Genome-Wide Analysis of the POD Gene Family in Avena sativa: Insights into Lignin Biosynthesis and Responding to Powdery Mildew
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Identification of AsPOD Genes
2.3. Characterization of AsPOD Proteins, Gene Structure and Phylogenetic Analyses
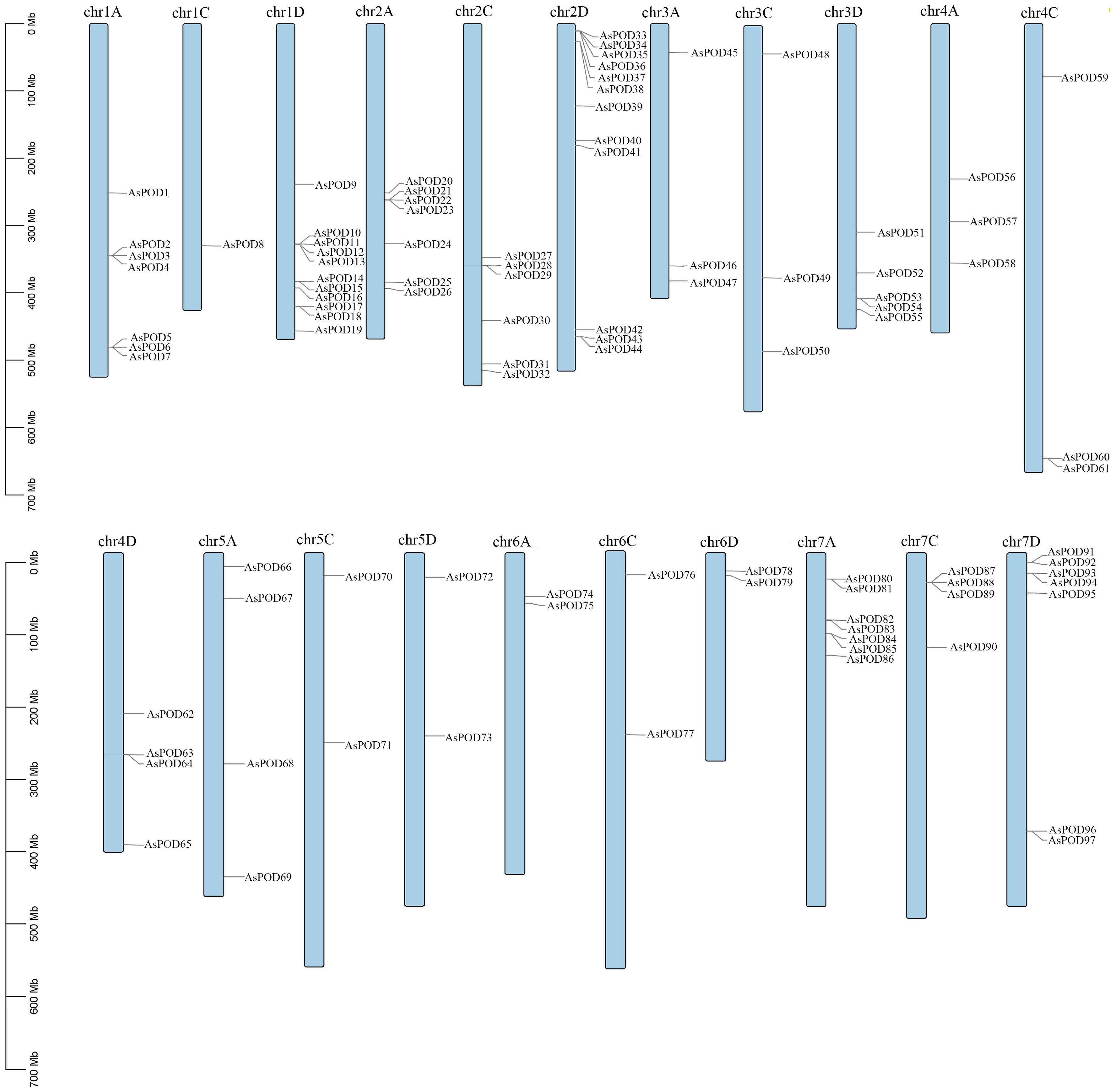
2.4. Cis-Acting Elements, Chromosome Distributions Analysis and Collinearity Analysis
2.5. Oat Growth Conditions and Blumeria graminis Infection
2.6. RNA Extraction and Quantitative PCR Analysis
2.7. Determination of Lignin Content and Correlation Analysis
2.8. Determination of H2O2 and POD Content
2.9. Data Analysis
3. Results
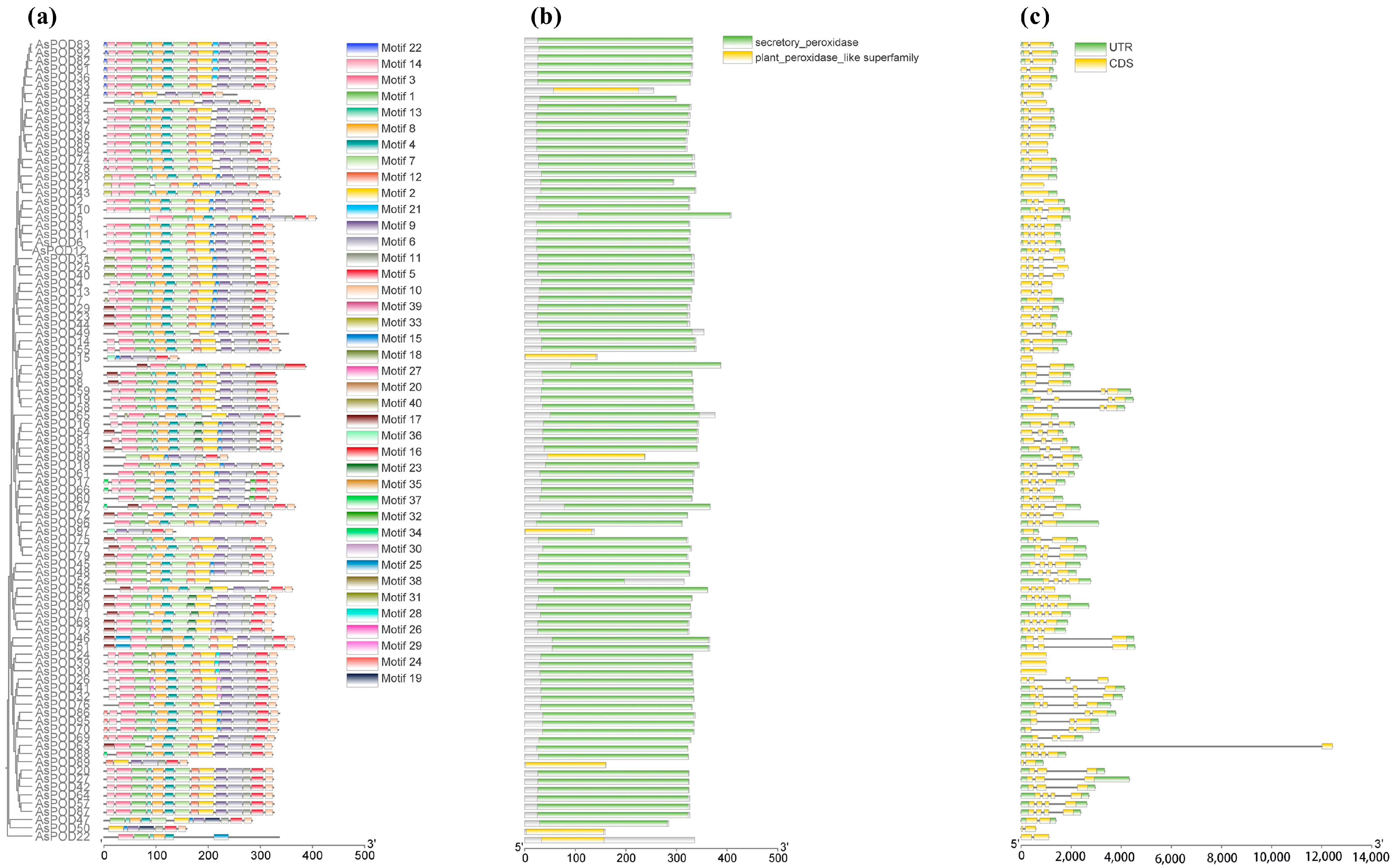
3.1. Identification and Physicochemical Properties of Oat POD Genes
3.2. Predictions of AsPOD Protein Secondary and Tertiary Structures
3.3. Conserved Motif Analysis of Oat AsPOD Proteins
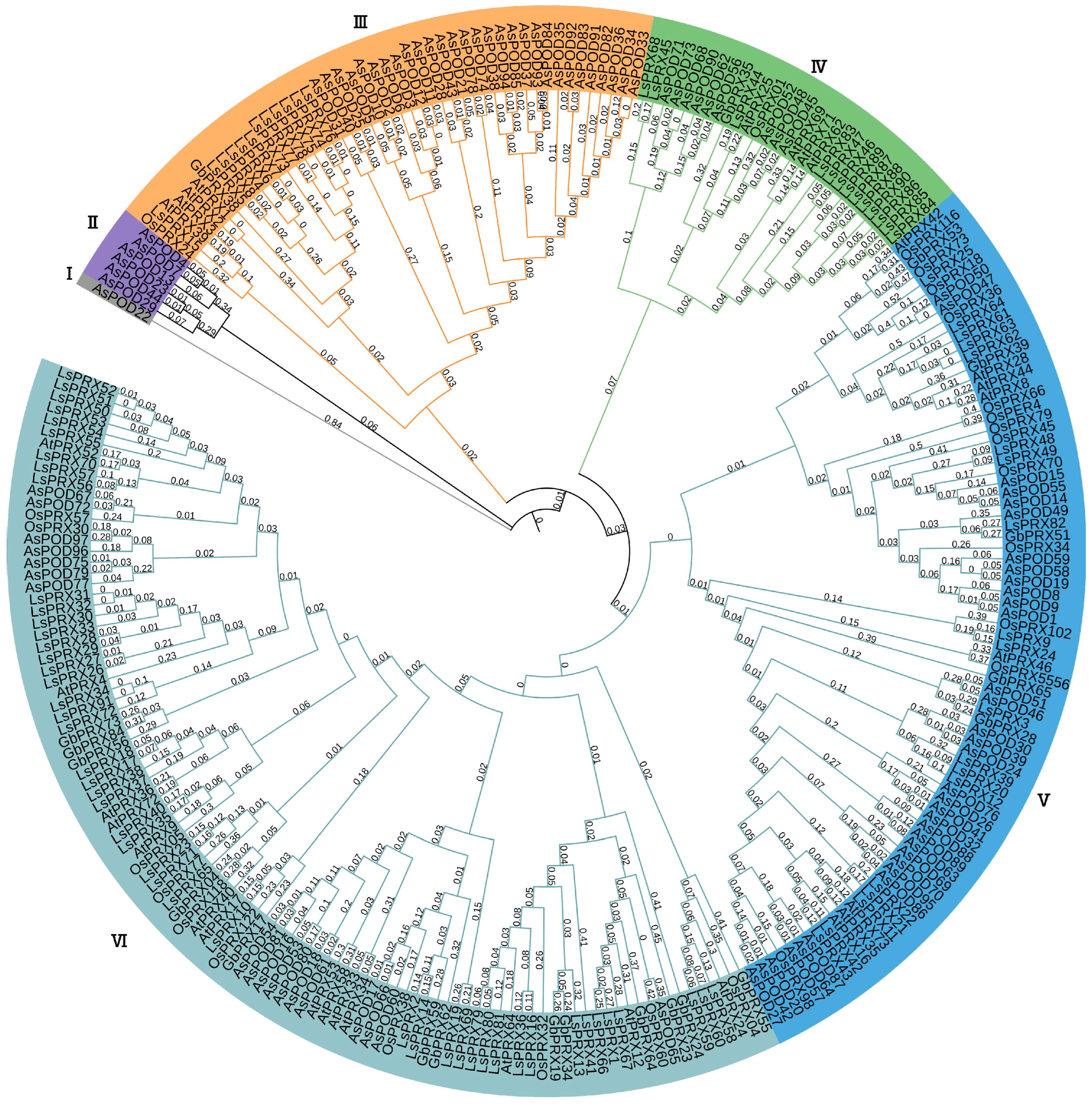
3.4. Phylogenetic Analysis of POD Proteins
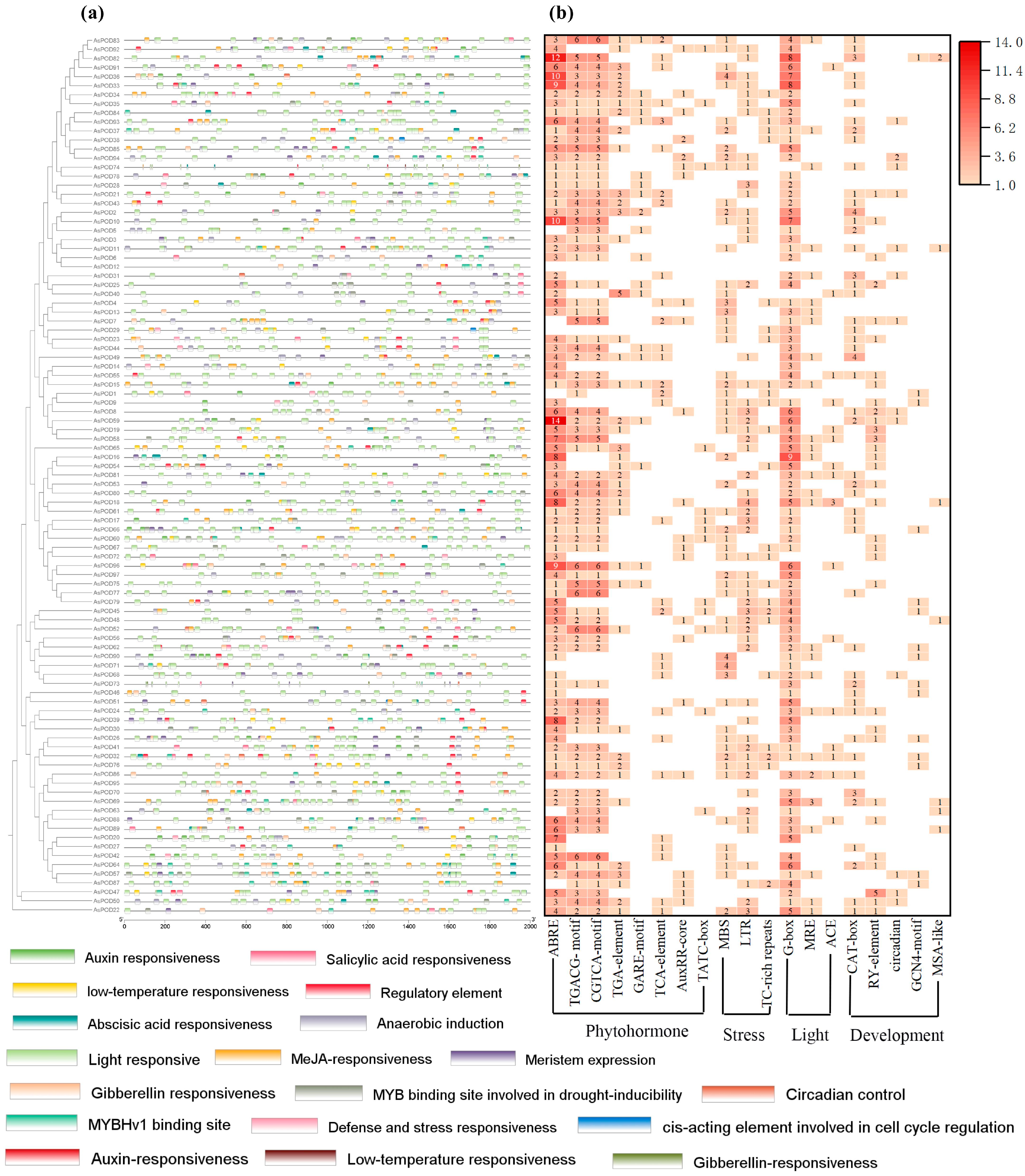
3.5. Identification of Cis-Acting Elements in AsPODs
3.6. GO and KEGG Pathway Enrichment Analysis of AsPODs
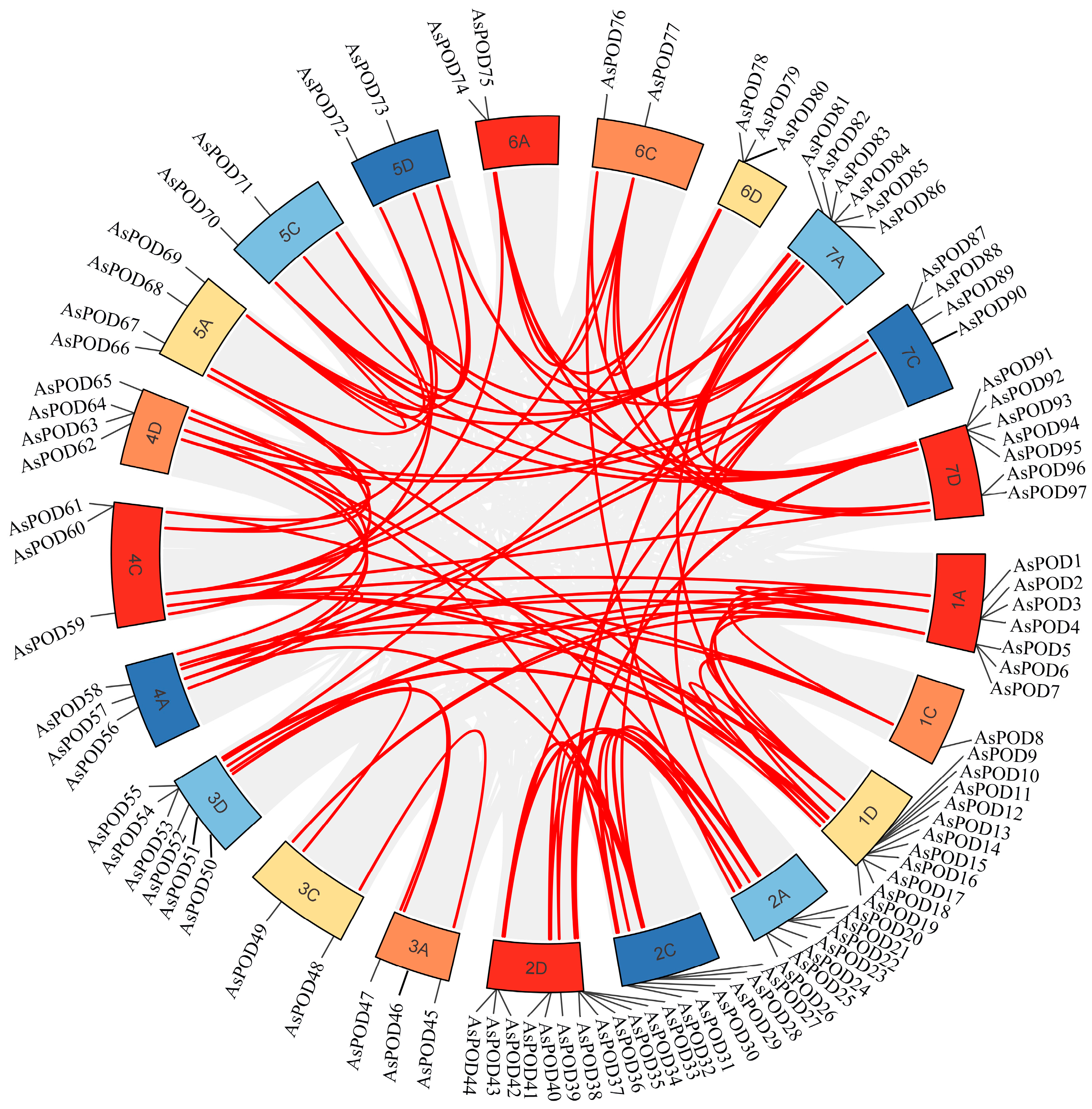
3.7. Synteny Analysis of POD Family Genes
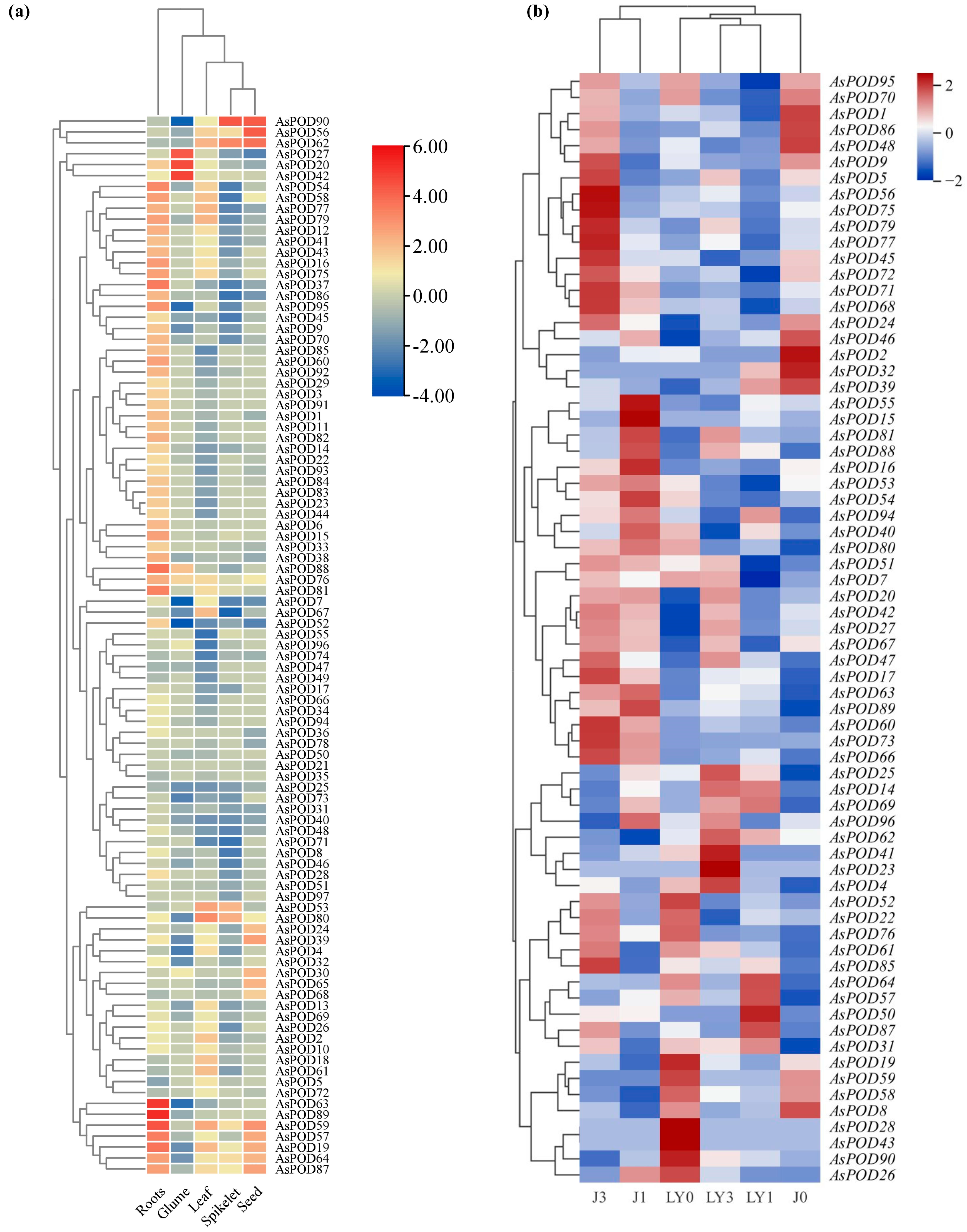
3.8. Expression Patterns of AsPOD Genes in Different Tissues
3.9. Response to Powdery Mildew
3.10. Changes in Lignin Content, H2O2 Content and POD Activity
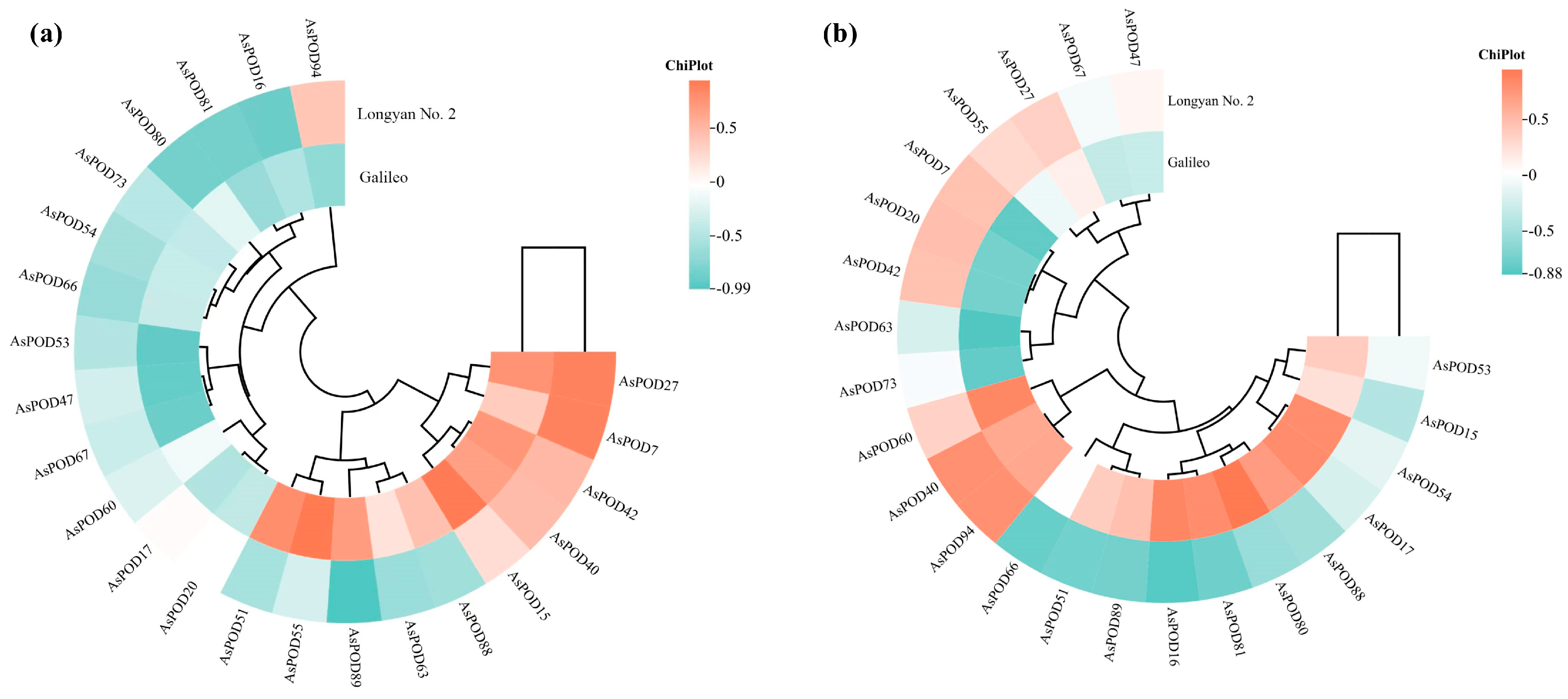
3.11. Correlation Between AsPOD Gene Expression and Lignin Content, POD Activity
4. Discussion
4.1. Class III Peroxidases and Their Roles in Plant Defense
4.2. Functional Insights into the Expression of AsPOD Genes and POD Activity, Lignin Content
4.3. Implications for the Roles of AsPOD Genes in Oat Disease Resistance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Youngs, V.L.; Forsberg, R.A. Oat. In Nutritional Quality of Cereal Grains: Genetic and Agronomic Improvement; American Society of Agronomy, Crop Science Society of America, Soil Science Society of America: Madison, WI, USA, 1987; Volume 28, pp. 457–499. [Google Scholar]
- Marshall, A.; Cowan, S.; Edwards, S.; Griffiths, I.; Howarth, C.; Langdon, T.; White, E. Crops That Feed the World 9. Oats—A Cereal Crop for Human and Livestock Feed with Industrial Applications. Food Secur. 2013, 5, 13–33. [Google Scholar] [CrossRef]
- Jackson, L.; Fernandez, B.; Meister, H.; Spiller, M.; Williams, J.; Kearney, T.; Marsh, B.; Wright, S.; Munier, D.; Pettygrove, G.S.; et al. Small Grain Production Manual; University of California Agriculture and Natural Resources: Oakland, CA, USA, 2006. [Google Scholar]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Wang, J.E.; Liu, K.K.; Li, D.W.; Zhang, Y.L.; Zhao, Q.; He, Y.M.; Gong, Z.H. A Novel Peroxidase CanPOD Gene of Pepper Is Involved in Defense Responses to Phytophthora capsici Infection as Well as Abiotic Stress Tolerance. Int. J. Mol. Sci. 2013, 14, 3158–3177. [Google Scholar] [CrossRef] [PubMed]
- Coego, A.; Ramirez, V.; Ellul, P.; Mayda, E.; Vera, P. The H2O2-Regulated Ep5C Gene Encodes a Peroxidase Required for Bacterial Speck Susceptibility in Tomato. Plant J. 2005, 42, 283–293. [Google Scholar]
- Choi, H.W.; Kim, Y.J.; Lee, S.C. Hydrogen Peroxide Generation by the Pepper Extracellular Peroxidase CaPO2 Activates Local and Systemic Cell Death and Defense Response to Bacterial Pathogens. Plant Physiol. 2007, 145, 890–904. [Google Scholar] [CrossRef]
- Neutelings, G. Lignin Variability in Plant Cell Walls: Contribution of New Models. Plant Sci. 2011, 181, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Florian, M.E. Deterioration of Organic Materials Other Than Wood. In Conservation of Marine Archaeological Objects; Butterworth-Heinemann: Oxford, UK, 1987; pp. 21–54. [Google Scholar]
- Andersen, E.J.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M.P. Disease Resistance Mechanisms in Plants. Genes 2018, 9, 339. [Google Scholar] [CrossRef]
- Kamal, N.; Tsardakas Renhuldt, N.; Bentzer, J.; Gundlach, H.; Haberer, G.; Juhász, A. The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 2022, 606, 113–119. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J. TBtools–II: A “one for all, all for one” bioinformatics platform for biological big–data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar]
- Jayhoon, A.S.; Kumar, P. In-silico characterization and expression analysis of the Waxy gene in rice (Oryza sativa L.). agriRxiv 2024, 20240292736. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R. SWISS–MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [PubMed]
- Bienert, S.; Waterhouse, A.; De Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS–MODEL Repository—New features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—Distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo–and hetero–oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 10480. [Google Scholar]
- Ghorbel, M.; Zribi, I.; Chihaoui, M.; Alghamidi, A.; Mseddi, K.; Brini, F. Genome-Wide Investigation and Expression Analysis of the Catalase Gene Family in Oat Plants (Avena sativa L.). Plants 2023, 12, 3694. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar]
- Pan, Y.; Niu, K.; Miao, P.; Zhao, G.; Zhang, Y.; Ju, Z.; Chai, J.; Yang, J.; Cui, X.; Zhang, R. Genome-wide analysis of the SWEET gene family and its response to powdery mildew and leaf spot infection in the common oat (Avena sativa L.). BMC Genom. 2024, 25, 995. [Google Scholar]
- Damgaard, M.V.; Treebak, J.T. Protocol for qPCR analysis that corrects for cDNA amplification efficiency. STAR Protoc. 2022, 3, 101515. [Google Scholar] [PubMed]
- Wang, Z.; Niu, K.; Zhao, G.; Zhang, Y.; Chai, J.; Ju, Z. Exogenous 24-epibrassinolide improves resistance to leaf spot disease through antioxidant regulation and phenylpropanoid metabolism in oats. Agronomy 2024, 14, 3035. [Google Scholar] [CrossRef]
- Chakrabarty, D.; Datta, S.K. Micropropagation of gerbera: Lipid peroxidation and antioxidant enzyme activities during acclimatization process. Acta Physiol. Plant. 2008, 30, 325–331. [Google Scholar]
- Muñoz-Muñoz, J.L.; García-Molina, F.; García-Ruiz, P.A.; Arribas, E.; Tudela, J.; García-Cánovas, F.; Rodríguez-López, J.N. Enzymatic and chemical oxidation of trihydroxylated phenols. Food Chem. 2009, 113, 435–444. [Google Scholar] [CrossRef]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 2002, 288, 129–138. [Google Scholar]
- Passardi, F.; Longet, D.; Penel, C.; Dunand, C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 2004, 65, 1879–1893. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar]
- Xiao, H.; Wang, C.; Khan, N.; Chen, M.; Fu, W.; Guan, L.; Leng, X. Genome-wide identification of the class III POD gene family and their expression profiling in grapevine (Vitis vinifera L.). BMC Genom. 2020, 21, 444. [Google Scholar]
- Wu, C.; Ding, X.; Ding, Z.; Tie, W.; Yan, Y.; Wang, Y.; Yang, H.; Hu, W. The class III peroxidase (POD) gene family in cassava: Identification, phylogeny, duplication, and expression. Int. J. Mol. Sci. 2019, 20, 2730. [Google Scholar] [CrossRef]
- Yan, J.; Su, P.; Li, W.; Xiao, G.; Zhao, Y.; Ma, X.; Wang, H.; Nevo, E.; Kong, L. Genome-wide and evolutionary analysis of the class III peroxidase gene family in wheat and Aegilops tauschii reveals that some members are involved in stress responses. BMC Genom. 2019, 20, 666. [Google Scholar]
- Fernández-Pérez, F.; Pomar, F.; Pedreño, M.A.; Novo-Uzal, E. The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiol. Plant. 2015, 154, 395–406. [Google Scholar] [CrossRef]
- Fernández-Pérez, F.; Vivar, T.; Pomar, F.; Pedreño, M.A.; Novo-Uzal, E. Peroxidase 4 is involved in syringyl lignin formation in Arabidopsis thaliana. J. Plant Physiol. 2015, 175, 86–94. [Google Scholar] [CrossRef]
- Herrero, J.; Fernández-Pérez, F.; Yebra, T.; Novo-Uzal, E.; Pomar, F.; Pedreño, M.Á.; Cuello, J.; Guéra, A.; Esteban-Carrasco, A.; Zapata, J.M. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 2013, 237, 1599–1612. [Google Scholar] [PubMed]
- Valério, L.; De Meyer, M.; Penel, C.; Dunand, C. Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry 2004, 65, 1331–1342. [Google Scholar]
- Jameson, P.E. Cytokinin Translocation to, and Biosynthesis and Metabolism within, Cereal and Legume Seeds: Looking Back to Inform the Future. Metabolites 2023, 13, 1076. [Google Scholar] [CrossRef]
- Jia, W.; Ma, M.; Chen, J.; Wu, S. Plant morphological, physiological and anatomical adaption to flooding stress and the underlying molecular mechanisms. Int. J. Mol. Sci. 2021, 22, 1088. [Google Scholar] [CrossRef] [PubMed]
- Almagro, L.; Gómez Ros, L.V.; Belchi-Navarro, S.; Bru, R.; Ros Barceló, A.; Pedreño, M.A. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2009, 60, 377–390. [Google Scholar]
- Zhang, Y.; Zhang, Y.; Gao, C.; Zhang, Z.; Yuan, Y.; Zeng, X.; Hu, W.; Yang, L.; Li, F.; Yang, Z. Uncovering genomic and transcriptional variations facilitates utilization of wild resources in cotton disease resistance improvement. Theor. Appl. Genet. 2023, 136, 204. [Google Scholar] [CrossRef]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef]
- Cosio, C.; Dunand, C. Specific functions of individual class III peroxidase genes. J. Exp. Bot. 2009, 60, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Hilaire, E.; Young, S.A.; Willard, L.H.; McGee, J.D. Vascular defense responses in rice: Peroxidase accumulation in xylem parenchyma cells and xylem wall thickening. Mol. Plant-Microbe Interact. 2001, 14, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Baştaş, K. Importance of reactive oxygen species in plants-pathogens interactions. Selçuk J. Agric. Food Sci. 2015, 28, 11–21. [Google Scholar]
- Lin, H.H.; Lin, K.H.; Tsai, Y.L.; Chen, R.J.; Lin, Y.C.; Chen, Y.C. Influences of Ipomoea batatas Anti-Cancer Peptide on Tomato Defense Genes. Curr. Protein Pept. Sci. 2024, 25, 651–665. [Google Scholar] [CrossRef]
- Ge, H.; Wang, M.; Wei, X.; Chen, X.L.; Wang, X. Copper-Based Nanozymes: Potential Therapies for Infectious Wounds. Small 2025, 2407195. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Magwanga, R.O.; Cai, X.; Lu, P.; Kirungu, J.N.; Zhou, Z.; Wang, X.; Wang, X.; Xu, Y.; Hou, Y.; et al. RNA-sequencing, physiological and RNAi analyses provide insights into the response mechanism of the ABC-mediated resistance to Verticillium dahliae infection in cotton. Genes. 2019, 10, 110. [Google Scholar] [CrossRef]
- Wu, F.; Chi, Y.; Jiang, Z.; Xu, Y.; Xie, L.; Huang, F.; Wan, D.; Ni, J.; Yuan, F.; Wu, X.; et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 2020, 578, 577–581. [Google Scholar] [CrossRef]
- Sánchez-Martín, J.; Montilla-Bascón, G.; Mur, L.A.J.; Rubiales, D.; Prats, E. Compromised photosynthetic electron flow and H2O2 generation correlate with genotype-specific stomatal dysfunctions during resistance against powdery mildew in oats. Front. Plant Sci. 2016, 7, 1660. [Google Scholar] [CrossRef]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef]
- Kuźniak, E.; Urbanek, H. The involvement of hydrogen peroxide in plant responses to stresses. Acta Physiol. Plant. 2000, 22, 195–203. [Google Scholar] [CrossRef]
- Li, D.; Limwachiranon, J.; Li, L.; Zhang, L.; Xu, Y.; Fu, M.; Luo, Z. Hydrogen peroxide accelerated the lignification process of bamboo shoots by activating the phenylpropanoid pathway and programmed cell death in postharvest storage. Postharvest Biol. Technol. 2019, 153, 79–86. [Google Scholar]
- Kawano, T. Roles of the reactive oxygen species-generating peroxidase reactions in plant defense and growth induction. Plant Cell Rep. 2003, 21, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Chittoor, J.M.; Leach, J.E.; White, F.F. Induction of peroxidase during defense against pathogens. In Pathogenesis-Related Proteins in Plants; CRC Press: Boca Raton, FL, USA, 1999; pp. 171–193. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.; Pan, Y.; Ju, Z.; Niu, K. Genome-Wide Analysis of the POD Gene Family in Avena sativa: Insights into Lignin Biosynthesis and Responding to Powdery Mildew. Agronomy 2025, 15, 852. https://doi.org/10.3390/agronomy15040852
Huang M, Pan Y, Ju Z, Niu K. Genome-Wide Analysis of the POD Gene Family in Avena sativa: Insights into Lignin Biosynthesis and Responding to Powdery Mildew. Agronomy. 2025; 15(4):852. https://doi.org/10.3390/agronomy15040852
Chicago/Turabian StyleHuang, Miaomiao, Yuanbo Pan, Zeliang Ju, and Kuiju Niu. 2025. "Genome-Wide Analysis of the POD Gene Family in Avena sativa: Insights into Lignin Biosynthesis and Responding to Powdery Mildew" Agronomy 15, no. 4: 852. https://doi.org/10.3390/agronomy15040852
APA StyleHuang, M., Pan, Y., Ju, Z., & Niu, K. (2025). Genome-Wide Analysis of the POD Gene Family in Avena sativa: Insights into Lignin Biosynthesis and Responding to Powdery Mildew. Agronomy, 15(4), 852. https://doi.org/10.3390/agronomy15040852





