Responses of Soybean Biomass and Bacterial Community Diversity of AMF Spore-Associated and Soybean Rhizosphere Soil to Microbial Inoculation and Chlorothalonil
Abstract
1. Introduction
2. Materials and Methods
2.1. Soybean Cultivar
2.2. AMF Inoculum and Phosphorus-Solubilizing Inoculum
2.3. Pesticides
2.4. Experimental Design
2.5. Sample Collection
2.6. Determination of the AMF Colonization Rate
2.7. Determination of the Spore Density of AMF and Spore Selection
2.8. Determination of Soybean Plant Biomass
2.9. Determination of Nodule Number in Soybean Plants
2.10. DNA Extraction, Amplification and 16S rRNA Gene Amplicon Sequencing
2.11. Sequence Analysis
2.12. Statistical Analysis
3. Results
3.1. Effects of Different Treatments on the AMF Colonization Rate
3.2. Effects of the Different Treatments on AMF Spore Density
3.3. Effects of Different Treatments on the Number of Soybean Root Nodules
3.4. Effects of Different Treatments on Soybean Plant Biomass
3.5. Diversity of the Bacterial Communities Associated with Rhizosphere Soil and AMF Spores
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, C.; Kong, F.J. Soybean. Curr. Biol. 2022, 32, R902–R904. [Google Scholar] [CrossRef] [PubMed]
- Van-Scoy, A.R.; Tjeerdema, R.S. Environmental fate and toxicology of chlorothalonil. Rev. Environ. Contam. Toxicol. 2014, 232, 89–105. [Google Scholar] [PubMed]
- Baćmaga, M.; Wyszkowska, J.; Kucharski, J. The effect of the Falcon 460 EC fungicide on soil microbial communities, enzyme activities and plant growth. Ecotoxicology 2016, 25, 1575–1587. [Google Scholar] [CrossRef]
- Jiang, J.L.; Yang, Y.W.; Wang, L.; Cao, S.H.; Long, T.; Liu, R.B. Effects of chlorothalonil application on the physio-biochemical properties and microbial community of a yellow-brown loam soil. Agriculture 2022, 12, 608. [Google Scholar] [CrossRef]
- Lin, Z.Q.; Pang, S.M.; Zhou, Z.; Wu, X.; Li, J.; Huang, Y.; Zhang, W.P.; Lei, Q.Q.; Bhatt, P.; Mishra, S.; et al. Novel pathway of acephate degradation by the microbial consortium ZQ01 and its potential for environmental bioremediation. J. Hazard. Mater. 2022, 426, 127841. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Wang, X.L.; Sun, S.; Jin, C.Z.; Su, J.M.; Wei, J.P.; Luo, X.Y.; Wen, J.W.; Wei, T.; Kumar, S.; et al. GWAS, MWAS and mGWAS provide insights into precision agriculture based on genotype-dependent microbial effects in foxtail millet. Nat. Commun. 2022, 13, 5913. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Zobel, M. How mycorrhizal associations drive plant population and community biology. Science 2020, 367, eaba1223. [Google Scholar] [CrossRef]
- Chang, X.; Yan, L.; Naeem, M.; Khaskheli, M.I.; Zhang, H.; Gong, G.; Zhang, M.; Song, C.; Yang, W.; Liu, T.; et al. Maize/soybean relay strip intercropping reduces the occurrence of Fusarium root rot and changes the diversity of the pathogenic Fusarium species. Pathogens 2020, 9, 211. [Google Scholar] [CrossRef]
- Shi, J.; Wang, X.; Wang, E. Mycorrhizal symbiosis in plant growth and stress adaptation: From genes to ecosystems. Annu. Rev. Plant Biol. 2023, 74, 569–607. [Google Scholar] [CrossRef]
- Jie, W.G.; Tan, Y.W.; Yang, D.Y.; Kan, L.B. Effects of Rhizophagus intraradices and Acinetobacter calcoaceticus on soybean growth and carbendazim residue. Sustainability 2023, 15, 10322. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.L.; Zhu, X.C.; Zhang, Y.; Yang, H.L.; Dobbie, S.; Zhang, X.; Deng, A.; Qian, H.Y.; Zhang, W.K. Effects of arbuscular mycorrhizal fungi on crop growth and soil N2O emissions in the legume system. Agric. Ecosyst. Environ. 2021, 322, 107641. [Google Scholar] [CrossRef]
- Zhang, R.; Mu, Y.; Li, X.; Li, S.; Sang, P.; Wang, X.; Wu, H.; Xu, N. Response of the arbuscular mycorrhizal fungi diversity and community in maize and soybean rhizosphere soil and roots to intercropping systems with different nitrogen application rates. Sci. Total Environ. 2020, 704, 139810. [Google Scholar] [CrossRef]
- Wang, X.L.; Feng, H.; Wang, Y.Y.; Wang, M.X.; Xie, X.G.; Chang, H.Z.; Wang, L.K.; Qu, J.C.; Sun, K.; He, W.; et al. Mycorrhizal symbiosis modulates the rhizosphere microbiota to promote rhizobia-legume symbiosis. Mol. Plant 2021, 14, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Liu, D. Root developmental responses to phosphorus nutrition. J. Integr. Plant Biol. 2021, 63, 1065–1090. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, V.; Monda, H.; Savy, D.; Meo, V.D.; Vinci, G.; Smalla, K. Cooperation among phosphate-solubilizing bacteria, humic acids and arbuscular mycorrhizal fungi induces soil microbiome shifts and enhances plant nutrient uptake. Chem. Biol. Technol. Agric. 2021, 8, 31. [Google Scholar] [CrossRef]
- Raliya, R.; Tarafdar, J.C.; Biswas, P. Enhancing the mobilization of native phosphorus in the mung bean rhizosphere using ZnO nanoparticles synthesized by soil fungi. J. Agric. Food Chem. 2016, 64, 3111–3118. [Google Scholar] [CrossRef]
- De-Zutter, N.D.; Ameye, M.; Bekaert, B.; Verwaerwn, J.; Gelder, L.D.; Audenaert, K. Uncovering new insights and misconceptions on the effectiveness of phosphate solubilizing rhizobacteria in plants: A meta-analysis. Front. Plant Sci. 2022, 13, 858804. [Google Scholar] [CrossRef]
- Arkhipova, T.N.; Veselov, S.U.; Melentiev, A.I.; Martynenko, E.V.; Kudoyarova, G.R. Ability of bacterium Bacillus subtilis to produce cytokinins and to influence the growth and endogenous hormone content of lettuce plants. Plant Soil 2005, 272, 201–209. [Google Scholar] [CrossRef]
- Foughalia, A.; Bouaoud, Y.; Chandeysson, C.; Djedidi, M.; Tahirine, M.; Aissat, K.; Nicot, P. Acinetobacter calcoaceticus SJ19 and Bacillus safensis SJ4, two Algerian rhizobacteria protecting tomato plants against Botrytis cinerea and promoting their growth. Egypt. J. Biol. Pest Control 2022, 32, 12. [Google Scholar] [CrossRef]
- Mueller, D.S.; Jeffers, S.N.; Buck, J.W. Toxicity of fungicides to urediniospores of six rust fungi that occur on ornamental crops. Plant Dis. 2005, 89, 255–261. [Google Scholar] [CrossRef]
- Jie, W.G.; Cai, B.Y.; Ge, J.P. Molecular detection and community analysis of arbuscular mycorrhizal fungi in the rhizosphere of Phellodendron amurense. Ann. Microbiol. 2012, 62, 1769–1777. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Kumutha, A. Standardization of the spore density of am fungal inoculum for effective colonization. Int. J. Agric. Sci. 2012, 4, 176–181. [Google Scholar] [CrossRef]
- Yang, Z.C.; Kang, J.; Ye, Z.M.; Qiu, W.; Liu, J.X.; Cao, X.B.; Ge, J.P.; Ping, W.X. Synergistic benefits of Funneliformis mosseae and Bacillus paramycoides: Enhancing soil health and soybean tolerance to root rot disease. Environ. Res. 2023, 238, 117219. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.C.; Zhu, S.X.; Li, W.J.; Tang, Q.; Luo, H.Y. Effects of chromium stress on the rhizosphere microbial community composition of Cyperus alternifolius. Ecotoxicol. Environ. Saf. 2021, 218, 112253. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Tarek, A.; Boussebough, I.; Chaoui, A.; Nouar, A.Z.; Chettah, M.C. Usearch: A meta search engine based on a new result merging strategy. In Proceedings of the 2015 7th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K), Lisbon, Portugal, 12–14 November 2015; Volume 1, pp. 531–536. [Google Scholar]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic Note: A Place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Hughes, J.B.; Hellmann, J.J.; Ricketts, T.H.; Bohannan, J.M. Counting the uncountable: Statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 2001, 67, 4399–4406. [Google Scholar] [CrossRef]
- Chao, A.; Chazdon, R.L.; Colwell, R.K.; Shen, T.J. A new statistical approach for assessing compositional similarity based on incidence and abundance data. Ecol. Lett. 2005, 8, 148–159. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ldeker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Rodrigues, V.D.; Torres, T.T.; Ottoboni, L.M.M. Bacterial diversity assessment in soil of an active brazilian copper mine using high-throughput sequencing of 16S rDNA amplicons. Antonie Van Leeuwenhoek 2014, 106, 879–890. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2006. [Google Scholar]
- Zhang, L.; Feng, G.; Declerck, S. Signal beyond nutrient, fructose, exuded by an arbuscular mycorrhizal fungus triggers phytate mineralization by a phosphate solubilizing bacterium. ISME J. 2018, 12, 2339–2351. [Google Scholar] [CrossRef]
- Zhang, C.F.; van der Heijden, M.G.A.; Dodds, B.K.; Nguyen, T.B.; Spooren, J.; Valzano-Held, A.; Cosme, M.; Berendsen, R.L. A tripartite bacterial-fungal-plant symbiosis in the mycorrhiza-shaped microbiome drives plant growth and mycorrhization. Microbiome 2024, 12, 13. [Google Scholar]
- Ordoñez, Y.M.; Fernandez, B.R.; Lara, L.S.; Rodriguez, A.; Uribe-Vélez, D.; Sanders, L.R. Bacteria with phosphate solubilizing capacity alter mycorrhizal fungal growth both inside and outside the root and in the presence of nativemicrobial communities. PLoS ONE 2016, 11, e0154438. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springer Plus 2013, 2, 587. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhang, L.; Zhou, J.C.; George, T.S.; Feng, G. Arbuscular mycorrhizal fungi enhance mineralisation of organic phosphorus by carrying bacteria along their extraradical hyphae. New Phytol. 2021, 230, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Huang, C.; Peng, F.; Zhang, W.J.; Luo, J.; Ma, S.X.; Xue, X. Effect of arbuscular mycorrhizal fungi (AMF) and plant growth-promoting bacteria (PGPR) inoculations on Elaeagnus angustifolia L. in saline soil. Appl. Sci. 2020, 10, 945. [Google Scholar] [CrossRef]
- Wu, Q.C.; Chen, Y.; Dou, X.H.; Liao, D.X.; Li, K.Y.; An, C.C.; Li, G.H.; Dong, Z. Microbial fertilizers improve soil quality and crop yield in coastal saline soils by regulating soil bacterial and fungal community structure. Sci. Total Environ. 2024, 949, 175127. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.F.; Li, Z.K.; Luo, X.; Wang, W.H.; Li, Y.K.; Li, R.; Zhang, B.; Qiao, Y.; Zhou, J.; Fan, J.Q.; et al. A predatory myxobacterium controls cucumber Fusarium wilt by regulating the soil microbial community. Microbiome 2020, 8, 49. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, C.; Wang, J.; Liu, M.; Wang, J.; Hu, J.; Liu, L.; Liu, X.; Xia, C.; Zhong, L.; et al. Predation of oomycetes by myxobacteria via a specialized CAZyme system arising from adaptive evolution. ISME J. 2023, 17, 1089–1103. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, J.; George, T.S.; Limpens, E.; Feng, G. Arbuscular mycorrhizal fungi conducting the hyphosphere bacterial orchestra. Trends Plant Sci. 2022, 27, 402–411. [Google Scholar] [CrossRef]
- Baćmaga, M.; Wyszkowska, J.; Kucharski, J. The influence of chlorothalonil on the activity of soil microorganisms and enzymes. Ecotoxicology 2018, 27, 1188–1202. [Google Scholar] [CrossRef]
- Lu, T.; Xu, N.; Lei, C.; Zhang, Q.; Zhang, Z.; Sun, L.; He, F.; Zhou, N.; Peñuelas, J.; Zhu, Y.; et al. Bacterial biogeography in China and its association to land use and soil organic carbon. Soil Ecol. Lett. 2023, 5, 230172. [Google Scholar] [CrossRef]
- Kouřilová, X.; Schwarzerová, J.; Pernicová, I.; Sedlář, K.; Mrázová, K.; Krzyžánek, V.; Nebesářová, J.; Obruča, S. The first insight into polyhydroxyalkanoates accumulation in multi-extremophilic Rubrobacter xylanophilus and Rubrobacter spartanus. Microorganisms 2021, 9, 909. [Google Scholar] [CrossRef]
- Ujvári, G.; Turrini, A.; Avio, L.; Agnolucci, M. Possible role of arbuscular mycorrhizal fungi and associated bacteria in the recruitment of endophytic bacterial communities by plant roots. Mycorrhiza 2021, 31, 527–544. [Google Scholar] [CrossRef]
- Sangwan, S.; Prasanna, R. Mycorrhizae helper bacteria: Unlocking their potential as bioenhancers of plant-arbuscular mycorrhizal fungal associations. Microb. Ecol. 2022, 84, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Feng, G.; Limpens, E.; Bonfante, P.; Xie, X.; Zhang, L. Cross-kingdom nutrient exchange in the plant-arbuscular mycorrhizal fungus-bacterium continuum. Nat. Rev. Microbiol. 2024, 22, 773–790. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Qiu, Y.Z.; Yao, T.; Ma, Y.C.; Zhang, H.R.; Yang, X.L. Effects of PGPR microbial inoculants on the growth and soil properties of Avena sativa, Medicago sativa, and Cucumis sativus seedlings. Soil Tillage Res. 2020, 199, 104577. [Google Scholar] [CrossRef]
- Gomila, M.; Bowien, B.; Falsen, E.; Moore, E.R.B.; Lalucat, J. Description of Pelomonas aquatica sp. nov. and Pelomonas puraquae sp. nov., isolated from industrial and haemodialysis water. Int. J. Syst. Evol. Microbiol. 2007, 57, 2629–2635. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
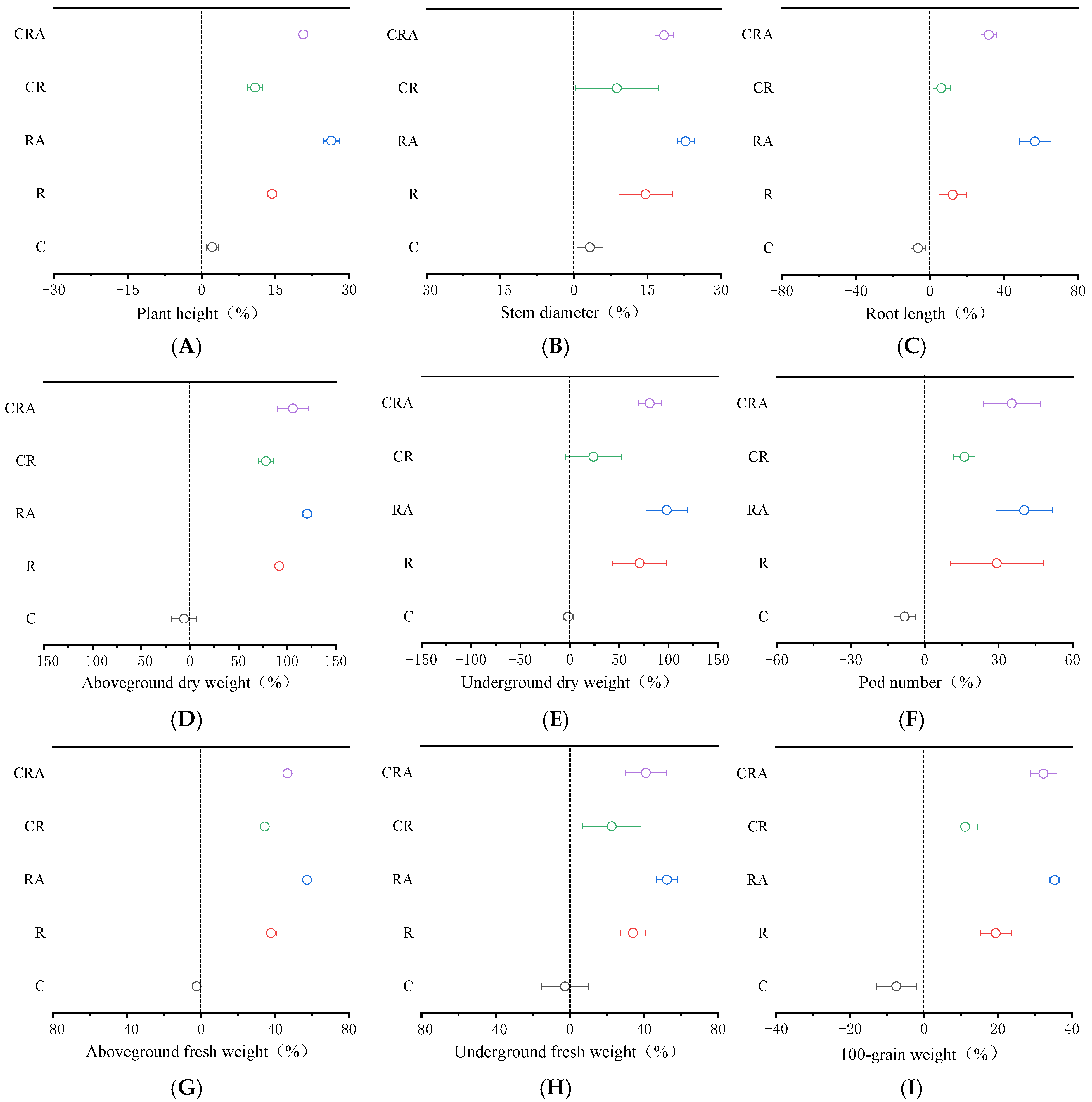


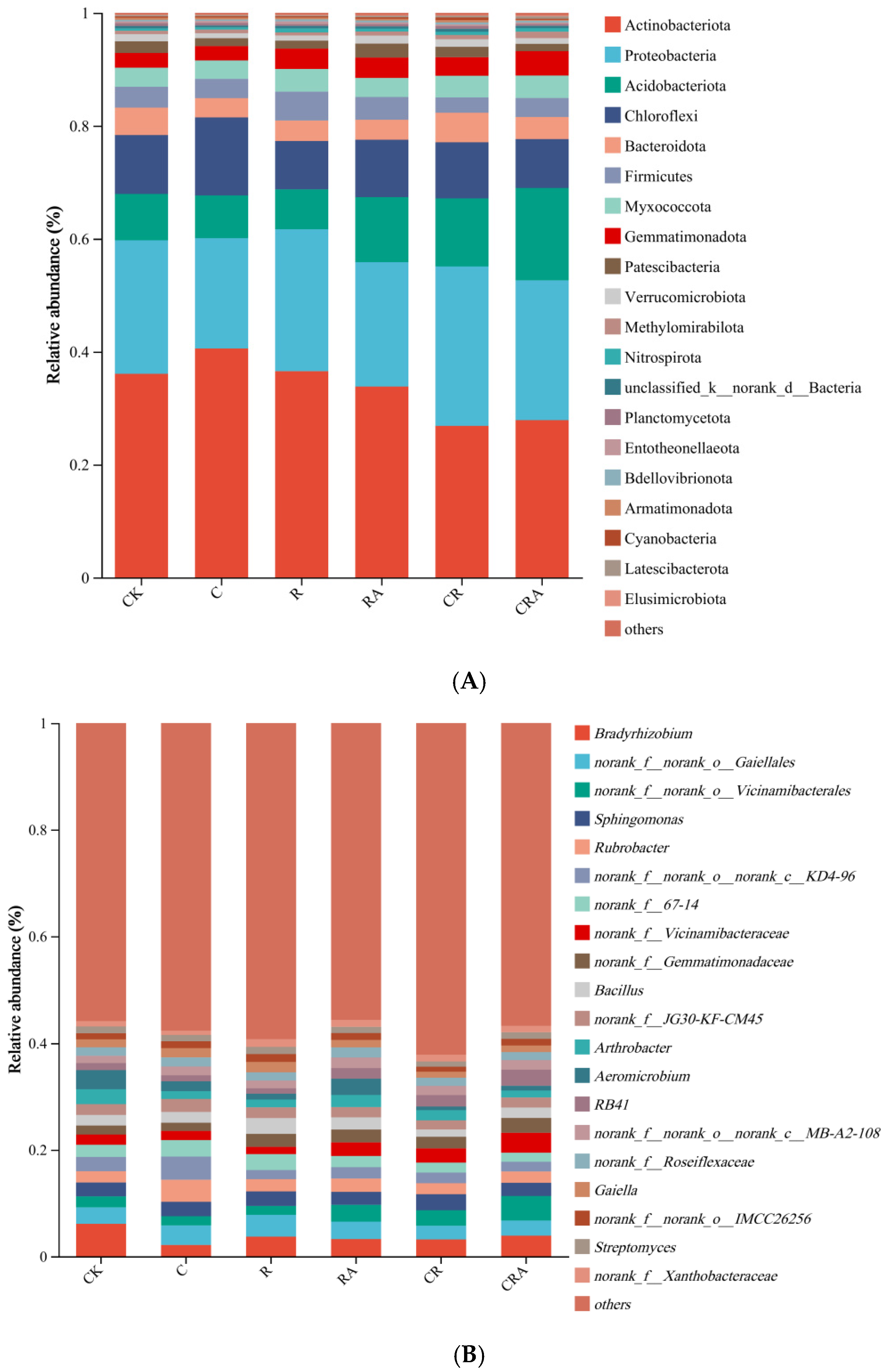
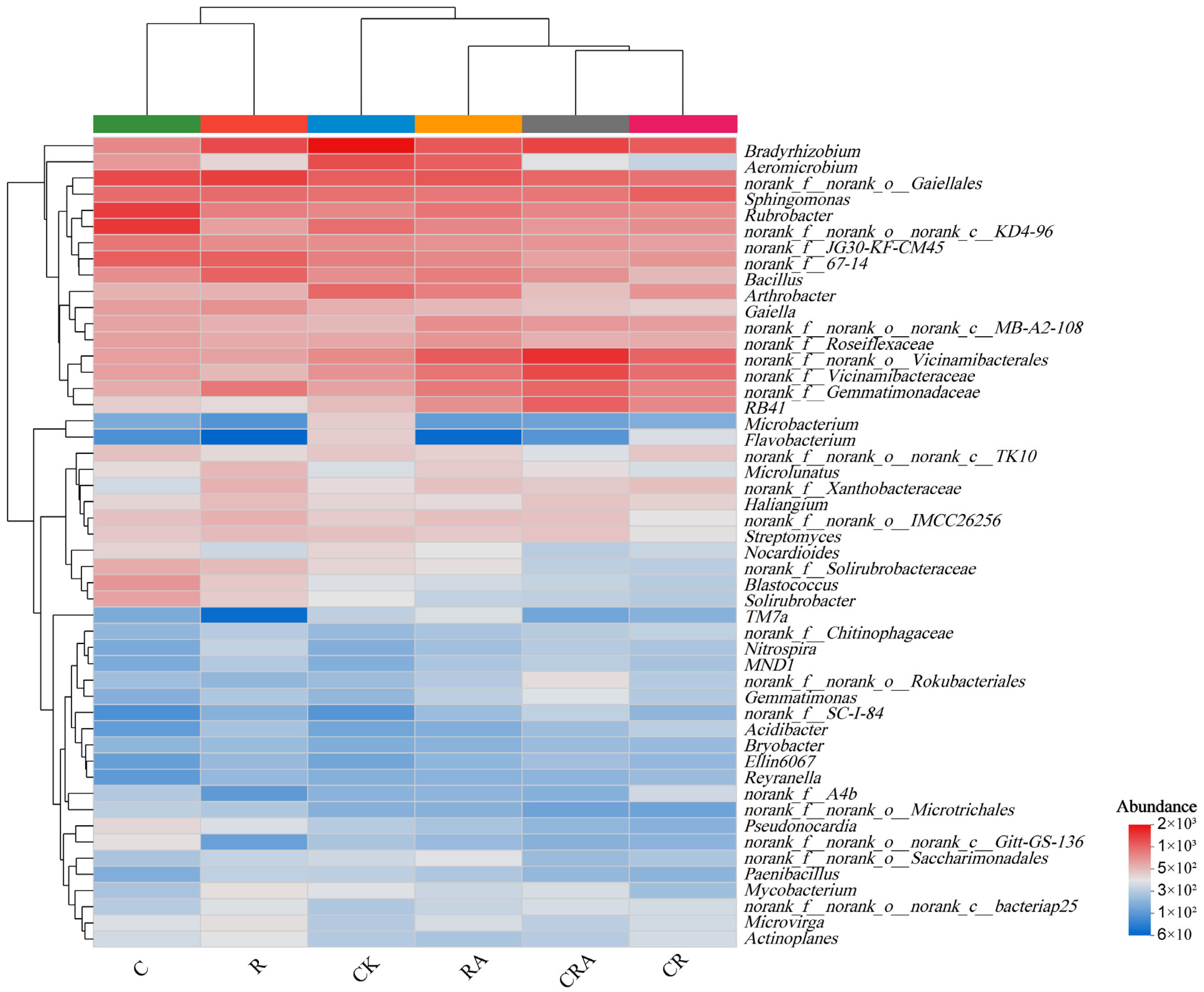


| Treatments | CK | C | R | RA | CR | CRA |
|---|---|---|---|---|---|---|
| Arbuscular mycorrhizal fungi colonization rate | 0.40 ± 0.06 de | 0.35 ± 0.02 e | 0.93 ± 0.06 ab | 0.96 ± 0.01 a | 0.87 ± 0.06 bc | 0.90 ± 0.03 abc |
| Arbuscular mycorrhizal fungi spore density per gram of soil | 4.05 ± 6.03 c | 3.61 ± 5.02 c | 23.70 ± 16.18 b | 32.22 ± 29.38 a | 7.37 ± 6.28 c | 9.54 ± 7.91 c |
| Number of soybean root nodules per plant | 64.33 ± 2.08 e | 53.00 ± 1.00 f | 82.67 ± 3.21 c | 111.67 ± 3.06 a | 76.67 ± 1.53 d | 101.00 ± 3.06 b |
| Treatments | CK | C | R | RA | CR | CRA |
|---|---|---|---|---|---|---|
| OTU | 3757 ± 61.44 de | 3693 ± 35.85 e | 3910 ± 27.62 b | 3849 ± 21.79 bc | 4123 ± 48.69 a | 3819 ± 28.83 cd |
| Chao 1 | 4498.59 ± 107.81 c | 4513.74 ± 54.92 bc | 4558.84 ± 31.18 bc | 4632.91 ± 82.06 ab | 4741.82 ± 56.07 a | 4562.80 ± 38.78 bc |
| Shannon | 6.5671 ± 0.0137 e | 6.7059 ± 0.0288 d | 6.7838 ± 0.0160 c | 6.7611 ± 0.0134 c | 6.9920 ± 0.0135 a | 6.8575 ± 0.0084 b |
| Ace | 4654.08 ± 108.86 bc | 4599.21 ± 71.57 c | 4749.24 ± 35.34 b | 4769.68 ± 79.85 b | 4908.67 ± 69.75 a | 4682.67 ± 15.42 bc |
| Coverage | 0.9761 ± 0.0004 a | 0.9784 ± 0.0019 a | 0.9776 ± 0.0009 a | 0.9764 ± 0.0013 a | 0.9762 ± 0.0013 a | 0.9769 ± 0.0014 a |
| Simpson | 0.0078 ± 0.0001 a | 0.0042 ± 0.0001 c | 0.0042 ± 0.0002 c | 0.0046 ± 0.0001 b | 0.0033 ± 0.0001 e | 0.0039 ± 0.0001 d |
| Treatments | SR | SRA | SCR | SCRA |
|---|---|---|---|---|
| OTU | 283 ± 105.19 a | 193 ± 52.94 a | 229 ± 96.96 a | 299 ± 154.98 a |
| Chao 1 | 290.72 ± 109.61 a | 201.70 ± 58.40 a | 238.92 ± 101.34 a | 316.46 ± 163.83 a |
| Shannon | 3.3961 ± 0.2992 a | 2.8866 ± 0.3387 a | 3.1025 ± 0.4394 a | 3.2732 ± 0.6613 a |
| Ace | 291.13 ± 108.69 a | 200.36 ± 54.74 a | 237.54 ± 99.81 a | 310.88 ± 155.06 a |
| Coverage | 0.9998 ± 0.0001 a | 0.9998 ± 0.0001 a | 0.9998 ± 0.0001 a | 0.9997 ± 0.0001 a |
| Simpson | 0.1010 ± 0.0295 a | 0.1530 ± 0.0625 a | 0.1259 ± 0.0355 a | 0.1354 ± 0.0420 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jie, W.; Zhang, M. Responses of Soybean Biomass and Bacterial Community Diversity of AMF Spore-Associated and Soybean Rhizosphere Soil to Microbial Inoculation and Chlorothalonil. Agronomy 2025, 15, 738. https://doi.org/10.3390/agronomy15030738
Jie W, Zhang M. Responses of Soybean Biomass and Bacterial Community Diversity of AMF Spore-Associated and Soybean Rhizosphere Soil to Microbial Inoculation and Chlorothalonil. Agronomy. 2025; 15(3):738. https://doi.org/10.3390/agronomy15030738
Chicago/Turabian StyleJie, Weiguang, and Min Zhang. 2025. "Responses of Soybean Biomass and Bacterial Community Diversity of AMF Spore-Associated and Soybean Rhizosphere Soil to Microbial Inoculation and Chlorothalonil" Agronomy 15, no. 3: 738. https://doi.org/10.3390/agronomy15030738
APA StyleJie, W., & Zhang, M. (2025). Responses of Soybean Biomass and Bacterial Community Diversity of AMF Spore-Associated and Soybean Rhizosphere Soil to Microbial Inoculation and Chlorothalonil. Agronomy, 15(3), 738. https://doi.org/10.3390/agronomy15030738






