicellulose, and lignin synthesis. Additionally, AtSND3, also a NAC transcription factor, has been identified as a key factor promoting fibrous secondary wall thickening [33,34].
Further studies highlight the functional conservation of NAC transcription factors across different plant species. For example, overexpression of FvNST1b, a NAC family member in wild strawberry, significantly upregulates the expression of secondary wall synthesis-related genes, resulting in thickened secondary walls in anthers, stamens, and leaves of Arabidopsis thaliana [53]. Likewise, overexpression of SND1 and GhNAC140/28/70/120, which are also NAC family members, in tobacco leaves leads to thickened secondary walls [54]. Moreover, in banana, the NAC transcription factor VND1-3 binds to SNBE sites in the promoters of downstream target genes, thereby increasing lignin and cellulose content [22]. In maize and Arabidopsis, overexpression of ZmNST3 and ZmNST4, both NAC family members, enhances secondary wall thickness in transgenic plants, whereas knockdown of these genes in maize reduces secondary wall thickness [55]. These findings collectively underscore the critical role of NAC transcription factors in regulating secondary wall development across diverse plant species.
3.2. Regulation of Root Development
The plant root system is an essential organ for nutrients and water absorption. The main root, the largest and fastest-growing root, gives rise to smaller lateral roots on its sides, which are vital for the plant’s healthy growth [56,57].
NAC transcription factors also play important roles in regulating root development in plants. For instance, in tobacco, the content of NtNAC-R1 and the number of lateral roots increase after mechanical injury, suggesting the involvement of jasmonic acid (JA) and indole-3-acetic acid (IAA) signalling pathways [58]. Similarly, the expression of StNAC262 results in a larger root system and an increased number of lateral roots [59]. In soybean, the expression of GmNAC109 promotes lateral root development [23]. Additionally, in Arabidopsis, the NAC structural domain transcription factor SMB negatively regulates FEZ activity, thereby inhibiting stem cell division in root crown cells [60]. AtNAC056 is primarily expressed in the root system and hypocotyls. It targets and induces NIA1 expression, promotes lateral root growth, and enhances root tolerance to nitrate stress [61]. Moreover, AtNAC075 reduces primary root sensitivity to nitrate [62]. Collectively, these findings highlight the critical roles of NAC transcription factors in regulating root development across diverse plant species [63].
3.3. Regulation of Leaf Senescence
Leaves are the primary organs for photosynthesis in plants, and leaf senescence marks the final stage of leaf development. The initiation and progression of leaf senescence are influenced by various internal and external factors, which induce changes in cellular structure, photosynthesis, and metabolic efficiency. Characterized by chlorophyll degradation, leaf senescence enhances plant adaptability [64,65].
Relevant studies have shown that hormones such as ethylene, salicylic acid (SA), and abscisic acid (ABA) play important roles in the involvement of NAC in plant leaf senescence. In Arabidopsis, three members of NAC transcription factors, AtNAC017, AtNAC082 and AtNAC090, play a negative role in the regulation of leaf senescence by regulating the contents of SA and reactive oxygen species (ROS), which are known as the NAC triad [66].
ZmNYE1 has been shown to accelerate chlorophyll degradation during leaf senescence, thus representing a major chlorophyll catabolic metabolism gene. In contrast, the function of ZmNAC132 in maize is to bind to and activate the ZmNYE1 promoter. The results of the study demonstrated that the knockdown of ZmNAC132 in maize resulted in delayed chlorophyll degradation and leaf senescence, whereas its overexpression had the opposite effect [29]. BrNAC029 alters senescence in cabbage leaves by regulating cytokinins [67]. It has been demonstrated that the ANAC059 and ANAC083 transcription factors positively regulate leaf senescence in Arabidopsis [68,69].
Trupkin et al. (2019) [70] conducted a screening process of 41 NAC family members in Arabidopsis thaliana, with the objective of identifying those that may respond to plant senescence. In addition, 10–15 PhNACs in P. hybrida were screened, including PhNAC024, with the aim of determining those that may be involved in the regulation of leaf and petal senescence. It is noteworthy that InEPH1 is the only NAC factor that is currently known to be involved in the regulation of petal senescence [70].
3.4. Regulation of Fruit Growth
Fruit quality is a major concern, and NAC transcription factors significantly regulate fruit growth and ripening in plants. These factors influence various characteristics, including fruit colour, taste, nutrients, and other components.
Fruit flavour is significantly influenced by organic acids, which also play a crucial role in determining fruit quality and nutritional value. MdNAC56, a transcription factor found in the nucleus of apple cells, has been shown to bind to the downstream target gene MdMa11. It has been hypothesised that the accumulation of malic acid in fruits may be regulated through the transcriptional control of MdMa11 expression. MdNAC029, which is also located in the nucleus of apple cells, is expressed in roots and fruits and regulates malic acid accumulation in apple fruits by regulating the expression of MdMYB1 or by binding to the promoter of MdMYB73 [17,18]. PpNAC050 expression in peach fruits has been found to correlate with fruit fructose accumulation, and it has been demonstrated to promote fruit fructose accumulation by suppressing the expression of the fructose content gene PpERDL6-1 [62].
It was determined that AaNAC2, AaNAC3 and AaNAC4 bind to the promoter of kiwifruit terpene synthase AaTPS1, thereby contributing to the increased accumulation of monoterpene volatiles in Actinidia arguta fruits [71]. In the case of plant species, Arabidopsis JUB1 has been observed to inhibit intracellular H2O2 levels, reduce the levels of senescence-related genes, and extend the lifespan of the plant [72]. The analysis further revealed that the ANAC087 and ANAC046 transcription factors, belonging to the ANAC family, play a regulatory role in various aspects of programmed cell death in the columnar layer and secondary roots [73,74,75].
3.5. Summary
NAC transcription factors have been demonstrated to play a multifaceted regulatory role in various processes of plant growth and development, including the formation of secondary cell walls, root development, leaf senescence, and the regulation of fruit quality. In terms of cell wall development, AtSND1, FvNST1b, etc. [33,53], enhance cell wall structure by activating genes related to cellulose and lignin synthesis (such as GhNAC140 and ZmNST3) [54,55]. It is noteworthy that the functions of these NAC transcription factors are highly conserved across various plant species. During the process of root development, StNAC262 and GmNAC109 promote the formation of lateral roots through the JA/IAA signalling pathway [23,59], while AtNAC056 optimises root adaptation by regulating the nitrate-responsive gene NIA1 [61]. In terms of leaf senescence, ZmNAC132 accelerates this process by activating the chlorophyll degradation gene ZmNYE1 [29], while AtNAC017/082/090 delays senescence by inhibiting SA/ROS accumulation, reflecting a collaborative regulatory mechanism of hormonal signalling and redox balance [66]. During fruit development, MdNAC56, PpNAC050, and other factors influence the accumulation of flavour substances by regulating malate synthase MdMa11 or fructose transporter PpERDL6-1, while AaNAC2-4 plays a role in the formation of fruit aroma by regulating terpene synthase AaTPS1 [62,72]. This further underscores the pivotal function of NAC in determining fruit quality.
With respect to future research directions, we need to improve our understanding of NAC by studying both upstream and downstream elements in its regulatory network. This includes exploring its synergistic mechanisms with MYB/bHLH transcription factors, as well as the interactions between specific promoters and NAC transcription factors. We should also comprehensively assess the phenotypic stability and potential ecological risks of NAC gene-edited crops under field conditions, especially in foods such as fruits and vegetables that are our primary research focus. We should also use a multifaceted approach to understand and maximize the role of NAC transcription factors in different crops.
4. Studies on the Involvement of NAC Transcription Factors in Plant Stresses
4.1. Biological Stress
It is evident that plants, including food crops and fruit trees, are susceptible to biotic stress damage on a global scale. To mitigate this damage and enhance yield and quality, it is crucial to enhance plant biotic stress resistance. As the global population expands and the demand for food increases, ensuring the quality of the food supply is vital [76].
Recent studies have shown that the activity of NAC transcription factors plays a key role in plant responses to biotic stresses. For example, by overexpressing SlNAP1 in tomato, it was found that SlNAP1 in transgenic tomato plants activates the initiation of downstream SlPAL3 by binding to the SlPAL3 promoter, which indirectly affects the accumulation of SA (salicylic acid), thereby enhancing the resistance of overexpressed transgenic tomato to bacterial stress. This increases the resistance of the overexpressed transgenic tomato to leaf spot and bacterial wilt. Notably, the overexpression of SlNAP1 resulted in a 10.7% increase in tomato yield, despite the initial shorter stature of the genetically modified plants compared to the wild-type control [38]. In addition to SlNAP1, ScATAF1, another member of the NAC family, was found to increase tobacco susceptibility to pathogens through overexpression. It has been hypothesized that ScATAF1 may induce the expression of other related genes to reduce ROS, which indirectly results in the synthesis of SA and JA. This leads to plants that are more susceptible to pathogens [26]. The SmNAC protein, a member of the NAC family found in aubergine, was found to reduce the resistance of transgenic plants to cycloidal blight by overexpression of the gene. It was hypothesized that it may regulate plant disease resistance through the SA signalling pathway [17].
The function of the StNAC103 gene, a member of the NAC family, has been elucidated in potato through a study of its role in plant defence mechanisms. It was demonstrated that the deposition of long-chain fatty groups in the cell wall could be increased by gene silencing of StNAC103, thereby enhancing plant antimicrobial resistance through the secondary cell wall (SCW) [18]. Furthermore, 74 VvNAC genes were identified in grapevine through bioinformatics analysis and in vitro experiments, with 11 of them being predicted to be associated with the grey mould stress response. Quantitative polymerase chain reaction (qPCR) analysis additionally confirmed that five genes, VvNAC08, VvNAC30, VvNAC36, VvNAC39, and VvNAC44, are involved in the defence response of grapevine against the pathogen. Despite the absence of significant changes in expression for VvNAC31, VvNAC41, VvNAC61, VvNAC73 and VvNAC74, this does not preclude their potential involvement in the regulation of plant defence against pathogens [46]. In tomato, the SNAC4 gene is involved in the response to biotic and abiotic stress by regulating the expression of downstream target genes. It was demonstrated that grey mould infection resulted in a notable reduction in the expression level of SNAC4 [39]. It was demonstrated that SNAC4 negatively regulates the expression of SRN1, JA2L and MKK4 genes. Furthermore, it was observed that SNAC4 combined with the promoter regions of grey mould defence-related genes, JA2L and SRN1, to enhance plant resistance to grey mould [77,78].
A review of the literature indicates that NAC members are involved in biotic stress responses across a diverse range of species. These include responses to leaf spot disease in tomato, grey mould in grapes, black sigatoka in sugarcane, and others. The involvement of NAC members in these processes is multifaceted, encompassing regulation of the SA signalling pathway, reduction of ROS, and modification of the cell wall to alter plant resistance to biotic stressors.
4.2. Abiotic Stresses
Compared to biotic stresses, the NAC family has been the subject of more comprehensive study with respect to abiotic stresses. This paper aims to present a detailed account of the recent advances in research and development within the field of abiotic stress responses, focusing on cold stress, drought stress, salt stress and heavy metal stress.
4.2.1. Cold Stress
Low temperature is a significant environmental factor influencing the growth and development of plants. Exposure to cold temperatures can result in reduced yields, impaired development, or even death in plants, a phenomenon known as cold damage. Low temperatures cause the condensation of water within plant cells, leading to the destruction of the internal cellular structure, damage to the cellular biofilms, a reduction in cellular stability, and an impairment of the activity of antioxidant enzymes; it is possible that this may result in further damage to the plant. Over the course of evolution, plants have developed a range of mechanisms to cope with cold damage, including the activation of responsive genes and alterations to physiological and biochemical processes, with transcription factors playing a pivotal role. In particular, NAC transcription factors have been identified as playing a role in this process [78].
An et al. (2018) [79] discovered that MdNAC029, a transcription factor in apple, responds to low-temperature stress. Overexpression of MdNAC029 in apple and Arabidopsis was observed to enhance cold tolerance. Further analysis revealed that MdNAC029 binds to the promoters of MdCBF1 and MdCBF4, repressing their expression and thereby regulating cold tolerance in plants [79].
Since the apple is a boreal plant and the banana is a tropical plant, in the case of the apple tree we are dealing with the frost resistance of the plant, whereas the banana is characterized by its resistance to cold. By means of quantitative fluorescence analysis, Shan Wei discovered that MaNAC1 expression in banana was prompted by low temperature and exogenous propylene. Furthermore, the binding capacity of MaICE1 to the MaNAC1 promoter was augmented under low-temperature stress, indicating that MaNAC1 plays a role in responding to low-temperature adversity and is associated with propylene-induced cold tolerance [80]. Additionally, in banana, MaNAC25 and MaNAC28 are implicated in the cold stress response, and they interact with other transcription factors, yet their functional roles diverge: MaNAC25 is primarily involved in the regulation of biological processes and the regulation of molecular functions, whereas MaNAC28 predominantly affects molecular functions [81].
CaNAC064 in chilli responds to cold stress by altering the levels of endophytic hormones, and overexpression and knockdown experiments showed that changes in the expression of CaNAC064 affected the tolerance and sensitivity of chilli to cold stress, respectively. It was shown that CaNAC064 was induced by temperature, SA and ABA, and interacted with low-temperature-induced haploid proteases [82].
Furthermore, plants enhance their resilience to cold damage by modifying lignin composition. The NAC transcription factor EjXND1 interacts with the lignin activator EjHB1, thereby attenuating the activation of the EjPRX12 promoter by EjHB1 and increasing the lignification of loquat fruit [83]. Ge Hang discovered that EjNAC3 is capable of activating the downstream lignin metabolism structural gene EjCAD-like in loquat, thereby altering cold tolerance [84]. SsNAC23, one of the most highly expressed NAC genes in sugarcane, is induced at 4 °C but remained unchanged at 12 °C. It was demonstrated that SsNAC23 mRNA levels were restored to their original levels within three hours in seedlings that were transferred to 26 °C after 48 h of cold treatment [85].
In conclusion, NAC transcription factors are crucial for the plant cold stress response, exerting their influence through a multitude of regulatory mechanisms. These include the alteration of hormone levels, the binding to stress-related genes, and the interaction with other transcription factors.
4.2.2. Drought Stress
Drought, a natural disaster caused by water scarcity, is the most common stress encountered by plants during the growth cycle of crops. Compared to other forms of natural disasters, drought is distinguished by its gradual onset and extensive impact. The phenomenon of global warming has resulted in an increase in the number of instances of drought-induced reduction in food production, which has had a significant impact on global food security. As evidenced in the Sixth Assessment Report, the global average surface temperature has increased by approximately 1 °C since the advent of the Industrial Revolution, as reported by the IPCC (Intergovernmental Panel on Climate Change), a joint initiative of the World Meteorological Organization and the United Nations Environment Programme. It is projected that the average temperature will increase by approximately 1.5 °C over the next 20 years [86]. It has been demonstrated that global wheat production is constrained by water scarcity. The findings indicate that a 40% deficit in precipitation results in a 20.6% decline in yield in arid and semiarid regions, which collectively account for approximately 43% of the total cultivated area [86].
Drought tolerance is the result of a complex interplay between multiple metabolic pathways, which are regulated by a network of genes and influenced by various external factors, including synthetic hormones, signal transduction, and plant physiological regulation [87,88]. To date, numerous NAC family members associated with drought tolerance have been identified in wheat, including TaNAC2a, TaNAC29, TaNAC48, and TaNAC071-A. The overexpression of TaNAC48 was observed to positively regulate drought tolerance in wheat seedlings through the ABA signalling pathway, as demonstrated by the control variable method [13,89]. Additionally, evidence indicates that OsNAC6 is a positive regulator of plant response to drought stress. Overexpression of OsNAC6 in rice plants has been shown to enhance tolerance to multiple stresses, including drought, high salinity, and rice blast [27]. While TdNACB was most closely related to TaNAC48 and OsNAC6, this suggests that TdNACB may be involved in the drought response. Subsequently, Gong et al. (2024) [90] overexpressed TdNACB and found that it could improve drought tolerance but not salt tolerance in rice. This indicates that TdNACB is a novel regulator that can enhance drought tolerance in wheat [27,90].
In addition to wheat, it has been established that NAC transcription factors participate in the response of various crops, including cotton, maize, and tomato, to environmental stress conditions, specifically drought. For example, GhNAC3, which is found in cotton, was demonstrated by protein structure analysis to possess a conserved NAC structural domain that is homologous to several stress-related NAC transcription factors [91]. Furthermore, overexpression of GhNAC3 was observed to enhance drought tolerance in Arabidopsis, while simultaneously decreasing sensitivity to ABA. Additionally, under drought stress, the germination rate of GhNAC3-overexpressing Arabidopsis transgenic plants demonstrated increased germination and longer roots [91]. In contrast, Kai Fan et al. (2024) [92] identified ZmNAC55 in maize, and observed that drought stress induced an up-regulation of ZmNAC55 expression levels in leaves. The overexpression of ZmNAC55 in Arabidopsis demonstrated that the plants exhibited heightened sensitivity to drought conditions, characterized by elevated water loss and a reduced survival rate. Conversely, the downregulation of ZmNAC55 in maize conferred enhanced drought tolerance. In other words, ZmNAC55 has been demonstrated to negatively regulate plant drought resistance [92]. The overexpression of SlNAC6 in tomato has been observed to increase ABA levels in the plant, which in turn affects the expression of ripening-related genes, thereby accelerating fruit ripening. Conversely, the SlNAC6-RNAi lines exhibited heightened oxidative damage in response to drought stress relative to the WT. This was attributed to the comparatively diminished antioxidant enzyme activity and antioxidant enzyme gene transcripts in the SlNAC6-RNAi lines. Conversely, the SlNAC6-OE lines demonstrated markedly elevated antioxidant enzyme activity and gene expression levels compared to the WT. These observations collectively indicate that SlNAC6 plays a pivotal role in tomato plants’ adaptive response to drought stress [93].
The FtNAC31 transcription factor, which is found in buckwheat, has been shown to enhance plant resistance to salt and drought stress when it is overexpressed in Arabidopsis. This result suggests that FtNAC31 may enhance drought resistance in plants in an ABA-dependent manner [94]. Furthermore, the NAC transcription factor PwNAC1 in spruce was observed to interact with the RNA-binding protein PwRBP1, thereby promoting plant salt and drought resistance. This interaction was hypothesized to positively regulate plant tolerance to drought and salt stresses, potentially through the ABA-dependent CBF pathway [20].
LpNAC13, a NAC transcription factor isolated from lily bulbs, demonstrated reduced drought tolerance but increased salt tolerance when overexpressed in tobacco. LpNAC13 overexpression plants exhibited reduced chlorophyll content, decreased antioxidant enzyme activity, and increased MDA content under drought conditions, whereas the results were opposite to those observed under salt stress conditions. This indicates that LpNAC13 functions as a negative regulator of the drought response in tobacco, while acting as a positive regulator of the salt response [95].
The overexpression of the apple NAC transcription factor, MdNAC1, has enhanced drought tolerance in transgenic apple plants. This is evidenced by an enhanced photosynthesis capability and an increased activity of ROS-scavenging enzymes in plant leaves. These findings suggest that MdNAC1 may positively regulate drought stress tolerance by maintaining photosynthetic activity and upregulating ROS-scavenging enzyme activity [14]. In banana, phylogenetic analyses identified 15 NAC genes associated with abiotic stresses. It was observed that the expression of the majority of the selected genes increased in conjunction with rising drought severity under both drought and high-temperature conditions. Of these, MaNAC100 and MaNAC136 were the most responsive to drought conditions [96].
The enhancement of drought resilience in crops, including fruits and vegetables, has constituted a significant area of investigation. The involvement of NAC transcription factors in related crops indicates that they play a pivotal role in the regulation of plant drought tolerance. For example, the overexpression of buckwheat FtNAC31 in Arabidopsis enhanced plant tolerance to salt and drought stress, which may be achieved through an ABA-dependent pathway [94]. The interaction of PwNAC1 with the RNA-binding protein PwRBP1 in spruce enhanced plant resistance to salt and drought, which may be positively regulated through the ABA-dependent CBF pathway [20]. The overexpression of LpNAC13 of lily bulbs in tobacco resulted in a reduction in drought tolerance but an enhancement in salt tolerance, indicating that LpNAC13 may play distinct roles under disparate stresses [95]. The overexpression of MdNAC1 in apple improved drought tolerance in transgenic apple plants by increasing photosynthetic capacity and upregulating ROS-scavenging enzyme activity [14]. The expression of NAC genes in banana was found to be altered under conditions of drought and high-temperature stress. Of particular note was the pronounced response of MaNAC100 and MaNAC136 to drought, which served to reinforce the crucial role of NAC transcription factors in the context of abiotic stress [96].
4.2.3. Salt Stress
NAC transcription factors also play a pivotal role in the process of coping with salt stress. Salt stress represents a significant abiotic stress factor that impedes crop growth and yield, thereby reducing crop yield by affecting physiological processes in plants. The global increase in saline soils represents a significant challenge to agricultural production. It has been demonstrated that NAC transcription factors regulate the plant’s response to salt stress through a number of different mechanisms.
In the case of Populus trichocarpa, the overexpression of PtNAC101 resulted in a reduction in plant salt tolerance, a decrease in lignin deposition, and a reduction in the capacity to scavenge ROS. This was achieved by inhibiting antioxidant enzyme activity [25]. The overexpression of ThNAC13 in Tamarix was observed to enhance the plant’s tolerance to salt stress by reducing ROS accumulation and enhancing antioxidant enzyme activities [97]. Overexpression of MpNAC1 in Millettia pinnata enhanced salt and drought tolerance in Arabidopsis and rice, and improved stress tolerance by upregulating stress-responsive genes and activating flavonoid biosynthesis [98]. The overexpression of ZmNAC84 in maize has been demonstrated to enhance salt stress tolerance by regulating ZmCAT1 expression, indicating that ZmNAC84 plays a positive role in regulating salt stress tolerance in plants [99].
In addition to modifying antioxidant enzyme activities, NAC transcription factors also enhance plant salt resistance by activating salt stress-related genes and inducing salt-resistant proteins. For example, the overexpression of NAC family members, including MbNAC25, PgNAC2, CaNAC46 and OsNAM2, has been evidenced to enhance salinity tolerance through the regulation of genes involved in ion transport and osmotic pressure regulation, as well as in antioxidant defences [100,101,102]. Similarly, overexpression of OsNAM2 in Arabidopsis has been demonstrated to facilitate the maintenance of ion homeostasis and the accumulation of key metabolites [103]. Overexpression of GhNAC4 has been observed to increase the expression of several stress-responsive marker genes and enhance salinity and drought resistance in transgenic tobacco [104]. SNAC1 in rice demonstrated enhanced tolerance to drought and salinity, as well as increased sensitivity to ABA, following overexpression in wheat [103,105]. Overexpression of OsNAC041 influenced the salt tolerance of plants and seed germination under salt stress, indicating that OsNAC041 plays a pivotal role in regulating salt tolerance [106].
4.2.4. Heavy Metal Stress
Soil heavy metal contamination represents a significant global environmental challenge, with the potential to negatively impact plant growth and human health. NAC transcription factors have been identified as crucial regulators of plant responses to heavy metal stress, enhancing tolerance through the modulation of diverse biological processes.
In rice, the overexpression of OsNAC300 was observed to enhance the tolerance of transgenic rice to Cd stress, whereas the knockdown of OsNAC300 resulted in increased sensitivity to Cd, confirming the critical role of OsNAC300 in the Cd stress response [76]. The SmNAC1 transcription factor in Salvia miltiorrhiza was observed to enhance the accumulation of cadmium in transgenic A. thaliana, indicating that SmNAC1 plays a role in the response to cadmium stress [107]. The expression of AemNAC2 and AemNAC3 in Aegilops markgrafii was observed to be upregulated under conditions of Cd stress. The overexpression of AemNAC2 resulted in a reduction in Cd concentration in wheat, thereby demonstrating its role in Cd stress [108]. The overexpression of ANAC004 in A. thaliana resulted in increased sensitivity to Cd and elevated Cd accumulation in roots and shoots, indicating that NAC004 plays a role in the Cd stress response [109].
In addition to Cd, NAC transcription factors have been demonstrated to regulate plant resistance to other heavy metals, including Zn, Pb and Al. The expression of AcNRZ1 in kiwifruit was found to increase in response to zinc stress, accompanied by a relocation to the nucleus from the endoplasmic reticulum (ER). It was demonstrated that the overexpression of AcNRZ1 enhanced Arabidopsis tolerance to zinc stress. The localization of SiNAC004 and SiNAC120 in willow to the nucleus was observed under lead stress, indicating their potential involvement in the lead stress response [110]. The expression of ANAC017 was found to be repressed under aluminium stress, while A. thaliana with a knockdown of ANAC017 demonstrated enhanced Al tolerance, suggesting that ANAC017 plays a positive role in regulating the Al stress response [111]. The expression of NAC32 was induced in response to nickel stress, and it was observed to reduce nickel compartmentalization in root cell vesicles by suppressing IREG2 expression [112].
5. Summary and Outlook
NAC transcription factors are one of the largest families of transcription factors in plants and have a major impact on a variety of plant growth processes. These processes include plant growth and development, responses to biotic and abiotic stresses, and many other biological functions. In this paper, we have elucidated the mechanism and rationale for the involvement of NAC transcription factors in stress response by synthesizing recent research advances in plant growth and development as well as stress.
The NAC family of transcription factors dominates the morphogenesis and tissue specialization of organs such as roots, stems, and leaves by coordinating the cell division and differentiation programs, and integrates light signals, endogenous hormones, and metabolic feedback in the regulation of developmental processes, controlling the key nodes such as leaf senescence, floral organ differentiation, and fruit ripening. In addition, NAC also dynamically regulates tissue mechanical strength and material transport efficiency by activating secondary metabolic pathways (lignin synthesis) and cell wall remodelling-related genes. The study demonstrated that NAC-mediated developmental regulation has a high degree of environmental plasticity, and its functional network realizes the dynamic balance between growth and development and ecological adaptation through the cross-interaction with stress-responsive signals (Figure 2).
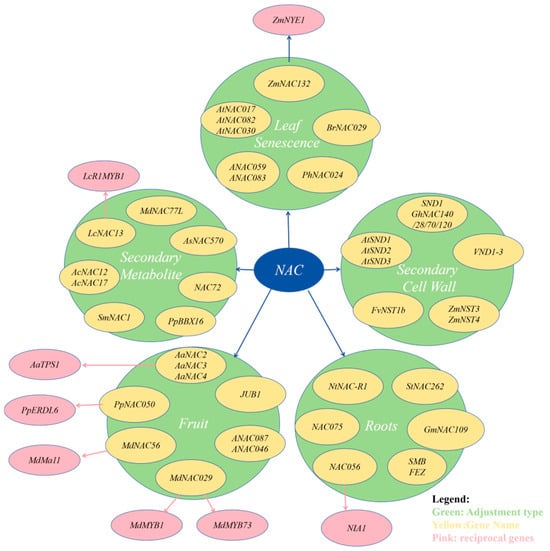
Figure 2.
Involvement of NAC transcription factors in plant growth and development.
The NAC transcription factor plays a central regulatory role in the plant’s response to biotic and abiotic stresses. It is widely involved in various response mechanisms of plants by regulating many genes. In terms of regulating biotic stress mechanisms, NAC transcription factors are affected by a range of biotic stresses in different plant species, including leaf spot, bacterial wilt, black mould and soft rot. NAC transcription factor can enhance or weaken the plant’s defence capacity by regulating the salicylic acid (SA) or jasmonic acid (JA) signalling pathway, secondary cell wall (SCW) synthesis, and other mechanisms (Table 2) [35,113,114,115,116,117,118,119].

Table 2.
Modes of regulation of NAC family members involved in biotic stress in different plants.
In terms of abiotic stresses, NAC transcription factors have been observed to respond to drought stress by maintaining photosynthesis and regulating phytohormones. Similarly, they also respond to salt stress by altering antioxidant enzyme activities, activating salt stress-related genes and regulating phytohormones. In addition, under heavy metal stress, NAC transcription factors were affected not only by cadmium stress, but also by heavy metal stresses such as zinc and lead, suggesting that they have a wide range of responses and involvement in plant stress tolerance (Table 3).

Table 3.
Involvement of NAC transcription factors in abiotic stresses.
Since the discovery of the first plant NAC (NAM, ATAF1/2, CUC2) transcription factor in 1996, significant progress has been made in the functional elucidation of this family member as a central regulator of plant growth, development and response to stress. Studies have shown that NAC transcription factors dynamically regulate the expression of related genes by integrating endogenous hormone signals with exogenous environmental stresses, thereby altering the functions of plant root development, leaf senescence, fruit ripening, secondary metabolite synthesis, response to stress, and other related functions.
However, most results have focused on model species such as Arabidopsis thaliana and rice, while the functional diversity of NAC in food crops and species adapted to extreme environments has not been fully explored, and the genetic basis and regulatory network of its evolution remain to be analysed.
The transcriptional regulation of NAC transcription factors is complex, and their interactions with downstream target genes depend on specific cis-acting elements and coactivators; even though they combine the same core NAC recognition sequences, they function differently [122].
As research goes on, we will face new development directions and challenges, and the identification of individual gene functions can no longer meet the demand for in-depth exploration of complex regulatory mechanisms in plants [123].
Therefore, research related to NAC transcription factors needs to go beyond individual transcription factors to the related research of multiple interacting genes [124]. A combination of approaches, including genomics and proteomics, was used to elucidate the functional differences of stress-responsive NAC factors and their relationship in transcriptional control at different developmental stages and under stress conditions [125,126].
NAC transcription factors play a pivotal role in the core of plant adaptive regulation, and the integration of knowledge and methods from different disciplines will help to further deepen the research related to NAC transcription factors.
Author Contributions
C.Z.: Conceptualization, Investigation, Writing—Original Draft, Writing—Review and Editing. Q.Y.: Data Curation, Formal Analysis, Visualization. X.W.: Data Curation, Formal Analysis. Y.C.: Methodology, Software, Validation. R.H.: Investigation, Resources, Data Curation. X.L.: Formal Analysis, Visualization, Writing—Original Draft. H.P.: Project Administration, Resources. R.Z.: Supervision, Funding Acquisition. T.Q.: Conceptualization, Supervision, Project Administration, Writing—Review and Editing (Corresponding Author). W.Q.: Conceptualization, Methodology, Funding Acquisition, Writing—Review and Editing (Corresponding Author). All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (2021YFD2200201).
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Souer, E.; van Houwelingen, A.; Kloos, D.; Jos, M.; Ronald, K. The no apical meristem gene of Petunia is required for pattern formation in embryosand flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Addie, N.; Heidi, E.; Leila, L.; Karen, S. NAC transcription factors:structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar]
- Ernst, H.A.; Olsen, A.N.; Larsen, S. Structure of theconserved domain of ANAC, a member of the NAC family oftranscription factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.J.; Fan, H.; Chen, Y. Integrative omic and transgenic analyses reveal the positive effect of ultraviolet-B irradiation.on salvianolic acid biosynthesis through upregulation of SmNAC1. Plant J. 2020, 104, 781–799. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gao, Y.; Wan, W.; Shi, J.; Xv, C.; Ma, D.; Su, Y.; Yang, X. AsNAC570 Gene Cloning Analysis and Requlates Alin Biosynthesis Preliminary Exploration. Mol. Plant Breed 2022, 1–8. Available online: http://kns.cnki.net/kcms/detail/46.1068.S.20220715.1426.004.html (accessed on 10 October 2024).
- Jiang, T.; Zhang, Y.; Zuo, Z.; Luo, T.; Wang, H.; Zhang, R.; Luo, Z. Transcription factor PgNAC72 activates DAMMARENEDIOL SYNTHASE expression to promote ginseng saponin biosynthesis. Plant Physiol. 2024, kiae202. [Google Scholar] [CrossRef]
- Li, G.; Li, S.; Liu, W. Transcription factor NAC and its role in plant growth and development. Mol. Plant Breed. 2019, 17, 811–826. [Google Scholar] [CrossRef]
- Wang, F.; Sun, L.; Zhao, X.; Wang, J.; Song, X. Research progress of plant NAC transcription factors. Biotechnol. Bull. 2019, 35, 88–93. [Google Scholar] [CrossRef]
- Zhao, C.; Huang, X.-Q.; Yin, F.-Y. Progress of rice NAC transcription factor family. J. Plant Sci. 2020, 38, 278–287. [Google Scholar]
- Liang, J.; Liu, X.; Xu, L.; Mu, R.; Shen, N.; Li, S.; Cheng, C.; Ren, Y.; Ma, L.; Wang, B.; et al. A novel NAC transcription factor from Haloxylon ammodendron promotes reproductive growth in Arabidopsis thaliana under drought stress. Environ. Exp. Bot. 2024, 228 Pt A, 106043. [Google Scholar] [CrossRef]
- Park, S.R.; Jeong, Y.; Son, S. Functions of transcription factor superfamilies in rice immunity. Crop J. 2025, 13, 5–22. [Google Scholar] [CrossRef]
- Summat, P.; Tongmark, K.; Chakhonkaen, S.; Sangarwut, N.; Panyawut, N.; Pinsupa, S.; Thanananta, T.; Muangprom, A. Investigating cold tolerance mechanisms in rice seedlings: Alternative splicing, promoter analysis, and their applications for marker development. Plant Stress 2024, 13, 100530. [Google Scholar] [CrossRef]
- Chen, J.; Gong, Y.; Gao, Y.; Zhou, Y.; Chen, M.; Xu, Z.; Guo, C.; Ma, Y. TaNAC48 positively regulates drought tolerance and ABA responses in wheat (Triticum aestivum L.). Crop J. 2021, 9, 9. [Google Scholar] [CrossRef]
- Jia, D.; Jiang, Q.; van Nocker, S.; Ma, F. An apple (Malus domestica) NAC transcription factor enhances drought tolerance in transgenic apple plants. Plant Physiol. Biochem. 2019, 139, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Gong, C. Progress in the study of biological functions of plant NAC transcription factors. Henan Sci. 2024, 1–13. [Google Scholar]
- Zhuo, M.; Wang, H. Regulatory role of NAC transcription factors in fruit ripening. J. Fruit Tree 2023, 40, 1455–1470. [Google Scholar] [CrossRef]
- Yan, S.; Wang, Y.; Yu, B.; Gan, Y.; Lei, J.; Chen, C.; Zhu, Z.; Qiu, Z.; Cao, B. A putative E3 ubiquitin ligase substrate receptor degrades transcription factor SmNAC to enhance bacterial wilt resistance in Eggplan. Hortic. Res. 2024, 11, uhad246. [Google Scholar] [CrossRef]
- Dastmalchi, K.; Chira, O.; Rodriguez, M.P.; Yoo, B.; Serra, O.; Figueras, M.; Stark, R. A chemical window into the impact of RNAi silencing of the StNAC103 gene in potato tuber periderms: Soluble metabolites, suberized cell walls, and antibacterial defense. Phytochemistry 2021, 190, 112885. [Google Scholar] [CrossRef]
- Ma, X.; Yin, Y.; Feng, J.X.; Chen, W.; Sun, L.; Xiao, Y. Progress in the study of plant NAC transcription factors. Plant Physiol. Lett. 2021, 57, 2225–2234. [Google Scholar] [CrossRef]
- Cui, X.; Cao, Y.; Zhang, H.; Zhang, L. A Picea wilsonii NAC transcription factor, PwNAC1, interacts with RNA-binding protein PwRBP1 and synergistically enhances drought and salt tolerance of transgenic Arabidopsis. Environ. Exp. Bot. 2023, 206, 105174. [Google Scholar] [CrossRef]
- Huang, Y.; Cui, L.; Chen, W.; Liu, Z.; Yuan, W.; Zhu, F.; Jiao, Z.; Zhang, X.; Deng, X.; Wang, L. Comprehensive analysis of NAC transcription factors and their expressions during taproot coloration in radish (Raphanus sativus L.). Sci. Hortic. 2022, 299, 111047. [Google Scholar] [CrossRef]
- Negi, S.; Tak, H.; Ganapathi, T. Native vascular related NAC transcription factors are efficient regulator of multiple classes of secondary wall associated genes in banana. Plant Sci. 2017, 265, 70–86. [Google Scholar] [CrossRef]
- Yang, X.; Kim, M.Y.; Ha, J.; Lee, S.H. Overexpression of the soybean NAC gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic Arabidopsis plants. Front. Plant Sci. 2019, 10, 1036. [Google Scholar] [CrossRef]
- Guo, F.; Siyuan, L.; Zhang, C.; Dong, T.; Meng, X.; Zhu, M. Genome-wide systematic survey and analysis of NAC transcription factor family and their response to abiotic stress in sweetpotato. Sci. Hortic. 2022, 299, 111048. [Google Scholar] [CrossRef]
- Qu, D.; Wu, F.; Yang, J.; Li, M.; Yang, L.; Xie, R.; Zhou, J.; Yang, J.; Wang, L.; Su, H.; et al. Transcription factor PtNAC101 negatively regulates the lignin synthesis and salt tolerance in Populus trichocarpa. Environ. Exp. Bot. 2023, 205, 105149. [Google Scholar] [CrossRef]
- Wang, H.; Qin, L.; Feng, C.; Wu, M.; Zhong, H.; Liu, J.; Wu, Q.; Que, Y. Pathogen resistance was negatively regulated by the NAC transcription factor ScATAF1 in sugarcane. Plant Physiol. Biochem. 2024, 213, 108828. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Chung, P.; Jeong, J.; Jang, G.; Bang, S.; Jung, H.; Kim, Y.; Ha, S.; Choi, Y.; Kim, J. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 2017, 15, 754–764. [Google Scholar] [CrossRef]
- Richa, S.; Lingaraj, S. Genome-wide analysis of cowpea NAC transcription factor family elucidating the genetic & molecular relationships that interface stress and growth regulatory signals. Plant Gene 2022, 31, 100363. [Google Scholar]
- Yuan, X.; Xu, J.; Yu, J.; Zhu, D.; Li, H.; Zhao, Q. The NAC transcription factor ZmNAC132 regulates leaf senescence and male fertility in maize. Plant Sci. 2023, 334, 111774. [Google Scholar] [CrossRef]
- Kang, C.; Guo, C.; Zhang, X.; Liu, Z. Genome-wide identification and analysis of the NAC gene family in walnut. J. Fruit Tree 2021, 38, 1444–1458. [Google Scholar]
- Nigarish, M.; Chen, Y.K.; Chen, X.H.; Muhammad, A.; Junaid, I.; Hafiz, M.; Shen, X.; Lin, Y.; Xu, X.; Lai, Z. Genome- wide identification and comprehensive analyses of NAC, transcription factor gene family and expression patterns during, somatic embryogenesis in Dimocarpus longan Lour. Plant Physiol. Biochem. 2020, 157, 169–184. [Google Scholar]
- Mohanta, T.K.; Yadav, D.; Khan, A. Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS ONE 2020, 15, e0231425. [Google Scholar] [CrossRef]
- Singh, S.; Koyama, H.; Bhati, K.K.; Alok, A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 2021, 134, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.G.; Jiang, S.H.; Zhang, T.L.; Xu, H.F.; Fang, H.C.; Zhang, J.; Su, M.Y.; Wang, Y.C.; Zhang, Z.Y.; Wang, N.; et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Lee, J.; Liu, Y.; Chen, H.; Hu, H. Distribution of heavy metal pollution in surface soil samples in China: A graphical review. Bull. Environ. Contam. Toxicol. 2016, 97, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, L.; Zhu, B.; Li, C. NAC transcription factors: Regulatory factors involved in many life processes in plants. Anhui Agric. Sci. 2024, 52, 22–29. [Google Scholar]
- Muskan, B.; Baljinder, S.; Avneesh, K.; Nandni, G.; Kashmir, S.; Ravindresh, C. Genome-wide identification of NAC transcription factors in Avena sativa under salinity stress. Plant Stress 2023, 10, 100276. [Google Scholar]
- Wang, J.; Zheng, C.; Shao, X.; Hu, Z.; Li, J.; Wang, P.; Wang, A.; Yu, J.; Shi, K. Transcriptomic and genetic approaches reveal an essential role of the NAC transcription factor SlNAP1 in the growth and defense response of tomato. Hortic. Res. 2020, 7, 209. [Google Scholar] [CrossRef]
- Zhao, X.; Xue, Z.; Liu, Y.; Huang, Z.; Sun, Y.; Wu, C.; Yan, S.; Kou, X. ChIP-seq revealed the role of tomato SNAC4 in response to biological and abiotic stresses and mediating auxin signaling response. Postharvest Biol. Technol. 2024, 209, 112696. [Google Scholar] [CrossRef]
- Xiang, C.; Gao, Q.; Yuan, C.; Zhang, X.; Liang, Y.; Zhao, Y. Identification and bioinformatics analysis of NAC transcription factor family in Calendula officinalis. Mol. Plant Breed. 2023, 12, 1518. [Google Scholar]
- Wang, Z.; Zhang, Z.; Wang, P.; Qin, C.; He, L.; Kong, L.; Ren, W.; Liu, X.; Ma, W. Genome-wide identification of the NAC transcription factors family and regulation of metabolites under salt stress in Isatis indigotica. Int. J. Biol. Macromol. 2023, 240, 124436. [Google Scholar] [CrossRef]
- Wang, Y.; Du, H.; Deng, P. Identification of NAC gene family in mulberry and analysis of expression pattern under rooting powder treatment. J. Fujian Agric. For. Univ. Nat. Sci. Ed. 2024, 53, 629–640. [Google Scholar] [CrossRef]
- Wan, F.; Gao, J.; Wang, G.; Niu, Y.; Wang, L.; Zhang, X.; Wang, Y.; Pan, Y. Genome-wide identification of NAC transcription factor family and expression analysis of ATAF subfamily members under abiotic stress in eggplant. Sci. Hortic. 2021, 289, 110424. [Google Scholar] [CrossRef]
- Liu, F.; Yang, C.; He, D.; Xing, B.; Li, M. Identification of pea NAC gene family and analysis of its response under drought stress. J. Agric. Biotechnol. 2024, 32, 1504–1517. [Google Scholar]
- Li, S.; Wang, C.; Li, X. Bioinformaticanalysis of the NAC transcription factor family in Punica granatum L. Mol. Plant Breed. 2021, 19, 88–99. [Google Scholar]
- Ribal, M.; Erzsébet, K. An integrative analysis of Vitis vinifera L. NAC Genes Response Botrytis Cinerea. Physiol. Mol. Plant Pathol. 2024, 131, 102247. [Google Scholar]
- Liu, H.; Jiang, B.; Zhang, J. Research progress of plant NAC transcription factors. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 2024, 1–12. [Google Scholar]
- Lin, S.; Longhui, L.; Jiang, Y.; Jing, Y. Advances in membrane-tethered NAC transcription factors in plants. Plant Sci. 2024, 342, 112034. [Google Scholar]
- Deng, Z.; Luo, L.; Yu, C.; Zhang, Q.; Sui, Y. Progress in the study of NAC transcription factors in ornamental plants. J. Plant Genet. Resour. 2024, 25, 737–750. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.H. Functional characterization of poplar wood-associated.NAC domain transcription factors. Plant Physiol. 2010, 152, 104–1055. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Long, G.; Wu, P.; Fu, J.; Lu, H.; Zhang, R. Progress of peroxidase-regulated lignin synthesis. Mod. Agric. Sci. Technol. 2021, 805, 47–49+54. [Google Scholar]
- Dang, X.; Zhang, B.; Li, C.; Nagawa, S. FvNST1b NAC protein induces secondary cell wall formation in strawberry. Int. J. Mol. Sci. 2022, 23, 13212. [Google Scholar] [CrossRef]
- Fang, S.; Shang, X.; Yao, Y. NST- and SND-sub group NAC proteins coordinately act to regulate second ary cell wall formation in cotton. Plant Sci. 2020, 301, 110657. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Yang, Y.; Yu, J. ZmNST3 and ZmNST4 are master switches for secondary wall deposition in maize (Zea mays L.). Plant Sci. 2018, 266, 83–94. [Google Scholar] [CrossRef]
- Yu, Y.; Ran, G.; Pi, L. Plant nitrogen signal perception and root development. J. Plant Sci. 2024, 42, 825–832. [Google Scholar]
- Meng, J.; Liu, H.; Wu, Z.; Zuo, W.; Wang, L. Effect of PsWOX11 gene on growth and development of lateral roots in Populus tremula. For. Sci. Res. 2024, 37, 54–61. [Google Scholar]
- Fu, Y.; Guo, H.; Cheng, Z.; Wang, R.; Li, G.; Huo, G.; Liu, W. NtNAC-R1, a novel NAC transcription factor gene in tobacco roots, responds to mechanical damage of shoot meristem. Plant Physiol. Biochem. 2013, 69, 74–81. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, L.; Zhang, N.; Yang, J.; Zhu, X.; Tang, X.; Calderon-Urrea, A.; Si, H. Lateral root development in potato is mediated by Stu-mi164 regulation of NAC transcription factor. Front. Plant Sci. 2018, 9, 383. [Google Scholar] [CrossRef]
- Viola, W.; Marion, B.; Tom, B.; Ana, C.; Harald, W.; Jian, X.; Jim, H.; Ben, S. The NAC Domain Transcription Factors FEZ and SOMBRERO Control the Orientation of Cell Division Plane in Arabidopsis Root Stem Cells. Dev. Cell 2008, 15, 913–922. [Google Scholar] [CrossRef]
- Xu, P.; Ma, W.; Hu, J.; Cai, W. The nitrate-inducible NAC transcription factor NAC056 controls nitrate assimilation and promotes lateral root growth in Arabidopsis thaliana. PLoS Genet. 2022, 18, e1010090. [Google Scholar] [CrossRef]
- Chenbo, D. Functional Analysis of MdNAC56 in the regulation of ACIDITY in Apple Fruit; Northwest Agriculture and Forestry University: Xianyang, China, 2024. [Google Scholar] [CrossRef]
- Xiao, H.; Hu, Y.; Wang, Y.; Cheng, J.; Wang, J.; Chen, G.; Li, Q.; Wang, S.; Wang, Y.; Wang, S. Nitrate availability controls translocation of the transcription factor NAC075 for cell-type-specific reprogramming of root growth. Dev. Cell 2022, 57, 2638–2651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Guo, P.R.; Xia, X.L.; Guo, H.W.; Li, Z.H. Multiple layers of regulation on leaf senescence: New advances and perspectives. Front. Plant Sci. 2021, 12, 788996. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Guo, Y. Research progress of leaf senescence related genes in tobacco. J. Plant Sci. 2018, 6, 87. [Google Scholar] [CrossRef]
- Wang, H.L.; Zhang, Y.; Wang, T.; Yang, Q.; Yang, Y.L.; Li, Z.; Li, B.S.; Wen, X.; Li, W.Y.; Yin, W.L.; et al. An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in Populus. Plant Cell 2021, 33, 1594–1614. [Google Scholar] [CrossRef]
- Li, F.; Shan, Y.; Wang, H.; Jiang, G.; Ding, X.; Liang, H.; Wang, C.; Kong, X.; Xie, L.; Jiang, Y. A NAC transcriptional factor BrNAC029 is involved in cytokinin-delayed leaf senescence in postharvest Chinese flowering cabbage. Food Chem. 2023, 404 Pt B, 134657. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.P.; Mueller-Roeber, B. ORS1, an H(2)O(2)-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef]
- Yang, S.D.; Seo, P.J.; Yoon, H.K.; Park, C.M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef]
- Trupkin, S.A.; Astigueta, F.H.; Baigorria, A.H.; García, M.N.; Delfosse, V.C.; González, S.A.; de la Torre, M.C.P.; Moschen, S.; Lía, V.V.; Fernández, P.; et al. Identification and expression analysis of NAC transcription factors potentially involved in leaf and petal senescence in Petunia hybrida. Plant Sci. 2019, 287, 110195. [Google Scholar] [CrossRef]
- Chang, W.J. Analysis of the Mechanism of MdNAC029 Regulating the Acidity of Apple Fruit; Northwest Agriculture and Forestry University: Xianyang, China, 2024. [Google Scholar] [CrossRef]
- Liu, J.; Jing, Y.; Liu, Y.; Xu, Y.; Yu, Y.; Ge, X.; Xie, H. Identification of peach NAC gene family and promotion of fruit fructose accumulation by PpNAC050. J. Hortic. 2024, 51, 1983–1996. [Google Scholar] [CrossRef]
- Nieuwenhuizen, N.J.; Chen, X.Y.; Wang, M.Y.; Matich, A.J.; Perez, R.L.; Allan, A.C.; Green, S.A.; Atkinson, R.G. Natural variation in monoterpene synthesis in kiwifruit: Transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol. 2015, 167, 1243–1258. [Google Scholar] [CrossRef]
- Wu, A.H.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.; Asensi-Fabado, M.; Munné-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 2, 482–506. [Google Scholar] [CrossRef]
- Huysmans, M.; Buono, R.A.; Skorzinski, N.; Radio, M.; De, W.; Parizot, B.; Mertens, J.; Karimi, M.; Fendrych, M.; Nowack, M. NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. Plant Cell 2018, 30, 2197–2213. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.; Yang, H.; Hu, R.; Wei, D.; Tang, Q.; Wang, Z. Role of NAC transcription factors in plant flower development. J. Biol. Eng. 2022, 38, 2687–2699. [Google Scholar]
- Cao, Y.; Hwarari, D.; Radani, Y.; Guan, Y.; Yang, L. Molecular Mechanism Underlying Plant Response to Cold Stress. Phyton-Int. J. Exp. Bot. 2023, 92, 2665–2682. [Google Scholar] [CrossRef]
- Li, S.; Ma, H.; Li, C. Progress in the study of bHLH transcription factors in response to low temperature stress in plants. Jiangsu Agric. Sci. 2024, 52, 11–19. [Google Scholar] [CrossRef]
- An, J.; Li, R.; Qu, F.; You, C.; Wang, X.; Hao, Y. An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 2018, 221, 74–80. [Google Scholar] [CrossRef]
- Shan, W. Mechanistic Analysis of NAC-Like Transcription Factors Regulating Banana fruit INDUCED Cold Tolerance and Ripening; South China Agricultural University: Guangzhou, China, 2016. [Google Scholar]
- Song, C. In-depth genome-wide characterization of MaNAC25 and MaNAC28 cold-responsive transcription factor binding sites in banana via DAP-Seq. Curr. Plant Biol. 2024, 40, 100389. [Google Scholar] [CrossRef]
- Hou, X.; Zhang, H.; Liu, S.; Wang, X.; Zhang, Y.; Meng, Y.; Luo, D.; Chen, R. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef]
- Liang, Z.; Shi, Y.; Huang, Y.; Lu, J.; Zhang, M.; Cao, X.; Hu, R.; Li, D.; Chen, W.; Zhu, C.; et al. YLEM NAC DOMAIN 1 (EjXND1) relieves cold-induced lignification by negatively regulating the EjHB1-EjPRX12 module in loquat fruit. J. Adv. Res. 2024, in press. [Google Scholar] [CrossRef]
- Ge, H. Transcription Factors NAC and MADS are Involved in the Regulation of Postharvest Loquat Fruit Lignification by Cold Damage; Zhejiang University: Hangzhou, China, 2018. [Google Scholar]
- Fábio, T.; Paulo, S.; Sandra, R.; Jorge, H.; Vicente, E.; Patrícia, P.; Paulo, A. SsNAC23, a member of the NAC domain protein family, is associated with cold, herbivory and water stress in sugarcane. Plant Sci. 2005, 169, 93–106. [Google Scholar]
- Zhaoqiang, Z.; Ping, W.; Linqi, L.; Qiang, F.; Yibo, D.; Peng, C.; Ping, X.; Tian, W.; Haiyun, S. Recent development on drought propagation: A comprehensive review. J. Hydrol. 2024, 645 Pt B, 132196. [Google Scholar]
- Du, L.; Huang, X.; Ding, L.; Wang, Z.; Tang, D.; Chen, B.; Ao, L.; Liu, Y.; Kang, Z.; Mao, H. TaERF87 and TaAKS1 synergistically regulate TaP5CS1/TaP5CR1-mediated proline biosynthesis to enhance drought tolerance in wheat. New Phytol. 2023, 237, 232–250. [Google Scholar] [CrossRef]
- Wang, J.; Li, C.; Hu, G.; Zhang, Y.; Reynolds, M.; Zhang, X.; Jia, J.; Mao, X.; Jing, R. DIW1 encoding a clade I PP2C phosphatase negatively regulates drought tolerance by de-phosphorylating TaSnRK1.1 in wheat. J. Integr. Plant Biol. 2023, 65, 1918–1936. [Google Scholar] [CrossRef]
- Mao, H.; Li, S.; Chen, B.; Jian, C.; Mei, F.; Zhang, Y.; Li, F.; Chen, N.; Li, T.; Du, L.; et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant 2022, 15, 276–292. [Google Scholar] [CrossRef]
- Gong, F.; Zhang, F.; Lu, Y.; Velu, G.; Liu, R.; Liu, J.; Wang, X.; Liu, D.; Zheng, Y.; Huang, L.; et al. Overexpression of TdNACB improves the drought resistance of rice. Plant Physiol. Biochem. 2024, 216, 109157. [Google Scholar] [CrossRef]
- Xia, L.; Sun, S.; Han, B.; Yang, X. NAC domain transcription factor gene GhNAC3 confers drought tolerance in plants. Plant Physiol. Biochem. 2023, 195, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Wu, Y.; Mao, Z.; Yin, K.; He, Y.; Pan, X.; Zhu, X.; Liao, C.; Cui, L.; Jia, Q.; et al. A novel NAC transcription factor ZmNAC55 negatively regulates drought stress in Zea mays. Plant Physiol. Biochem. 2024, 214, 108938. [Google Scholar] [CrossRef] [PubMed]
- Jian, W.; Zheng, Y.; Yu, T.; Cao, H.; Chen, Y.; Cui, Q.; Xu, C.; Li, Z. SlNAC6, A NAC transcription factor, is involved in drought stress response and reproductive process in tomato. J. Plant Physiol. 2021, 264, 153483. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, Q.; Wu, H.; Wang, A.; Wang, X.; Li, C.; Zhao, H.; Wu, Q. FtNAC31, a Tartary buckwheat NAC transcription factor, enhances salt and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2022, 191, 20–33. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, S.; Guan, C.; Kong, X.; Wang, Y.; Cui, Y.; Liu, B.; Zho, Y.; Zhang, Y. Overexpressing the NAC transcription factor LpNAC13 from Lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 2020, 149, 96–110. [Google Scholar] [CrossRef]
- Rakesh, S.; Bhavesh, L.; Abdul, A.; Bal, K.; Prafullachandra, V.; Aniruddha, P. Differential regulation of the banana stress NAC family by individual and combined stresses of drought and heat in susceptible and resistant genotypes. Plant Physiol. Biochem. 2019, 145, 184–194. [Google Scholar]
- Liu, R.; Meng, J.; Zuo, W.; Zuo, W.; Wang, L.; Sun, T. The phloem protein 2 (PP2) is positively regulated by ThNAC13 that enhances salt tolerance of Tamarix. Environ. Exp. Bot. 2024, 224, 105784. [Google Scholar] [CrossRef]
- Yang, H.; Yi, Z.; Shanwu, L.; Liu, Y.; Jian, S.; Deng, S. MpNAC1, a transcription factor from the mangrove associate Millettia pinnata, confers salt and drought stress tolerance in transgenic Arabidopsis and rice. Plant Physiol. Biochem. 2024, 211, 108721. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Han, T.; Xiang, Y.; Wang, C.; Zhang, A. The transcription factor ZmNAC84 increases maize salt tolerance by regulating ZmCAT1 expression. Crop J. 2024, 12, 1344–1356. [Google Scholar] [CrossRef]
- Gong, C.; Zheng, Y.; Pan, V. Salt tolerance analysis of Capsicum annuum CaNAC36. Northwest J. Botany 2024, 44, 593–601. [Google Scholar]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. An NAC transcription factor gene from Malus baccata, MbNAC29, increases cold and high salinity tolerance in Arabidopsis. Vitr. Cell. Dev. Biol. Plant 2020, 56, 588–599. [Google Scholar] [CrossRef]
- Li, P.; Peng, Z.; Xu, P.; Tang, G.; Ma, C.; Zhu, J.; Shan, L.; Wan, S. Genome-wide identification of NAC transcription factors and their functional prediction of abiotic stress response in peanut. Front. Genet. 2021, 12, 630292. [Google Scholar] [CrossRef]
- Harshita, J.; Klaus, H.; Leander, R.; Shashank, K.; Puneet, S. Elucidation of PGPR-responsive OsNAM2 regulates salt tolerance in Arabidopsis by AFP2 and SUS protein interaction. Microbiol. Res. 2024, 289, 127890. [Google Scholar] [CrossRef]
- Trishla, V.S.; Kirti, P.B. KirtiStructure-function relationship of Gossypium hirsutum NAC transcription factor, GhNAC4 with regard to ABA and abiotic stress responses. Plant Sci. 2021, 302, 110718. [Google Scholar] [CrossRef]
- Abu, S.; Li, X.; Li, H.; Huang, T.; Gao, C.; Guo, M.; Cheng, W.; Zhao, G.; Liao, Y. A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci. 2013, 203–204, 33–40. [Google Scholar]
- Wang, D.; Lliu, Z.; Lu, X.; Gao, Y.; Sun, S.; Guo, H.; Tian, W.; Wang, L.; Li, Z.; Li, L.; et al. Progress and prospect of salt tolerance mechanism in plants. North China J. Agric. 2024, 39, 80–92. [Google Scholar]
- Hu, S.; Shinwari, K.I.; Song, Y.; Xia, J.; Xu, H.; Du, B.; Luo, L.; Zheng, L. OsNAC300 positively regulates cadmium stress responses and tolerance in rice roots. Agronomy 2021, 11, 95. [Google Scholar] [CrossRef]
- Zhu, B.; Huo, D.A.; Hong, X.X.; Guo, J.; Peng, T.; Liu, J.; Huang, X.; Yan, H.; Weng, Q.; Zhang, X.; et al. The Salvia miltiorrhiza NAC transcription factor SmNAC1 enhances zinc content in transgenic Arabidopsis. Gene 2019, 688, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; He, F.; Zhu, B.; Ren, M.; Tang, H. NAC transcription factors from Aegilops markgrafii reduce cadmium concentration in transgenic wheat. Plant Soil 2020, 449, 39–50. [Google Scholar] [CrossRef]
- Meng, Y.; Zhang, X.; Wu, Q.; Fang, S.; Fang, Z. Transcription factor ANAC004 enhances Cd tolerance in Arabidopsis thaliana by regulating cell wall fixation, translocation and vacuolar detoxification of Cd, ABA accumulation and antioxidant capacity. J. Hazard. Mater. 2022, 436, 129121. [Google Scholar] [CrossRef]
- Xin, Y.; Huang, R.; Xu, M.; Xu, L. Transcriptome-Wide Identification and Response Pattern Analysis of the Salix integra NAC Transcription Factor in Response to Pb Stress. Int. J. Mol. Sci. 2023, 24, 11334. [Google Scholar] [CrossRef]
- Tao, Y.; Wan, J.X.; Liu, J.X.; Zheng, Y.; Fang, S.; Fang, Z. The NAC transcription factor ANAC017 regulates aluminum tolerance by regulating the cell wall-modifying genes. Plant Physiol. 2022, 189, 2517–2534. [Google Scholar] [CrossRef]
- Zheng, Y.; Lu, Y.; Feng, H.; Feng, J.; Chen, C.; Xu, C.; Li, H. Progress on the response mechanism of mustard to salt stress. Vegetables 2024, 17–23. [Google Scholar]
- Ou, X.; Sun, L.; Chen, Y.; Zhao, Z.; Jian, W. Characteristics of NAC transcription factors in Solanaceae crops and their roles in responding to abiotic and biotic stresses. Biochem. Biophys. Res. Commun. 2024, 709, 149840. [Google Scholar] [CrossRef]
- Wei, Y.; Tian, H. Progress of research on environmental heavy metal pollution. Mod. Agric. Sci. Technol. 2024, 104–106+120. [Google Scholar]
- Wang, Y.F. Research progress on the effects of heavy metal stress on alfalfa. Contemp. Anim. Husb. 2024, 45–47. [Google Scholar]
- Xu, S. Hazards of soil heavy metal pollution on agricultural cultivation and management strategy. Heilongjiang Grain 2024, 119–121. [Google Scholar]
- Angon, P.B.; Islam, M.S.; Das, A.; Anjum, N.; Poudel, A.; Suchi, S.A. Sources, effects and present perspectives of heavy metals contamination: Soil, plants and human food chain. Heliyon 2024, 10, e28357. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Tao, C.; Cao, L.; Liu, L.; Zheng, J.; Zhu, W.; Xiang, M.; Wang, L.; Liu, Y.; Zhang, C.; et al. AcNRZ1, an endoplasmic reticulum-localized NAC transcription factor in kiwifruit, enhances zinc stress tolerance in Arabidopsis. Plant Stress 2024, 13, 100559. [Google Scholar] [CrossRef]
- Wang, B.; Zhong, Z.; Zhang, H.; Wang, X.; Liu, B.; Yang, L.; Han, X.; Yu, D.; Zheng, X.; Wang, C. Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice. Rice Sci. 2019, 26, 98–108. [Google Scholar]
- Sun, L.; Zhang, P.; Li, W. The bifunctional transcription factor NAC32 modulates nickel toxicity responses through repression of root-nickel compartmentalization and activation of auxin biosynthesis. J. Hazard. Mater. 2024, 480, 135925. [Google Scholar] [CrossRef]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genom. 2010, 284, 173–183. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Chen, X.; Zhang, B.; Xin, Y.; Li, L.; Cao, S.; Liu, F.; Wang, Z.; Huang, H. A NAC Transcription Factor OsNAC3 Positively Regulates ABA Response and Salt Tolerance in Rice. BMC Plant Biol. 2021, 21, 546. [Google Scholar] [CrossRef]
- Chen, Y.; Xia, P. NAC transcription factors as biological macromolecules responded to abiotic stress: A comprehensive review. Int. J. Biol. Macromol. 2025, 308 Pt 1, 142400. [Google Scholar] [CrossRef]
- Kurowska, M.; Daszkowska-Golec, A. Molecular mechanisms of SNAC1 (Stress-responsive NAC1) in conferring the abiotic stress tolerance. Plant Sci. 2023, 337, 111894. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).