MicroRNA-Mediated Changes in DNA Methylation Affect the Expression of Genes Involved in the Thickness-of-Pod-Canopy Trait in Brassica napus L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Bisulfite Sequencing
2.2. Quality Control
2.3. Aligning the Reads to the Reference Genome
2.4. Analysis of Differentially Methylated Regions
2.5. GO Analysis of DMGs
2.6. Data Verification Using Traditional Bisulfite Sequencing PCR
3. Results
3.1. Analysis of Clean Reads
3.2. Types and Distribution of Methylation in the High- and Low-TPC Lines
3.3. Methylation Density on Chromosomes and Distribution of Methylation Levels in Genes
3.4. Comparison of the Overall Methylation Levels of High- and Low-TPC Lines
3.5. DMR Analysis of High- vs. Low-TPC Lines
3.6. DMR Analysis of the Promoters of High- vs. Low-TPC Lines
3.7. DMRs in Promoters Underlie Differences in Gene Expression Between High- and Low-TPC Lines
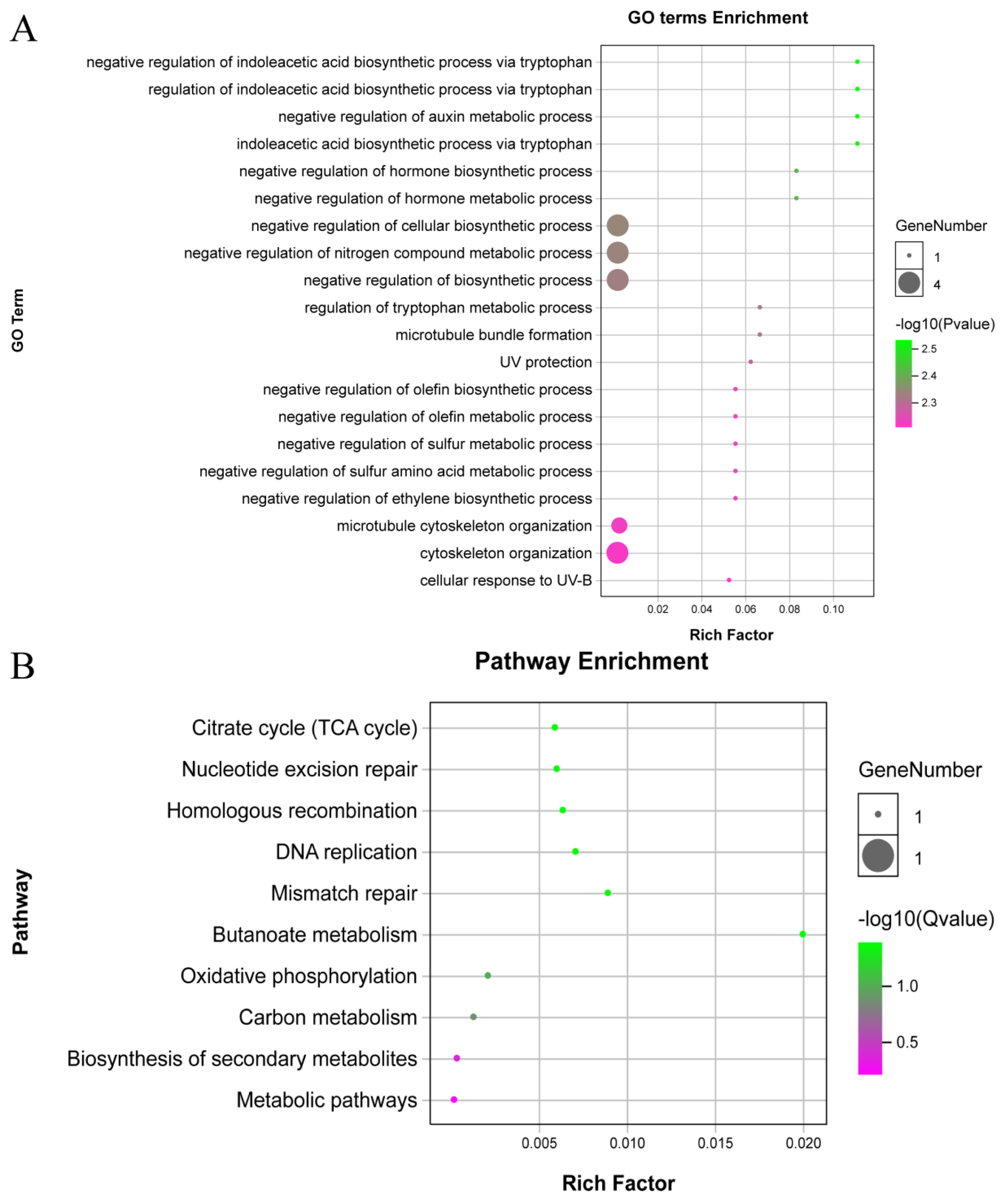
3.8. GO and KEGG Analysis of Differentially Methylated Genes
3.9. MiRNAs and Methylation Jointly Regulate the TPC Trait
3.10. Verification of the Methylation Sequencing Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, Z.; Huo, Q.; Yang, H.; Jian, H.; Qu, C.; Lu, K.; Li, J. Joint RNA-Seq and miRNA Profiling Analyses to Reveal Molecular Mechanisms in Regulating Thickness of Pod Canopy in Brassica napus. Genes 2019, 10, 591. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, C.; Lin, J.; Liu, J.; Liu, B.; Wang, J.; Huang, A.; Li, H.; Zhao, T. OsMPH1 regulates plant height and improves grain yield in rice. PLoS ONE 2017, 12, e0180825. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-T.; Chen, P.-W.; Chen, L.-C.; Yang, C.-C.; Chen, S.-Y.; Huang, G.; Lin, T.C.; Ku, H.-M.; Chen, J.J.W. Suppressive effect of microRNA319 expression on rice plant height. Theor. Appl. Genet. 2017, 130, 1507–1518. [Google Scholar] [CrossRef]
- Li, F.; Chen, B.; Xu, K.; Gao, G.; Yan, G.; Qiao, J.; Li, J.; Li, H.; Li, L.; Xiao, X.; et al. A genome-wide association study of plant height and primary branch number in rapeseed (Brassica napus). Plant Sci. 2016, 242, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Peng, C.; Liu, H.; Tang, M.; Yang, H.; Li, X.; Liu, J.; Sun, X.; Wang, X.; Xu, J.; et al. Genome-Wide Association Study Reveals Candidate Genes for Control of Plant Height, Branch Initiation Height and Branch Number in Rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 1246. [Google Scholar] [CrossRef]
- Satyaki, P.R.V.; Gehring, M. DNA methylation and imprinting in plants: Machinery and mechanisms. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Naveh-Many, T.; Cedar, H.; Razin, A. Sequence specificity of methylation in higher plant DNA. Nature 1981, 292, 860–862. [Google Scholar] [CrossRef]
- Meyer, P.; Niedenhof, I.; Lohuis, M.T. Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrida. EMBO J. 1994, 13, 2084–2088. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Gehring, M.; Huh, J.H.; Hsieh, T.-F.; Penterman, J.; Choi, Y.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DEMETER DNA Glycosylase Establishes MEDEA Polycomb Gene Self-Imprinting by Allele-Specific Demethylation. Cell 2006, 124, 495–506. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Silva, P.; Rodrigues, J.A.; Dotson, B.; Brooks, M.D.; Zilberman, D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 2010, 107, 18729–18734. [Google Scholar] [CrossRef]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.-K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, E4511–E4519. [Google Scholar] [CrossRef]
- Huang, H.; Liu, R.; Niu, Q.; Tang, K.; Zhang, B.; Zhang, H.; Chen, K.; Zhu, J.-K.; Lang, Z. Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. USA 2019, 116, 1430–1436. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qin, Q.; Sun, F.; Wang, Y.; Xu, D.; Li, Z.; Fu, B. Genome-Wide Differences in DNA Methylation Changes in Two Contrasting Rice Genotypes in Response to Drought Conditions. Front. Plant Sci. 2016, 7, 1675. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.J.; Liu, X.S.; Tao, H.; Tan, S.K.; Chu, S.S.; Oono, Y.; Zhang, X.D.; Chen, J.; Yang, Z.M. Variation of DNA methylation patterns associated with gene expression in rice (Oryza sativa) exposed to cadmium. Plant Cell Environ. 2016, 39, 2629–2649. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Mathioni, S.M.; Johnson, S.; Tucker, D.; Bewick, A.J.; Do Kim, K.; Daron, J.; Slotkin, R.K.; Jackson, S.A.; Parrott, W.A.; et al. Genome-Wide Reinforcement of DNA Methylation Occurs during Somatic Embryogenesis in Soybean. Plant Cell 2019, 31, 2315–2331. [Google Scholar] [CrossRef] [PubMed]
- Gupta, O.P.; Dahuja, A.; Sachdev, A.; Jain, P.K.; Kumari, S.; Vinutha, T.; Praveen, S. Cytosine Methylation of Isoflavone Synthase Gene in the Genic Region Positively Regulates Its Expression and Isoflavone Biosynthesis in Soybean Seeds. DNA Cell Biol. 2019, 38, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Mager, S.; Schönberger, B.; Ludewig, U. The transcriptome of zinc deficient maize roots and its relationship to DNA methylation loss. BMC Plant Biol. 2018, 18, 372. [Google Scholar] [CrossRef]
- Qian, Y.; Hu, W.; Liao, J.; Zhang, J.; Ren, Q. The Dynamics of DNA methylation in the maize (Zea mays L.) inbred line B73 response to heat stress at the seedling stage. Biochem. Biophys. Res. Commun. 2019, 512, 742–749. [Google Scholar] [CrossRef]
- Shafiq, S.; Zeb, Q.; Ali, A.; Sajjad, Y.; Nazir, R.; Widemann, E.; Liu, L. Lead, Cadmium and Zinc Phytotoxicity Alter DNA Methylation Levels to Confer Heavy Metal Tolerance in Wheat. Int. J. Mol. Sci. 2019, 20, 4676. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, X.; Wu, Z.; An, H.; Yi, B.; Wen, J.; Ma, C.; Shen, J.; Fu, T.; Tu, J. Genome-Wide DNA Methylation Comparison between Brassica napus Genic Male Sterile Line and Restorer Line. Int. J. Mol. Sci. 2018, 19, 2689. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, Q.; Sun, M.; Zhang, T.; Li, H.; Chen, B.; Xu, K.; Gao, G.; Li, F.; Yan, G.; et al. Global DNA methylation variations after short-term heat shock treatment in cultured microspores of Brassica napus cv. Topas. Sci. Rep. 2016, 6, 38401. [Google Scholar] [CrossRef]
- Jiang, P.; Wang, S.; Jiang, H.; Cheng, B.; Wu, K.; Ding, Y. The COMPASS-Like Complex Promotes Flowering and Panicle Branching in Rice. Plant Physiol. 2018, 176, 2761–2771. [Google Scholar] [CrossRef]
- Liu, X.; Wei, X.; Sheng, Z.; Jiao, G.; Tang, S.; Luo, J.; Hu, P. Polycomb Protein OsFIE2 Affects Plant Height and Grain Yield in Rice. PLoS ONE 2016, 11, e0164748. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Morota, G.; Rosa, G.J.M.; Gianola, D. Prediction of Plant Height in Arabidopsis thaliana Using DNA Methylation Data. Genetics 2015, 201, 779–793. [Google Scholar] [CrossRef] [PubMed]
- Loreti, E.; Betti, F.; Ladera-Carmona, M.J.; Fontana, F.; Novi, G.; Valeri, M.C.; Perata, P.P. ARGONAUTE1 and ARGONAUTE4 Regulate Gene Expression and Hypoxia Tolerance. Plant Physiol. 2020, 182, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Liu, J.; Chu, L.; Jing, X.; Wang, H.; Guo, J.; Yi, B. Exogenous Promoter Triggers APETALA3 Silencing through RNA-Directed DNA Methylation Pathway in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 4478. [Google Scholar] [CrossRef]
- Zhang, Y.; Ramming, A.; Heinke, L.; Altschmied, L.; Slotkin, R.K.; Becker, J.D.; Kappel, C.; Lenhard, M. The poly(A) polymerase PAPS1 interacts with the RNA-directed DNA-methylation pathway in sporophyte and pollen development. Plant J. 2019, 99, 655–672. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Hyvärinen, L.; Piskurewicz, U.; Lopez-Molina, L. Non-canonical RNA-directed DNA methylation participates in maternal and environmental control of seed dormancy. eLife 2019, 8, e37434. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Wang, Z.Q.; Xiao, R.; Wang, Y.; Xie, Y.; Zhou, X. iTRAQ analysis of the tobacco leaf proteome reveals that RNA-directed DNA methylation (RdDM) has important roles in defense against geminivirus-betasatellite infection. J. Proteom. 2017, 152, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Kumaki, Y.; Oda, M.; Okano, M. QUMA: Quantification tool for methylation analysis. Nucleic Acids Res. 2008, 36, W170–W175. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, C.; Long, Y.; Hopkins, C.; Kurup, S.; Liu, K.; King, G.J.; Meng, J. Universal endogenous gene controls for bisulphite conversion in analysis of plant DNA methylation. Plant Methods 2011, 7, 39. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Zhong, S.; Fei, Z.; Chen, Y.-R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J.; et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Heath, S.; Bibikova, M.; Queirós, A.C.; Navarro, A.; Clot, G.; Martínez-Trillos, A.; Castellano, G.; Brun-Heath, I.; Pinyol, M.; et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012, 44, 1236–1242. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Conneely, K.N.; Wu, H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014, 42, e69. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Xu, T.; Feng, H.; Chen, L.; Li, B.; Yao, B.; Qin, Z.; Jin, P.; Conneely, K.N. Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res. 2015, 43, e141. [Google Scholar] [CrossRef]
- Park, Y.; Wu, H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 2016, 32, 1446–1453. [Google Scholar] [CrossRef]
- Bartels, A.; Han, Q.; Nair, P.; Stacey, L.; Gaynier, H.; Mosley, M.; Huang, Q.; Pearson, J.; Hsieh, T.-F.; An, Y.-Q.; et al. Dynamic DNA Methylation in Plant Growth and Development. Int. J. Mol. Sci. 2018, 19, 2144. [Google Scholar] [CrossRef]
- Smallwood, S.A.; Lee, H.J.; Angermueller, C.; Krueger, F.; Saadeh, H.; Peat, J.; Andrews, S.R.; Stegle, O.; Reik, W.; Kelsey, G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 2014, 11, 817–820. [Google Scholar] [CrossRef]
- Lister, R.; O’Malley, R.C.; Tonti-Filippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.H.; Ecker, J.R. Highly Integrated Single-Base Resolution Maps of the Epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Carol Huang, S.-S.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Castanon, R.; Nery, J.R.; Barragan, C.; He, Y.; et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Niederhuth, C.E.; Bewick, A.J.; Ji, L.; Alabady, M.S.; Kim, K.D.; Li, Q.; Rohr, N.A.; Rambani, A.; Burke, J.M.; Udall, J.A.; et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016, 17, 194. [Google Scholar] [CrossRef] [PubMed]
- Regulski, M.; Lu, Z.; Kendall, J.; Donoghue, M.T.A.; Reinders, J.; Llaca, V.; Deschamps, S.; Smith, A.; Levy, D.; McCombie, W.R.; et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 2013, 23, 1651–1662. [Google Scholar] [CrossRef]
- Schmitz, R.J.; He, Y.; Valdés-López, O.; Khan, S.M.; Joshi, T.; Urich, M.A.; Nery, J.R.; Diers, B.; Xu, D.; Stacey, G.; et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013, 23, 1663–1674. [Google Scholar] [CrossRef] [PubMed]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Turco, G.M.; Kajala, K.; Kunde-Ramamoorthy, G.; Ngan, C.Y.; Olson, A.; Deshphande, S.; Tolkunov, D.; Waring, B.; Stelpflug, S.; Klein, P.; et al. DNA methylation and gene expression regulation associated with vascularization in Sorghum bicolor. New Phytol. 2017, 214, 1213–1229. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Nagaharu, U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilisation. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Allender, C.J.; King, G.J. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 2010, 10, 54. [Google Scholar] [CrossRef]
- Parkin, I.A.; Koh, C.; Tang, H.; Robinson, S.J.; Kagale, S.; Clarke, W.E.; Town, C.D.; Nixon, J.; Krishnakumar, V.; Bidwell, S.L.; et al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014, 15, R77. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, C.; Tang, X.; Pei, S.; Jin, D.; Guo, M.; Yang, M.; Zhang, Y. Genomic methylation and transcriptomic profiling provides insights into heading depression in inbred Brassica rapa L. ssp. pekinensis. Gene 2018, 665, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Eprintsev, A.T.; Fedorin, D.N.; Karabutova, L.A.; Igamberdiev, A.U. Expression of genes encoding subunits A and B of succinate dehydrogenase in germinating maize seeds is regulated by methylation of their promoters. J. Plant Physiol. 2016, 205, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-L.; Song, H.-X.; Liao, Q.; Yu, Y.; Jian, S.-F.; Lepo, J.E.; Liu, Q.; Rong, X.-M.; Tian, C.; Zeng, J.; et al. Nitrogen Use Efficiency Is Mediated by Vacuolar Nitrate Sequestration Capacity in Roots of Brassica napus. Plant Physiol. 2016, 170, 1684–1698. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Anwar, S.; Kuai, J.; Ullah, S.; Fahad, S.; Zhou, G. Optimization of Nitrogen Rate and Planting Density for Improving Yield, Nitrogen Use Efficiency, and Lodging Resistance in Oilseed Rape. Front. Plant Sci. 2017, 8, 532. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Saito, K. Subcellular localization of spinach cysteine synthase isoforms and regulation of their gene expression by nitrogen and sulfur. Plant Physiol. 1996, 112, 273–280. [Google Scholar] [CrossRef]
- Cui, F.; Liu, L.; Zhao, Q.; Zhang, Z.; Li, Q.; Lin, B.; Wu, Y.; Tang, S.; Xie, Q. Arabidopsis Ubiquitin Conjugase UBC32 Is an ERAD Component That Functions in Brassinosteroid-Mediated Salt Stress Tolerance. Plant Cell 2012, 24, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Eom, H.; Park, S.J.; Kim, M.K.; Kim, H.; Kang, H.; Lee, I. TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant J. 2017, 93, 79–91. [Google Scholar] [CrossRef]
- Eom, H.; Lee, I. Role of TAF15b in transcriptional regulation of autonomous pathway for flowering. Plant Signal. Behav. 2018, 13, e1471300. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Wang, Z.; Yang, C. MicroRNA Regulation of Epigenetic Modifiers in Breast Cancer. Cancers 2019, 11, 897. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhang, X.; Guan, H.; Huang, F.; Wu, L.; Hou, D.; Zheng, Z.; Yu, M.; Huang, L.; Ge, L. miR-140-5p regulates T cell differentiation and attenuates experimental autoimmune encephalomyelitis by affecting CD4+T cell metabolism and DNA methylation. Int. Immunopharmacol. 2019, 75, 105778. [Google Scholar] [CrossRef]
- Kelly, A.A.; Shaw, E.; Powers, S.J.; Kurup, S.; Eastmond, P.J. Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol. J. 2012, 11, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, B.; Sasabe, M.; Deng, X.; Machida, Y.; Lin, H.; Julie Lee, Y.R.; Liu, B. Arabidopsis MAP65-4 plays a role in phragmoplast microtubule organization and marks the cortical cell division site. New Phytol. 2017, 215, 187–201. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Ecker, J.R. Diversity and dynamics of DNA methylation: Epigenomic resources and tools for crop breeding. Breed. Sci. 2019, 69, 191–204. [Google Scholar] [CrossRef] [PubMed]
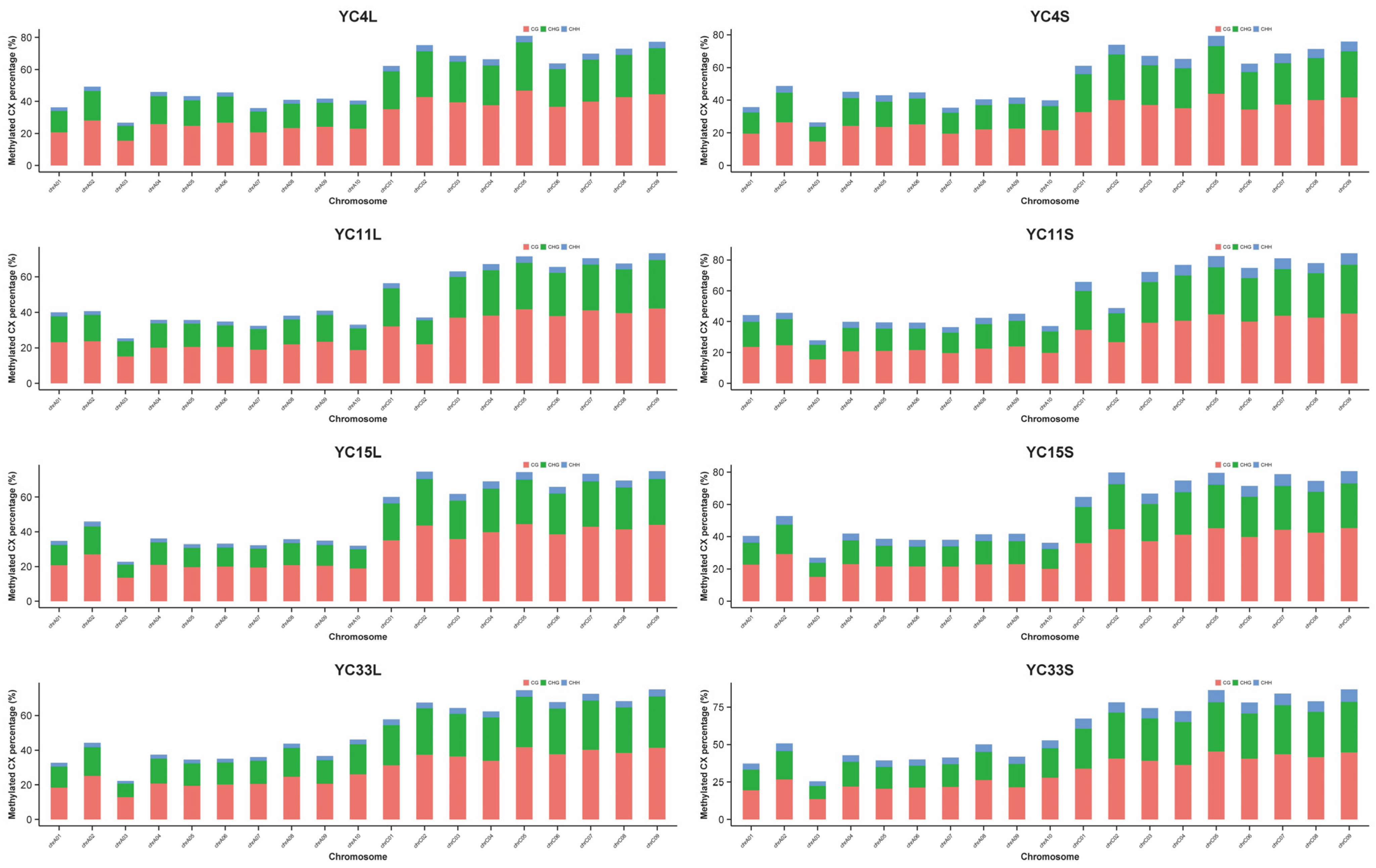
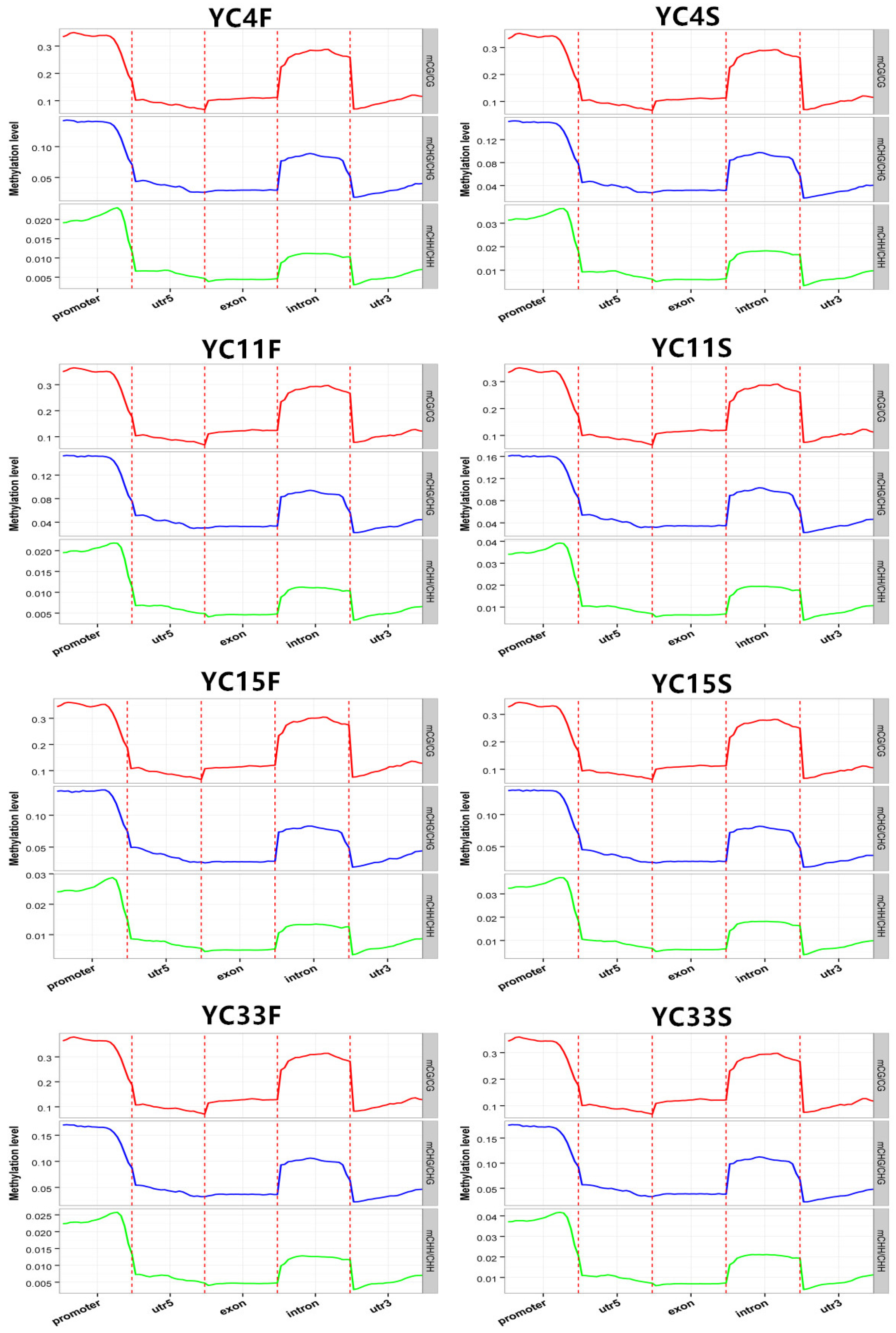

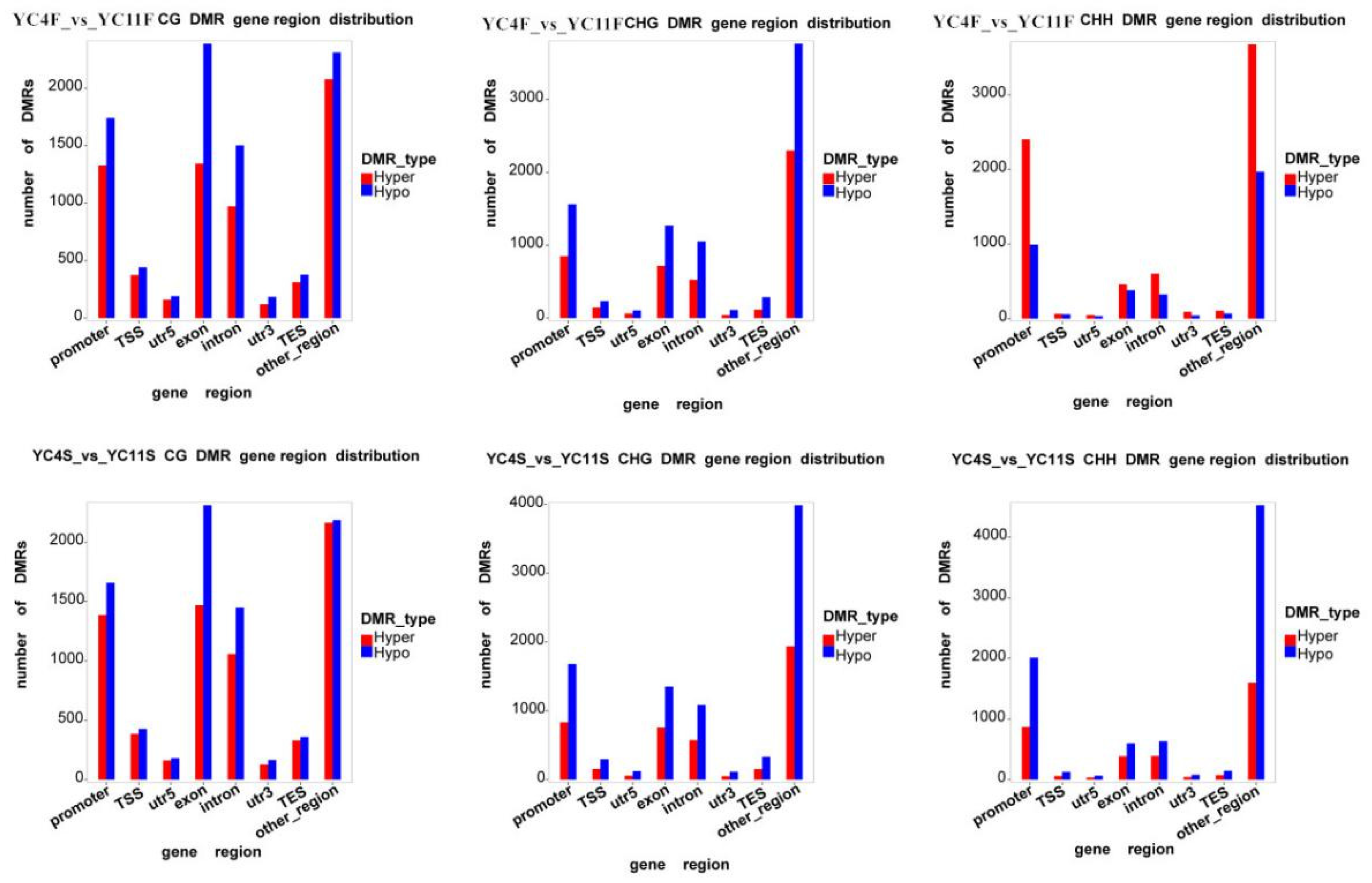
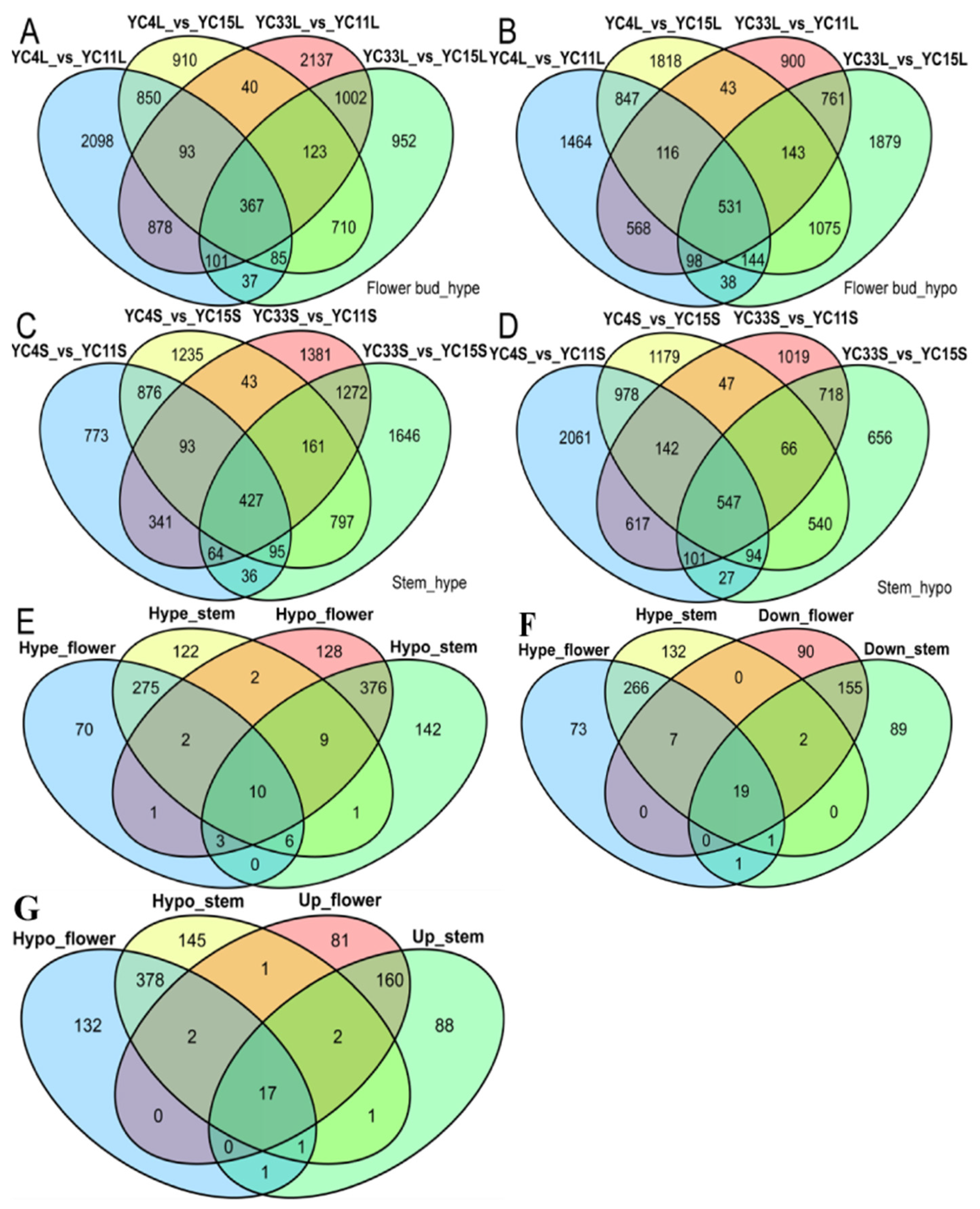
| Samples | TPC Trait | mC (%) | mCpG (%) | mCHG (%) | mCHH (%) |
|---|---|---|---|---|---|
| YC4F | High | 11.31% | 37.15% | 23.92% | 3.42% |
| YC33F | High | 10.61% | 33.59% | 23.43% | 3.26% |
| YC11F | Low | 9.96% | 33.32% | 21.05% | 2.88% |
| YC15F | Low | 10.88% | 35.98% | 21.55% | 3.54% |
| YC4S | High | 12.1% | 34.88% | 23.29% | 5.12% |
| YC33S | High | 13.78% | 36.2% | 26.69% | 6.52% |
| YC11S | Low | 12.8% | 35.64% | 24.51% | 5.71% |
| YC15S | Low | 13.11% | 37.32% | 22.72% | 6.17% |
| Gene_ID | MiRNAs | Methylation | MiRNAs_ Expression | Gene_ Expression | Sites | Comparison with Arabidopsis Genes |
|---|---|---|---|---|---|---|
| BnaAnng30250D | miR319 | hypermethylation | up | down | common | - |
| BnaC04g55380D | miR9409/miR6029 | hypermethylation | up | down | common | microtubule-associated protein 65-4 (MAP65-4) |
| BnaC08g03460D | miR5726/miR9409 | hypermethylation | up | down | common | FUNCTIONS IN: sequence-specific DNA binding transcription factor activity |
| BnaCnng50740D | miR9563 | hypermethylation | up | down | common | RNA-directed DNA polymerase (reverse transcriptase)-related family protein |
| BnaA09g11170D | miR9409 | hypermethylation | up | down | flower bud | pentatricopeptide (PPR) repeat-containing protein |
| BnaCnng64040D | miR159 | hypermethylation | up | down | common | Lipase family protein |
| BnaA07g07800D | miR319 | hypermethylation | up | down | stem apex | - |
| BnaC02g22110D | miR9409 | hypermethylation | up | down | stem apex | - |
| BnaC02g22120D | miR319 | hypermethylation | up | down | common | Spc97 family of spindle pole body (SBP) component |
| BnaA04g14930D | miR159/miR319/miR5726 | hypomethylation | down | up | common | S-locus lectin protein kinase family protein |
| BnaCnng56050D | miR122/miR5726/miR9410 | hypomethylation | down | up | common | Pyridoxal phosphate (PLP)-dependent transferases superfamily protein |
| BnaA06g09210D | miR395/miR827 | hypomethylation | down | up | flower bud | IQ-domain 28 (IQD28) |
| BnaC03g09180D | miR827 | hypomethylation | down | up | flower bud | - |
| BnaC09g30490D | miR167 | hypomethylation | down | up | flower bud | TBP-associated factor 15B (TAF15b) |
| Chromosome | Start | End | Length | Context | Gene_ID | Region | Methylation | Materials |
|---|---|---|---|---|---|---|---|---|
| chrC04 | 28,932,484 | 28,933,278 | 795 | CG | BnaC04g27680D | promoter | hypermethylation | YC4F/YC11F |
| chrC05 | 35,959,735 | 35,961,483 | 1749 | CG | BnaC05g36710D | promoter | hypermethylation | YC4F/YC11F |
| chrC06 | 3,994,751 | 3,995,435 | 685 | CG | BnaC06g03310D | promoter | hypermethylation | YC4F/YC11F |
| chrA07 | 8,074,819 | 8,075,354 | 536 | CHG | BnaA07g07800D | promoter | hypermethylation | YC4F/YC11F |
| chrC08 | 3,215,209 | 3,217,065 | 1857 | CG | BnaC08g03460D | promoter | hypermethylation | YC4F/YC11F |
| chrCnn_random | 63,802,915 | 63,803,399 | 485 | CG | BnaCnng64040D | promoter | hypermethylation | YC4F/YC11F |
| chrC03 | 60,388,542 | 60,389,391 | 850 | CG | BnaC03g70780D | promoter | hypermethylation | YC4F/YC11F |
| chrA05 | 16,370,918 | 16,372,104 | 1187 | CHG | BnaA05g21120D | promoter | hypermethylation | YC4F/YC11F |
| chrC03 | 54,554,408 | 54,555,203 | 796 | CG | BnaC03g65070D | promoter | hypermethylation | YC4F/YC11F |
| chrA09 | 20,870,973 | 20,871,783 | 811 | CG | BnaA09g27840D | promoter | hypermethylation | YC4F/YC11F |
| chrC03 | 14,444,574 | 14,445,925 | 1352 | CG | BnaC03g25740D | promoter | hypermethylation | YC4F/YC11F |
| chrA09 | 5,783,775 | 5,784,177 | 403 | CHG | BnaA09g11170D | promoter | hypermethylation | YC4F/YC11F |
| chrC04 | 29,430,769 | 29,431,783 | 1015 | CG | BnaC04g28000D | promoter | hypermethylation | YC4F/YC11F |
| chrC04 | 47,080,059 | 47,080,491 | 433 | CHG | BnaC04g48480D | promoter | hypomethylation | YC4F/YC11F |
| chrC03 | 59,224,107 | 59,224,481 | 375 | CHG | BnaC03g69480D | promoter | hypomethylation | YC4F/YC11F |
| chrA04 | 12,508,634 | 12,509,001 | 368 | CHG | BnaA04g14930D | promoter | hypomethylation | YC4F/YC11F |
| chrA05 | 19,453,849 | 19,454,171 | 323 | CHG | BnaA05g26660D | promoter | hypomethylation | YC4F/YC11F |
| chrA10 | 6,400,443 | 6,401,340 | 898 | CG | BnaA10g07880D | promoter | hypomethylation | YC4F/YC11F |
| chrC09 | 33,390,641 | 33,391,416 | 776 | CG | BnaC09g30490D | promoter | hypomethylation | YC4F/YC11F |
| chrC03 | 4,369,774 | 4,371,039 | 1266 | CG | BnaC03g09180D | promoter | hypomethylation | YC4F/YC11F |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, L.; Cao, L.; Zeng, L.; Lu, K.; Qu, C.; Li, J.; Chen, Z. MicroRNA-Mediated Changes in DNA Methylation Affect the Expression of Genes Involved in the Thickness-of-Pod-Canopy Trait in Brassica napus L. Agronomy 2025, 15, 398. https://doi.org/10.3390/agronomy15020398
Jia L, Cao L, Zeng L, Lu K, Qu C, Li J, Chen Z. MicroRNA-Mediated Changes in DNA Methylation Affect the Expression of Genes Involved in the Thickness-of-Pod-Canopy Trait in Brassica napus L. Agronomy. 2025; 15(2):398. https://doi.org/10.3390/agronomy15020398
Chicago/Turabian StyleJia, Ledong, Lu Cao, Lijun Zeng, Kun Lu, Cunmin Qu, Jiana Li, and Zhiyou Chen. 2025. "MicroRNA-Mediated Changes in DNA Methylation Affect the Expression of Genes Involved in the Thickness-of-Pod-Canopy Trait in Brassica napus L." Agronomy 15, no. 2: 398. https://doi.org/10.3390/agronomy15020398
APA StyleJia, L., Cao, L., Zeng, L., Lu, K., Qu, C., Li, J., & Chen, Z. (2025). MicroRNA-Mediated Changes in DNA Methylation Affect the Expression of Genes Involved in the Thickness-of-Pod-Canopy Trait in Brassica napus L. Agronomy, 15(2), 398. https://doi.org/10.3390/agronomy15020398








