sRNA Sequencing of Dahlia Bicolor Petals Revealed the Post-Transcriptional Regulation of Anthocyanin Biosynthetic Pathway
Abstract
1. Introduction
2. Materials and Methods
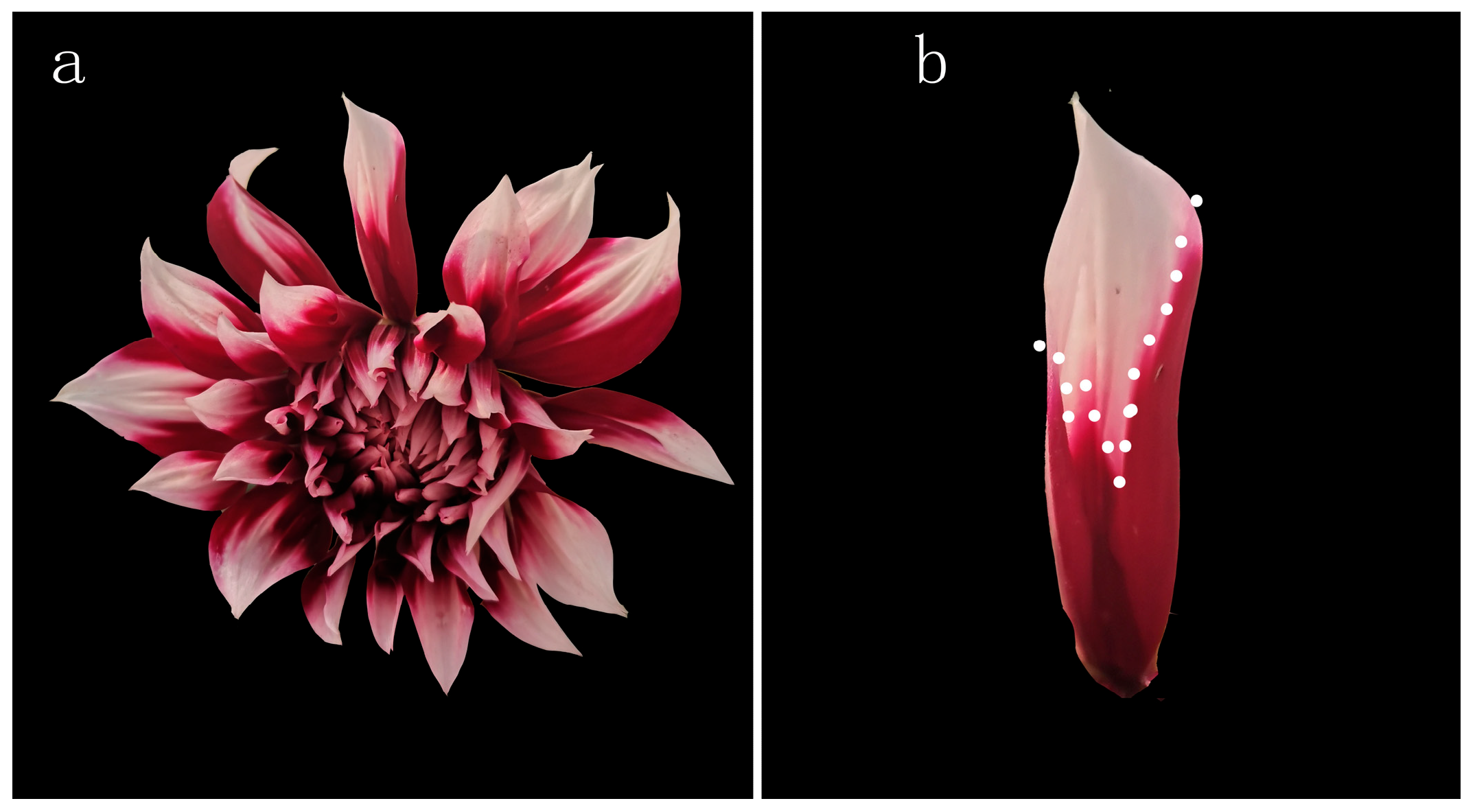
2.1. Plant Materials
2.2. Construction and Sequencing of sRNA Libraries
2.3. Identification and Quantification of miRNAs of Dahlias
2.4. Identification of Target Genes and Differentially Expressed miRNAs (DEMs)
2.5. Construction for the Post-Transcriptional Regulation Network of ABPs
2.6. Validation of Expression Levels of miRNA-TF Pairs
3. Results
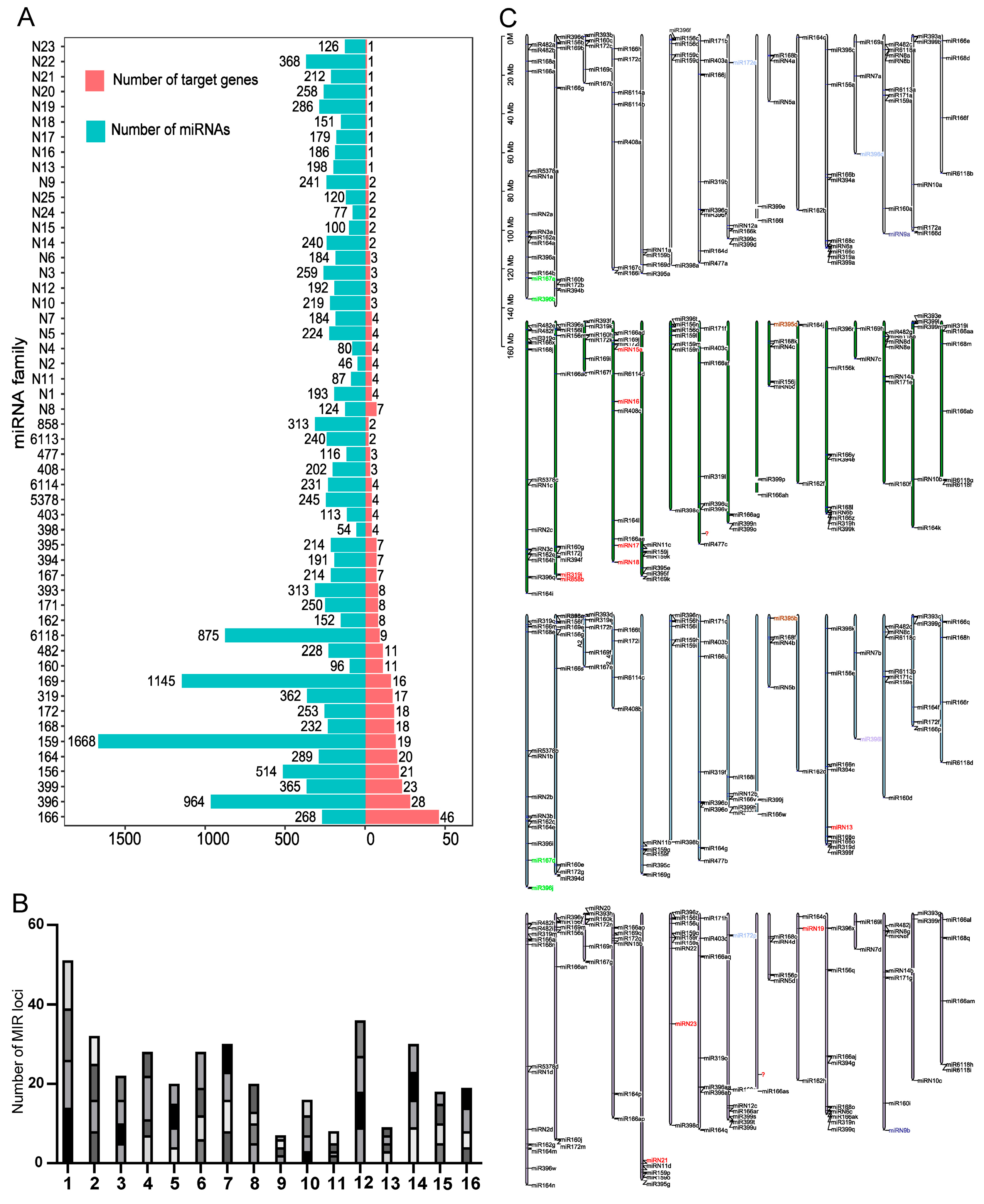
3.1. Identification of miRNA and Target Genes of Dahlias
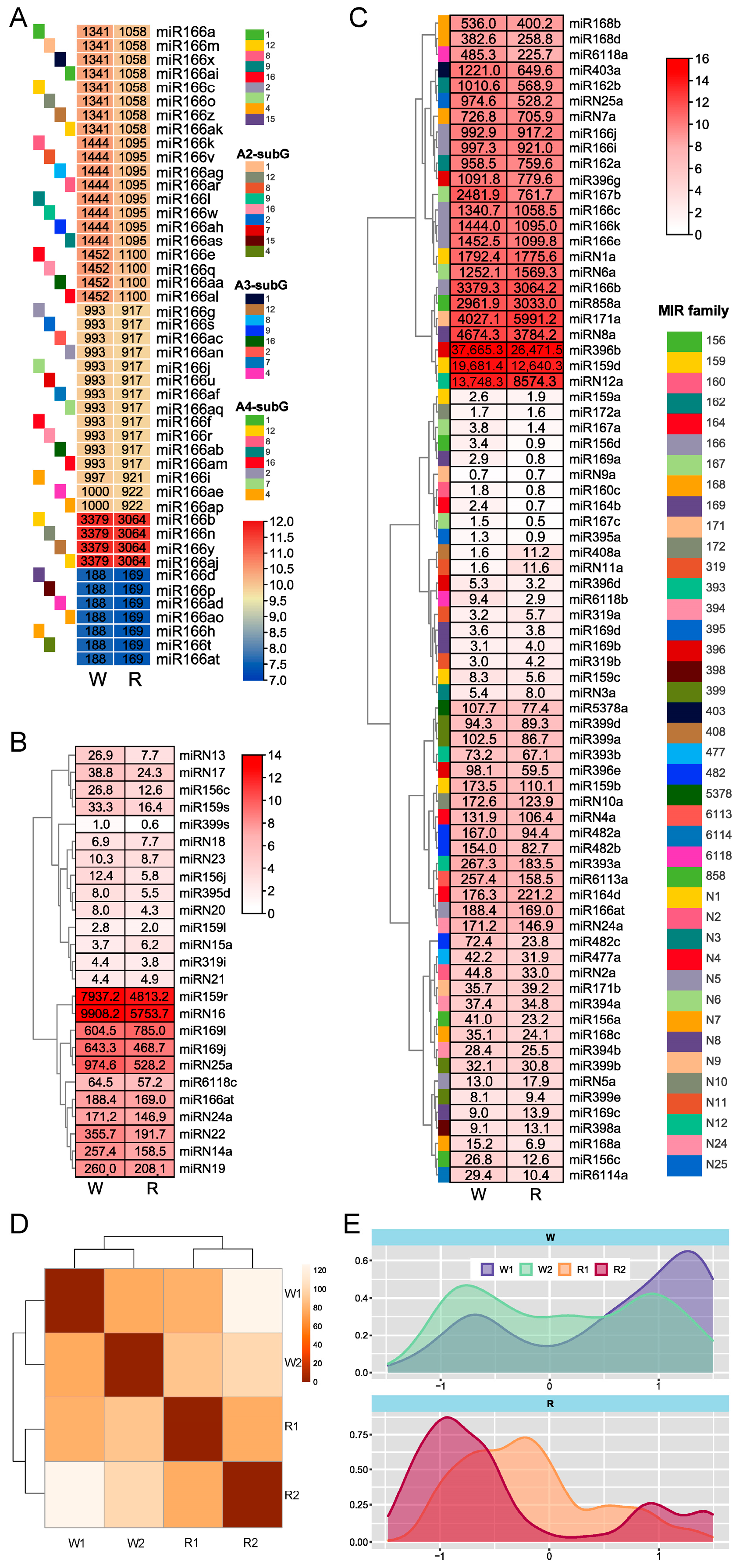
3.2. MiRNA Expression Profiles of Dahlia Petals
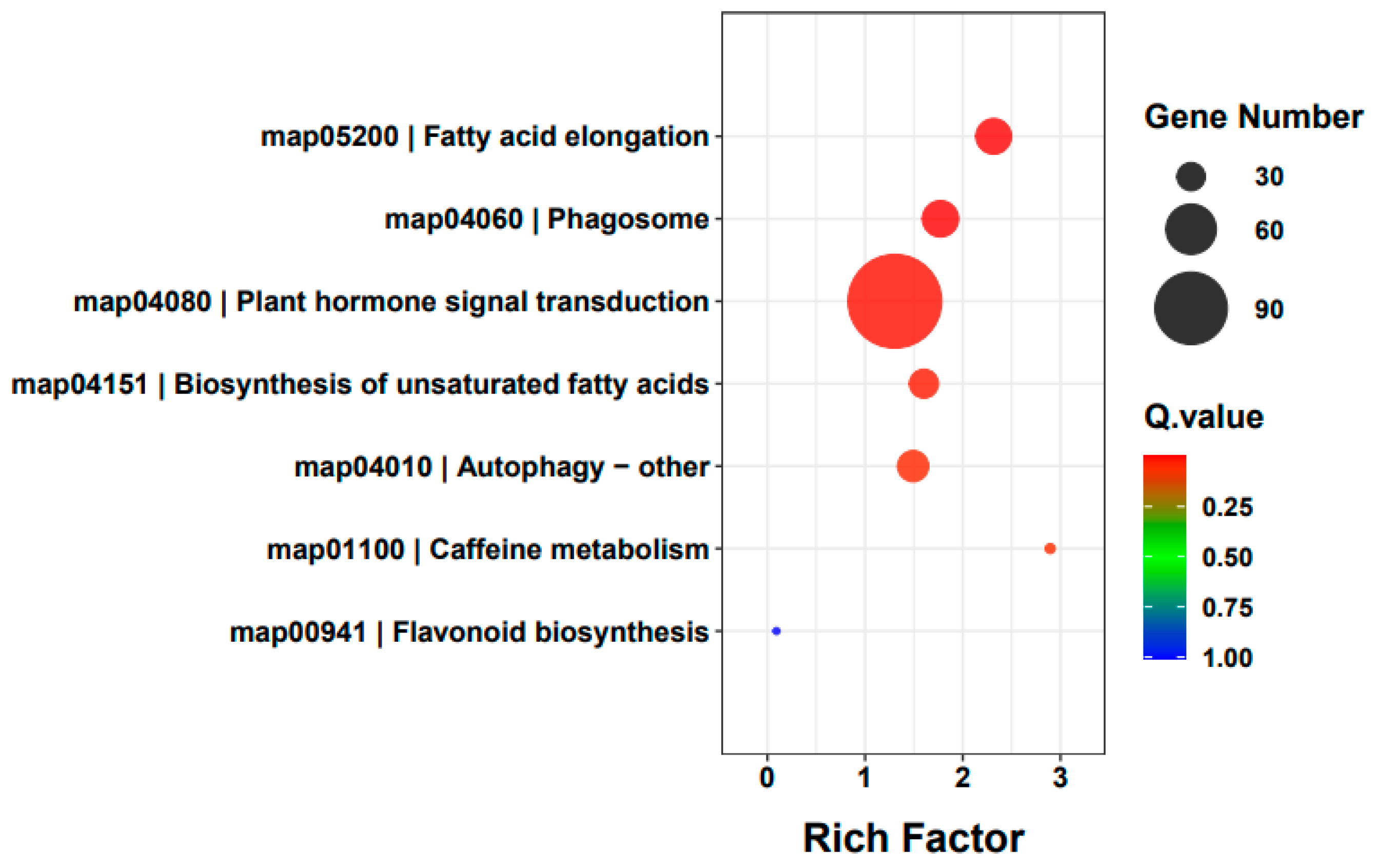
3.3. DEMs Identification and Enrichment Analysis
3.4. Analysis of the Post-Transcriptional Regulation Network Involved in ABP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Walliser, B.; Lucaciu, C.R.; Molitor, C.; Marinovic, S.; Nitarska, D.A.; Aktaş, D.; Rattei, T.; Kampatsikas, I.; Stich, K.; Haselmair-Gosch, C.; et al. Dahlia variabilis cultivar ‘Seattle’ as a model plant for anthochlor biosynthesis. Plant Physiol. Biochem. 2021, 159, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Hosokawa, M.; Kojima, M.; Kitamura, Y.; Hoshino, A.; Tatsuzawa, F.; Doi, M.; Yazawa, S. Simultaneous post-transcriptional gene silencing of two different chalcone synthase genes resulting in pure white flowers in the octoploid dahlia. Planta 2011, 234, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.D.H. Polyploidy and Evolution in Wild and Cultivated Dahlia Species. Ann. Bot. 1998, 81, 647–656. [Google Scholar]
- Lehnert, E.M.; Walbot, V. Sequencing and de novo assembly of a Dahlia hybrid cultivar transcriptome. Front. Plant Sci. 2014, 5, 340. [Google Scholar] [CrossRef][Green Version]
- Almeyda, C.V.; Raikhy, G.; Pappu, H.R. Characterization and comparative analysis of promoters from three plant pararetroviruses associated with Dahlia (Dahlia variabilis). Virus Genes 2015, 51, 96–104. [Google Scholar] [CrossRef]
- Deguchi, A.; Ohno, S.; Hosokawa, M.; Tatsuzawa, F.; Doi, M. Endogenous post-transcriptional gene silencing of flavone synthase resulting in high accumulation of anthocyanins in black Dahlia cultivars. Planta 2013, 237, 1325–1335. [Google Scholar] [CrossRef]
- Wang, H.; Xu, D.; Jiang, F.; Wang, S.; Wang, A.; Liu, H.; Lei, L.; Qian, W.; Fan, W. The genomes of Dahlia pinnata, Cosmos bipinnatus, and Bidens alba in tribe Coreopsideae provide insights into polyploid evolution and inulin biosynthesis. Gigascience 2024, 13, giae032. [Google Scholar] [CrossRef]
- Ohno, S.; Hosokawa, M.; Hoshino, A.; Kitamura, Y.; Morita, Y.; Park, K.I.; Nakashima, A.; Deguchi, A.; Tatsuzawa, F.; Doi, M.; et al. A bHLH transcription factor, DvIVS, is involved in regulation of anthocyanin synthesis in dahlia (Dahlia variabilis). J. Exp. Bot. 2011, 62, 5105–5116. [Google Scholar] [CrossRef]
- Zou, J.; Ran, L.; Zhou, R.; Wang, Z. The Transcriptome of Dahlia pinnata Provides Comprehensive Insight into the Formation Mechanism of Polychromatic Petals. Agronomy 2024, 14, 2748. [Google Scholar] [CrossRef]
- Ohno, S.; Hori, W.; Hosokawa, M.; Tatsuzawa, F.; Doi, M. Post-transcriptional silencing of chalcone synthase is involved in phenotypic lability in petals and leaves of bicolor dahlia (Dahlia variabilis) Yuino. Planta 2018, 247, 413–428. [Google Scholar] [CrossRef]
- Onozaki, T.; Mato, M.; Shibata, M.; Ikeda, H. Differences in Flower Color and Pigment Composition Among White Carnation (Dianthus caryophyllus L.) Cultivars. Sci. Hortic. 1999, 82, 103–111. [Google Scholar] [CrossRef]
- Schlangen, K.; Miosic, S.; Halbwirth, H. Allelic variants from Dahlia variabilis encode flavonoid 3′-hydroxylases with functional differences in chalcone 3-hydroxylase activity. Arch. Biochem. Biophys. 2010, 494, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Mato, M.; Onozaki, T.; Ozeki, Y.; Higeta, D.; Itoh, Y.; Yoshimoto, Y.; Ikeda, H.; Yoshida, H.; Shibata, M. Flavonoid biosynthesis in white-flowered Sim carnations (Dianthus caryophyllus). Sci. Hortic. 2000, 84, 333–347. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Schwinn, K.E. Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signal Behav. 2014, 9, e29526. [Google Scholar] [CrossRef]
- Ohno, S.; Deguchi, A.; Hosokawa, M.; Tatsuzawa, F.; Doi, M. A basic helix-loop-helix transcription factor DvIVS determines flower color intensity in cyanic dahlia cultivars. Planta 2013, 238, 331–343. [Google Scholar] [CrossRef]
- Wang, N.; Liu, W.; Mei, Z.; Zhang, S.; Zou, Q.; Yu, L.; Jiang, S.; Fang, H.; Zhang, Z.; Chen, Z.; et al. A Functional InDel in the WRKY10 Promoter Controls the Degree of Flesh Red Pigmentation in Apple. Adv. Sci. 2024, 11, e2400998. [Google Scholar] [CrossRef]
- Wang, J.; Lian, W.; Cao, Y.; Wang, X.; Wang, G.; Qi, C.; Liu, L.; Qin, S.; Yuan, X.; Li, X. Overexpression of BoNAC019, a NAC transcription factor from Brassica oleracea, negatively regulates the dehydration response and anthocyanin biosynthesis in Arabidopsis. Sci. Rep. 2018, 8, 13349. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Li, S.; Zachgo, S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013, 76, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.F.; Fitzsimmons, K.; Khandelwal, A.; Kranz, R.G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2009, 2, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Appelhagen, I.; Lu, G.H.; Huep, G.; Schmelzer, E.; Weisshaar, B.; Sagasser, M. TRANSPARENT TESTA1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. Plant J. 2011, 67, 406–419. [Google Scholar] [CrossRef] [PubMed]
- LaFountain, A.M.; Yuan, Y.W. Repressors of anthocyanin biosynthesis. New Phytol. 2021, 231, 933–949. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Song, Z.; Zhang, H. Repression of MYBL2 by Both microRNA858a and HY5 Leads to the Activation of Anthocyanin Biosynthetic Pathway in Arabidopsis. Mol. Plant 2016, 9, 1395–1405. [Google Scholar] [CrossRef]
- Tirumalai, V.; Swetha, C.; Nair, A.; Pandit, A.; Shivaprasad, P.V. miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J. Exp. Bot. 2019, 70, 4775–4792. [Google Scholar] [CrossRef]
- Li, Z.; Liu, W.; Chen, Q.; Zhang, S.; Mei, Z.; Yu, L.; Wang, C.; Mao, Z.; Chen, Z.; Chen, X. Mdm-miR858 targets MdMYB9 and MdMYBPA1 to participate anthocyanin biosynthesis in red-fleshed apple. Plant J. 2023, 113, 1295–1309. [Google Scholar] [CrossRef]
- Qu, D.; Yan, F.; Meng, R.; Jiang, X.; Yang, H.; Gao, Z.; Dong, Y.; Yang, Y.; Zhao, Z. Identification of microRNAs and their targets associated with fruit-bagging and subsequent sunlight re-exposure in the “Granny Smith” apple exocarp using high-throughput sequencing. Front. Plant Sci. 2016, 7, 27. [Google Scholar] [CrossRef]
- Hsieh, L.-C.; Lin, S.-I.; Shih, A.C.-C.; Chen, J.-W.; Lin, W.-Y.; Tseng, C.-Y.; Li, W.-H.; Chiou, T.-J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef]
- Peng, Z.; Tian, J.; Luo, R.; Kang, Y.; Lu, Y.; Hu, Y.; Liu, N.; Zhang, J.; Cheng, H.; Niu, S.; et al. MiR399d and epigenetic modification comodulate anthocyanin accumulation in Malus leaves suffering from phosphorus deficiency. Plant Cell Environ. 2020, 43, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; He, H.; Wang, X.; Wang, X.; Yang, X.; Li, L.; Deng, X.W. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011, 65, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.J.; Mittal, A.; Jia, F.; Rock, C.D. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 2012, 80, 117–129. [Google Scholar] [CrossRef]
- Vale, M.; Badim, H.; Gerós, H.; Conde, A. Non-Mature miRNA-Encoded Micropeptide miPEP166c Stimulates Anthocyanin and Proanthocyanidin Synthesis in Grape Berry Cells. Int. J. Mol. Sci. 2024, 25, 1539. [Google Scholar] [CrossRef] [PubMed]
- Fairnie, A.L.M.; Yeo, M.T.S.; Gatti, S.; Chan, E.; Travaglia, V.; Walker, J.F.; Moyroud, E. Eco-Evo-Devo of petal pigmentation patterning. Essays Biochem. 2022, 66, 753–768. [Google Scholar] [CrossRef]
- Li, G.; Chen, C.; Chen, P.; Meyers, B.C.; Xia, R. sRNAminer: A multifunctional toolkit for next-generation sequencing small RNA data mining in plants. Sci. Bull. 2024, 69, 784–791. [Google Scholar] [CrossRef]
- Axtell, M.J.; Meyers, B.C. Revisiting Criteria for Plant MicroRNA Annotation in the Era of Big Data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef]
- Wei, G.; Xu, M.; Shi, X.; Wang, Y.; Shi, Y.; Wang, J.; Feng, L. Integrative analysis of miRNA profile and degradome reveals post-transcription regulation involved in fragrance formation of Rosa rugosa. Int. J. Biol. Macromol. 2024, 279, 135266. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, C.; Wang, S.; Qian, K.; Feng, Y.; Yu, F.; Wang, J. Genome-wide analysis of cuticle protein family genes in rice stem borer Chilo suppressalis: Insights into their role in environmental adaptation and insecticidal stress response. Int. J. Biol. Macromol. 2023, 242, 124989. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, J.; Feng, J.; Liu, B.; Feng, L.; Yu, X.; Li, G.; Zhai, J.; Meyers, B.C.; Xia, R. sRNAanno-a database repository of uniformly annotated small RNAs in plants. Hortic Res. 2021, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef] [PubMed]
- Inukai, T.; Kim, H.; Matsunaga, W.; Masuta, C. Battle for control of anthocyanin biosynthesis in two Brassicaceae species infected with turnip mosaic virus. J. Exp. Bot. 2023, 74, 1659–1674. [Google Scholar] [CrossRef]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.A.; Rothstein, S.J. The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.-I.; Asensi-Fabado, M.A.; Munne-Bosch, S.; Antonio, C.; Tohge, T. JUNGBRUNNEN1, a reactive oxygen species–responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef]
- Hu, X.; Yin, G.; Zhang, Y.; Zhu, L.; Huang, H.; Lv, K. Recent advances in the functional explorations of nuclear microRNAs. Front. Immunol. 2023, 14, 1097491. [Google Scholar] [CrossRef]
- Kubo, H.; Peeters, A.J.; Aarts, M.G.; Pereira, A.; Koornneef, M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 1999, 11, 1217–1226. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Hu, Q.; Dai, X.; Tian, H.; Zheng, K.; Wang, X.; Mao, T.; Chen, J.G.; Wang, S. Characterization of an activation-tagged mutant uncovers a role of GLABRA 2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 2015, 83, 300–311. [Google Scholar] [CrossRef]
- Rerie, W.G.; Feldmann, K.A.; Marks, M.D. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994, 8, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, X.; Sun, F.; Dong, H. Tobacco TTG 2 and ARF 8 function concomitantly to control flower colouring by regulating anthocyanin synthesis genes. Plant Biol. 2017, 19, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Wang, W.; Chang, M.; Yang, H.; Yin, F.; Liu, Y. The PpIAA5-ARF8 Module Regulates Fruit Ripening and Softening in Peach. Horticulturae 2023, 9, 1149. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Wang, N.; Xu, H.-F.; Jiang, S.-H.; Fang, H.-C.; Su, M.-Y.; Zhang, Z.-Y.; Zhang, T.-L.; Chen, X.-S. Auxin regulates anthocyanin biosynthesis through the Aux/IAA–ARF signaling pathway in apple. Hortic. Res. 2018, 5, 59. [Google Scholar] [CrossRef]
- Davies, K.M.; Albert, N.W.; Schwinn, K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 2012, 39, 619–638. [Google Scholar] [CrossRef]
- Yamagishi, M. MicroRNA828/MYB12 module mediated bicolor flower development in Lilium dauricum. Hortic. J. 2022, 91, 399–407. [Google Scholar] [CrossRef]
- Barrera-Rojas, C.H.; Nogueira, F.T.S.; van den Berg, C. Painting the plant body: Pigment biosynthetic pathways regulated by small RNAs. New Phytol. 2025, 245, 1411–1420. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, J.; Wu, X.; Li, S.; Liu, M.; Chen, Y.; Wang, H.; Tao, X. sRNA Sequencing of Dahlia Bicolor Petals Revealed the Post-Transcriptional Regulation of Anthocyanin Biosynthetic Pathway. Agronomy 2025, 15, 495. https://doi.org/10.3390/agronomy15020495
Zou J, Wu X, Li S, Liu M, Chen Y, Wang H, Tao X. sRNA Sequencing of Dahlia Bicolor Petals Revealed the Post-Transcriptional Regulation of Anthocyanin Biosynthetic Pathway. Agronomy. 2025; 15(2):495. https://doi.org/10.3390/agronomy15020495
Chicago/Turabian StyleZou, Jiuchun, Xiaoshuang Wu, Shuyan Li, Mengqing Liu, Yuyu Chen, Haoran Wang, and Xue Tao. 2025. "sRNA Sequencing of Dahlia Bicolor Petals Revealed the Post-Transcriptional Regulation of Anthocyanin Biosynthetic Pathway" Agronomy 15, no. 2: 495. https://doi.org/10.3390/agronomy15020495
APA StyleZou, J., Wu, X., Li, S., Liu, M., Chen, Y., Wang, H., & Tao, X. (2025). sRNA Sequencing of Dahlia Bicolor Petals Revealed the Post-Transcriptional Regulation of Anthocyanin Biosynthetic Pathway. Agronomy, 15(2), 495. https://doi.org/10.3390/agronomy15020495






