Genome-Wide Identification, Characterization, and Expression Profiles of the CCO Gene Family in Raphanus sativus L. Response to Fusarium oxysporum Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of CCO Gene Family Members
2.2. Phylogenetic and Gene Structure Analysis
2.3. Chromosomal Localization and Synteny Analysis
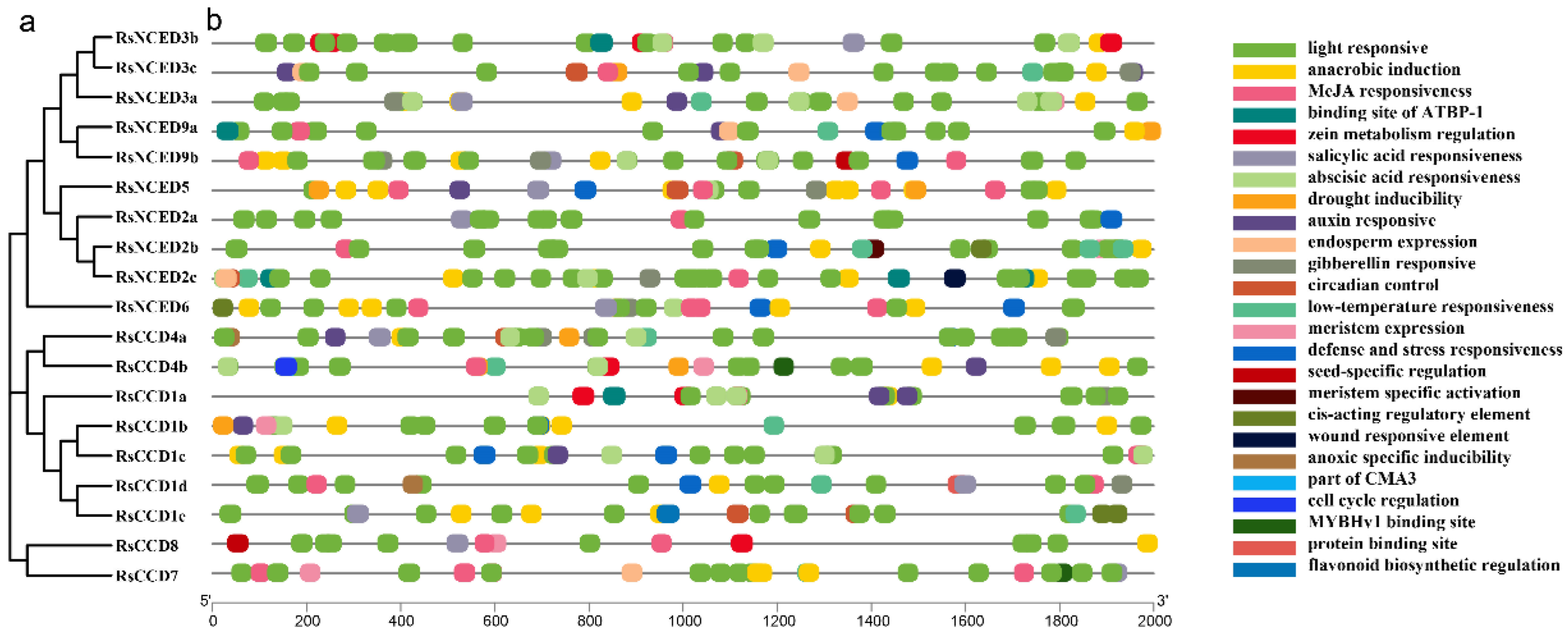
2.4. Promoter and Cis-Acting Element Analysis
2.5. Plant Growth and F. oxysporum Inoculations
2.6. Total RNA Extraction and Gene Expression Analysis
3. Results
3.1. Identification and Physicochemical Properties of CCO Gene Family Members
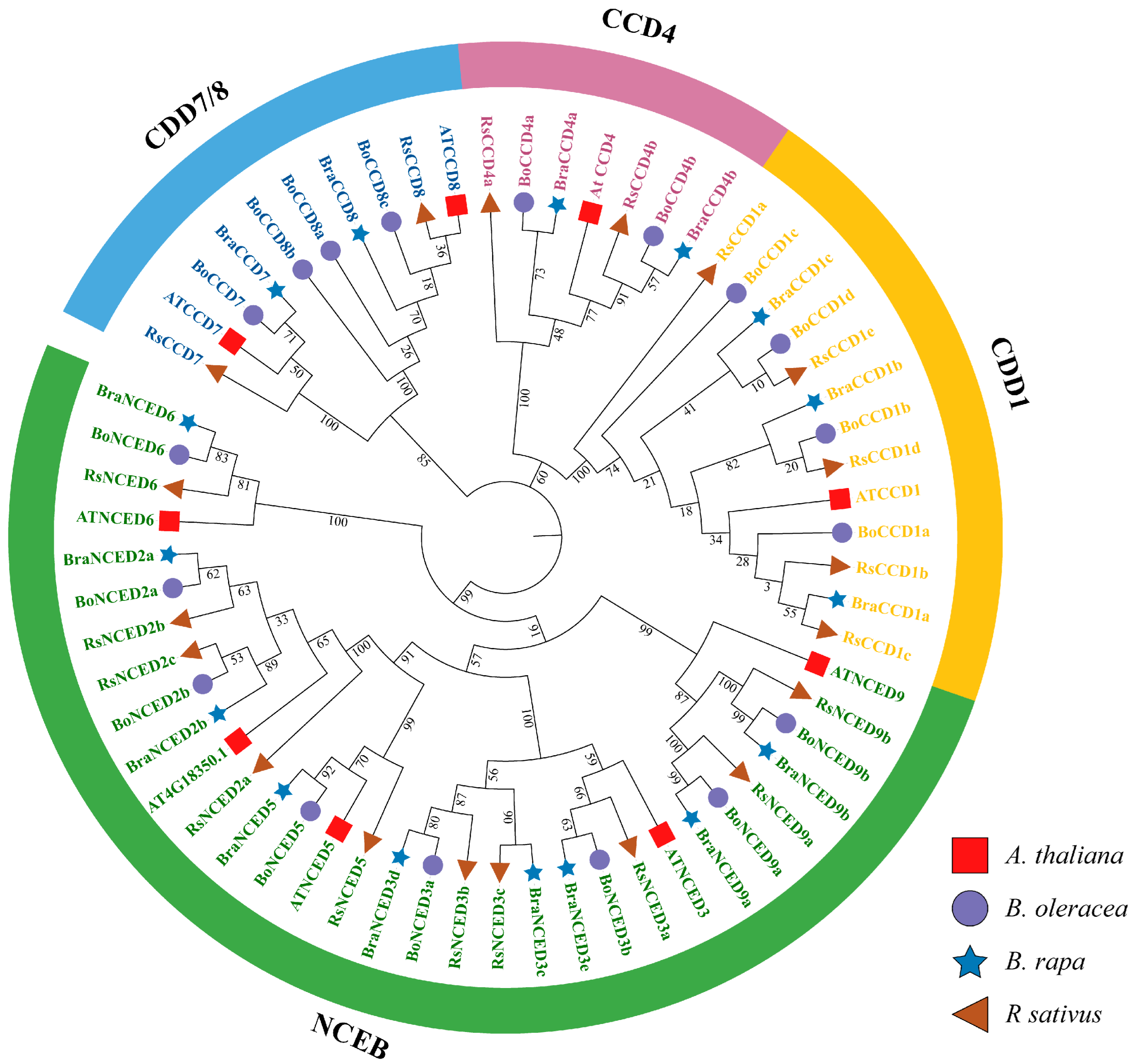
3.2. Phylogenetic Analysis and Classification of the CCO Family
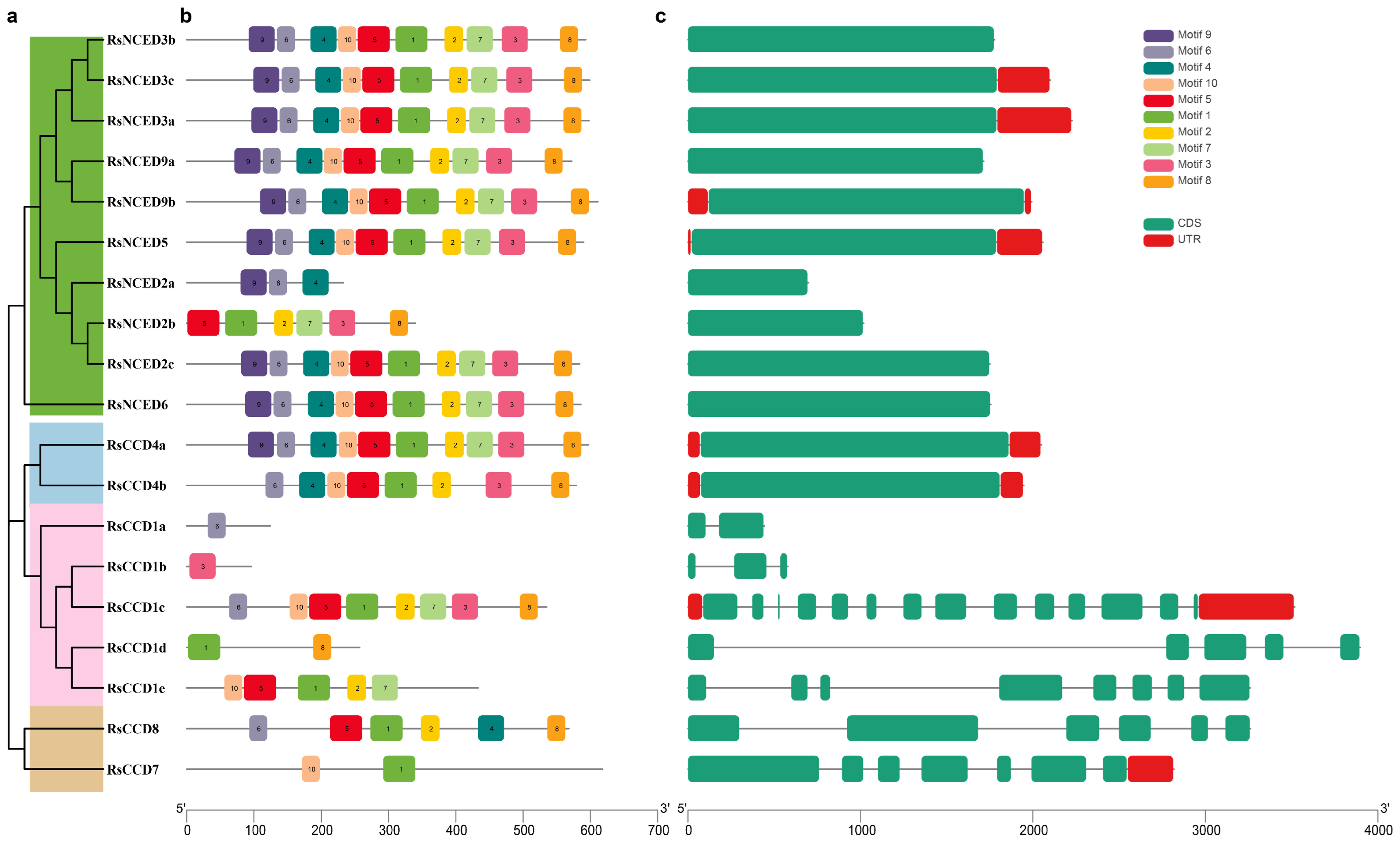
3.3. Gene Structure and Conserved Motif Analysis of the Radish CCO Gene Family
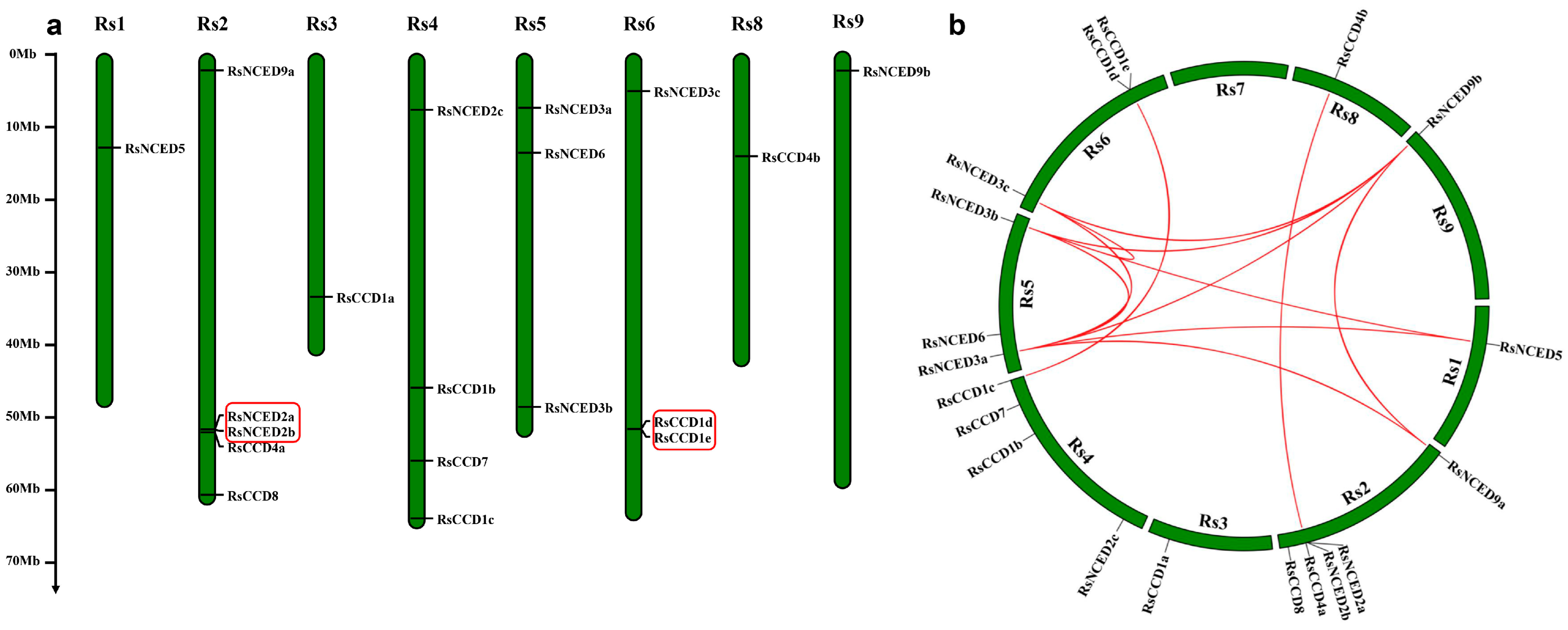
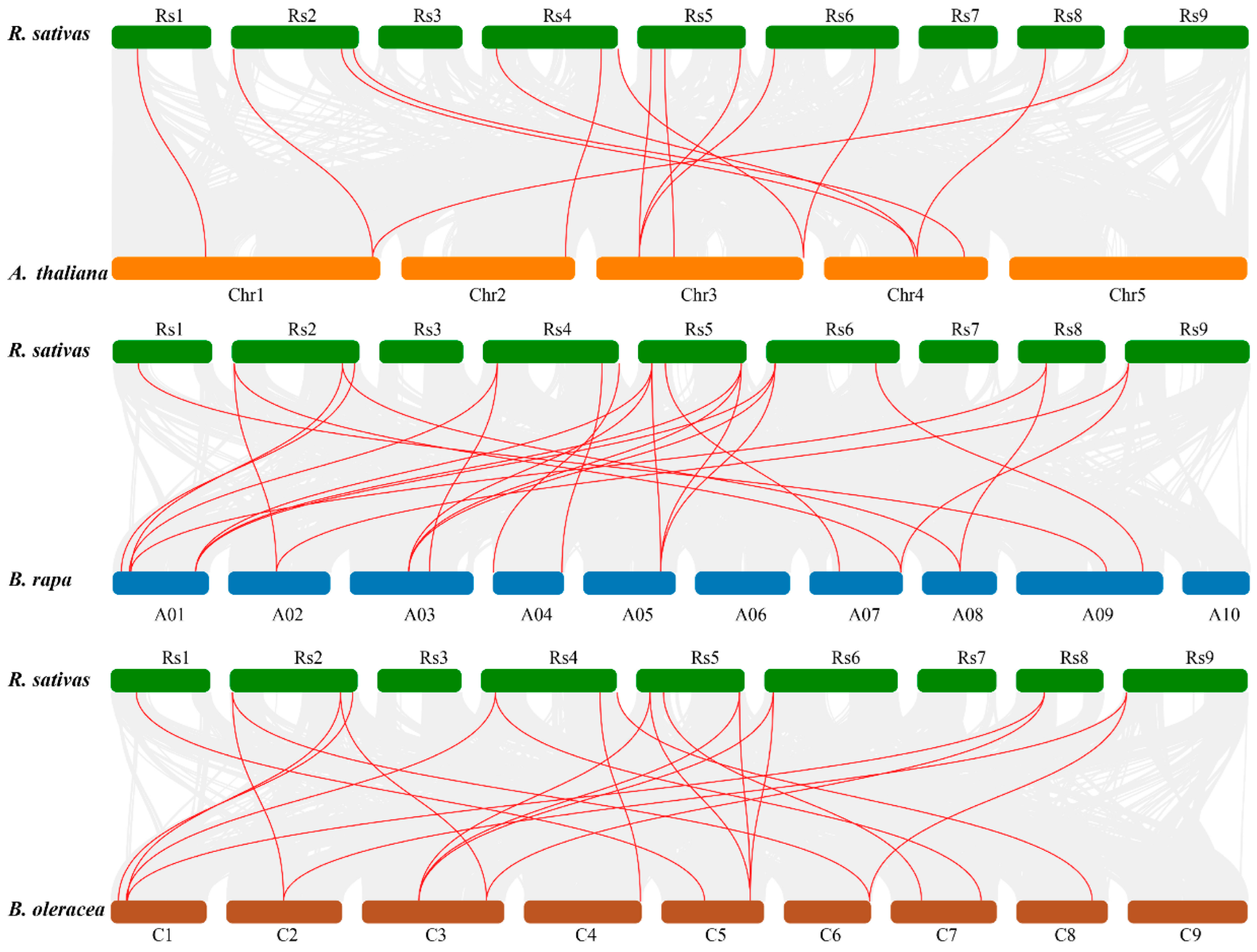
3.4. Chromosomal Localization and Synteny Analysis
3.5. Prediction of Cis-Acting Elements in RsCCO Promoter Regions
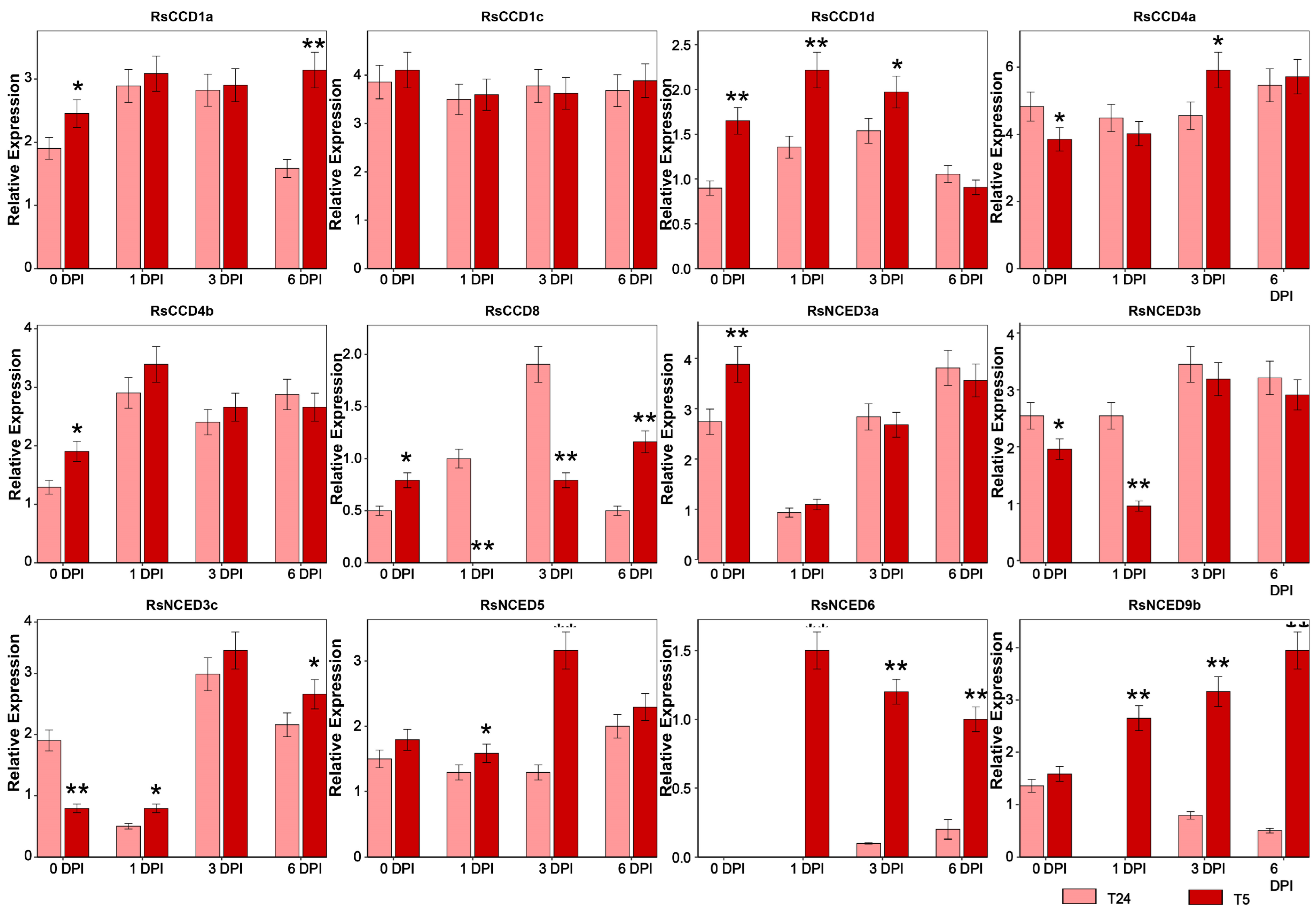
3.6. Expression Pattern Analysis of RsCCO Genes Under Fusarium Wilt Stress
4. Discussion
4.1. Genome-Wide Identification and Classification of the RsCCO Gene Family
4.2. Conserved Motifs, Gene Structure, and Implications for Enzymatic Function
4.3. Chromosomal Distribution, Synteny, and Evolutionary Dynamics in Brassicaceae
4.4. Functional Roles of the CCO Family: Integrating ABA and Plant Defense
4.5. Limitations and Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nisar, N.; Li, L.; Lu, S.; Khin, N.; Pogson, B. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef]
- Al-Babili, S.; Bouwmeester, H. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.-P.; Li, C.; Bouwmeester, H.J.; Al-Babili, S. Strigolactone biosynthesis and signal transduction. In Strigolactones-Biology and Applications; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–45. [Google Scholar]
- Rella, A.; Farnoud, A.M.; Del Poeta, M. Plasma membrane lipids and their role in fungal virulence. Prog. Lipid Res. 2016, 61, 63–72. [Google Scholar] [CrossRef]
- Auldridge, M.E.; McCarty, D.R.; Klee, H.J. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 2006, 9, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.J.; Bugg, T.D. Enzymology of the carotenoid cleavage dioxygenases: Reaction mechanisms, inhibition and biochemical roles. Arch. Biochem. Biophys. 2014, 544, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M. Carotenoid oxidation products as stress signals in plants. Plant J. 2014, 79, 597–606. [Google Scholar] [CrossRef]
- Walter, M.H.; Floss, D.S.; Strack, D. Apocarotenoids: Hormones, mycorrhizal metabolites and aroma volatiles. Planta 2010, 232, 1–17. [Google Scholar] [CrossRef]
- Heier, C.; Haemmerle, G. Fat in the heart: The enzymatic machinery regulating cardiac triacylglycerol metabolism. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2016, 1861, 1500–1512. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, G.; Cao, C.; Liu, P. Metabolism, signaling, and transport of jasmonates. Plant Commun. 2021, 2, 100231. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, A.; Kishimoto, S.; Aida, R.; Yoshioka, S.; Sumitomo, K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006, 142, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Floss, D.S.; Walter, M.H. Role of carotenoid cleavage dioxygenase 1 (CCD1) in apocarotenoid biogenesis revisited. Plant Signal. Behav. 2009, 4, 172–175. [Google Scholar] [CrossRef]
- Bruno, M.; Koschmieder, J.; Wuest, F.; Schaub, P.; Fehling-Kaschek, M.; Timmer, J.; Beyer, P.; Al-Babili, S. Enzymatic study on AtCCD4 and AtCCD7 and their potential to form acyclic regulatory metabolites. J. Exp. Bot. 2016, 67, 5993–6005. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Simkin, A.J.; Schwartz, S.H.; Auldridge, M.; Taylor, M.G.; Klee, H.J. The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J. 2004, 40, 882–892. [Google Scholar] [CrossRef]
- Liu, H.; Micic, N.; Miller, S.; Crocoll, C.; Bjarnholt, N. Species-specific dynamics of specialized metabolism in germinating sorghum grain revealed by temporal and tissue-resolved transcriptomics and metabolomics. Plant Physiol. Biochem. 2023, 196, 807–882. [Google Scholar] [CrossRef]
- Su, W.; Zhang, C.; Feng, J.; Feng, A.; You, C.; Ren, Y.; Wang, D.; Sun, T.; Su, Y.; Xu, L. Genome-wide identification, characterization and expression analysis of the carotenoid cleavage oxygenase (CCO) gene family in Saccharum. Plant Physiol. Biochem. 2021, 162, 196–210. [Google Scholar] [CrossRef]
- Hao, G.; Usgaard, T.; Tiley, H.; McCormick, S.; Vaughan, M.; Ward, T. Trichothecene NX-3 is required for Fusarium graminearum initial infection and disease spread. In Meeting Abstract; National Center for Agricultural Utilization Research: Peoria, IL, USA, 2021. [Google Scholar]
- Singh, N.K.; Anand, S.; Jain, A.; Das, S. Comparative genomics and synteny analysis of KCS17-KCS18 cluster across different genomes and sub-genomes of brassicaceae for analysis of its evolutionary history. Plant Mol. Biol. Report. 2017, 35, 237–251. [Google Scholar] [CrossRef]
- Sun, G.; Liao, J.; Kurze, E.; Hoffmann, T.D.; Steinchen, W.; McGraphery, K.; Habegger, R.; Marek, L.; Catici, D.A.; Ludwig, C. Apocarotenoids are allosteric effectors of a dimeric plant glycosyltransferase involved in defense and lignin formation. New Phytol. 2023, 238, 2080–2098. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, Y.; Shimomura, M.; Komatsu, K.; Namiki, N.; Shibata-Hatta, M.; Imai, M.; Katayose, Y.; Mukai, Y.; Kanamori, H.; Kurita, K. The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci. Rep. 2015, 5, 1083. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.H.; Strack, D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 2011, 28, 663–692. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, Y.; Dong, J.; Zhang, W.; Tang, M.; Zhang, W.; Wang, K.; Chen, Y.; Zhang, X.; He, Q. A chromosome-level genome assembly of radish (Raphanus sativus L.) reveals insights into genome adaptation and differential bolting regulation. Plant Biotechnol. J. 2023, 21, 990–1004. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Choi, S.R.; Ramchiary, N.; Miao, X.; Lee, S.H.; Sun, H.J.; Kim, S.; Ahn, C.H.; Lim, Y.P. Comparative mapping of Raphanus sativus genome using Brassica markers and quantitative trait loci analysis for the Fusarium wilt resistance trait. Theor. Appl. Genet. 2013, 126, 2553–2562. [Google Scholar] [CrossRef]
- Zhou, X.-T.; Jia, L.-D.; Duan, M.-Z.; Chen, X.; Qiao, C.-L.; Ma, J.-Q.; Zhang, C.; Jing, F.-Y.; Zhang, S.-S.; Yang, B. Genome-wide identification and expression profiling of the carotenoid cleavage dioxygenase (CCD) gene family in Brassica napus L. PLoS ONE 2020, 15, e0238179. PLoS ONE 2020, 15, e0238179. [Google Scholar]
- Zhou, Q.; Li, Q.; Li, P.; Zhang, S.; Liu, C.; Jin, J.; Cao, P.; Yang, Y. Carotenoid cleavage dioxygenases: Identification, expression, and evolutionary analysis of this gene family in tobacco. Int. J. Mol. Sci. 2019, 20, 5796. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yao, X.; He, A.; Xue, G.; Yang, H.; Fan, Y.; Yang, S.; Ruan, J. Genome-wide identification and gene expression pattern analysis of the carotenoid cleavage oxygenase gene family in Fagopyrum tataricum. BMC Plant Biol. 2025, 25, 466. [Google Scholar] [CrossRef]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11178–E11187. [Google Scholar] [CrossRef]
- Cheng, C.; Yang, R.; Yin, L.; Zhang, J.; Gao, L.; Lu, R.; Yang, Y.; Wang, P.; Mu, X.; Zhang, S. Characterization of carotenoid cleavage oxygenase genes in Cerasus humilis and functional analysis of ChCCD1. Plants 2023, 12, 2114. [Google Scholar] [CrossRef]
- Zheng, X.; Yang, Y.; Al-Babili, S. Exploring the diversity and regulation of apocarotenoid metabolic pathways in plants. Front. Plant Sci. 2021, 12, 787049. [Google Scholar] [CrossRef]
- Jeong, Y.-M.; Chung, W.-H.; Chung, H.; Kim, N.; Park, B.-S.; Lim, K.-B.; Yu, H.-J.; Mun, J.-H. Comparative analysis of the radish genome based on a conserved ortholog set (COS) of Brassica. Theor. Appl. Genet. 2014, 127, 1975–1989. [Google Scholar] [CrossRef]
- Walden, N.; Schranz, M.E. Synteny identifies reliable orthologs for phylogenomics and comparative genomics of the Brassicaceae. Genome Biol. Evol. 2023, 15, evad034. [Google Scholar] [CrossRef]
- Zeng, L.; Zeng, L.; Wang, Y.; Xie, Z.; Zhao, M.; Chen, J.; Ye, X.; Tie, W.; Li, M.; Shang, S. Identification and expression of the CCO family during development, ripening and stress response in banana. Genetica 2023, 151, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Boba, A.; Kostyn, K.; Kozak, B.; Wojtasik, W.; Preisner, M.; Prescha, A.; Gola, E.M.; Lysh, D.; Dudek, B.; Szopa, J. Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 2020, 251, 50. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Chang, Y.; Wang, J. Identification of genes responsible for stress resistance in Fusarium oxysporum-inoculated flax seedlings using weighted gene co-expression network analysis. Eur. J. Plant Pathol. 2022, 163, 513–528. [Google Scholar] [CrossRef]
- Kaushal, M.; Mahuku, G.; Swennen, R. Comparative transcriptome and expression profiling of resistant and susceptible banana cultivars during infection by Fusarium oxysporum. Int. J. Mol. Sci. 2021, 22, 3002. [Google Scholar] [CrossRef]

| Sequence ID | Rename | Gene Position | Amino NO. | CDS Length | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Rsa1g020070.1 | RsNCED5 | Rs1 | 11432111 | 11434169 | 589 | 1770 | 65,319.91 | 5.59 | 44.45 | 81.09 |
| Rsa2g001310.1 | RsNCED9a | Rs2 | 874445 | 876160 | 571 | 1716 | 63,884.73 | 6.00 | 42.70 | 80.49 |
| Rsa2g034050.1 | RsNCED2a | Rs2 | 49901379 | 49902077 | 232 | 699 | 25,757.87 | 9.88 | 49.28 | 87.07 |
| Rsa2g034060.1 | RsNCED2b | Rs2 | 49902091 | 49903110 | 339 | 1020 | 38,160.35 | 4.89 | 43.30 | 87.58 |
| Rsa2g034800.1 | RsCCD4a | Rs2 | 50306503 | 50308552 | 596 | 1791 | 65,701.83 | 6.34 | 46.70 | 80.79 |
| Rsa2g044970.1 | RsCCD8 | Rs2 | 55823959 | 55827221 | 567 | 1704 | 63,959.94 | 7.14 | 32.12 | 80.95 |
| Rsa3g018990.1 | RsCCD1a | Rs3 | 31801220 | 31801663 | 123 | 372 | 13,990.17 | 6.00 | 47.07 | 76.75 |
| Rsa4g010680.1 | RsNCED2c | Rs4 | 6306354 | 6308105 | 583 | 1752 | 64,952.98 | 5.38 | 47.50 | 86.28 |
| Rsa4g035880.1 | RsCCD1b | Rs4 | 44228908 | 44229488 | 95 | 288 | 10,682.19 | 5.41 | 22.84 | 83.05 |
| Rsa4g050050.1 | RsCCD7 | Rs4 | 54180447 | 54183265 | 617 | 1854 | 69,335.66 | 6.01 | 45.89 | 73.61 |
| Rsa4g059070.1 | RsCCD1c | Rs4 | 62045500 | 62049018 | 534 | 1605 | 60,191.09 | 5.67 | 35.34 | 83.18 |
| Rsa5g009680.1 | RsNCED3a | Rs5 | 6008373 | 6010600 | 597 | 1794 | 65,683.13 | 5.68 | 42.67 | 78.56 |
| Rsa5g017550.1 | RsNCED6 | Rs5 | 12157040 | 12158797 | 585 | 1758 | 65,152.36 | 5.72 | 39.17 | 82.91 |
| Rsa5g051130.1 | RsNCED3b | Rs5 | 46819839 | 46821617 | 592 | 1779 | 65,437.04 | 5.70 | 44.90 | 78.72 |
| Rsa6g006820.1 | RsNCED3c | Rs6 | 3721915 | 3724016 | 598 | 1797 | 65,802.37 | 5.90 | 44.27 | 76.62 |
| Rsa6g044170.1 | RsCCD1d | Rs6 | 49813437 | 49817335 | 256 | 771 | 29,094.44 | 6.50 | 35.27 | 75.00 |
| Rsa6g044210.1 | RsCCD1e | Rs6 | 49873388 | 49876648 | 432 | 1299 | 49,328.98 | 6.29 | 30.57 | 83.03 |
| Rsa8g021180.1 | RsCCD4b | Rs8 | 12612204 | 12614151 | 578 | 1737 | 63,266.25 | 6.72 | 42.76 | 87.35 |
| Rsa9g001690.1 | RsNCED9b | Rs9 | 1244782 | 1246776 | 610 | 1833 | 67,759.82 | 5.97 | 49.34 | 79.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Zuo, W.; Shi, P.; Cao, X.; Xia, Y.; Tan, R.; Hao, S.; Sun, H.; Hu, W. Genome-Wide Identification, Characterization, and Expression Profiles of the CCO Gene Family in Raphanus sativus L. Response to Fusarium oxysporum Resistance. Agronomy 2025, 15, 2722. https://doi.org/10.3390/agronomy15122722
Ma Y, Zuo W, Shi P, Cao X, Xia Y, Tan R, Hao S, Sun H, Hu W. Genome-Wide Identification, Characterization, and Expression Profiles of the CCO Gene Family in Raphanus sativus L. Response to Fusarium oxysporum Resistance. Agronomy. 2025; 15(12):2722. https://doi.org/10.3390/agronomy15122722
Chicago/Turabian StyleMa, Yinbo, Wei Zuo, Piaopiao Shi, Xinyi Cao, Yuchen Xia, Run Tan, Shuai Hao, Hanyan Sun, and Wenxing Hu. 2025. "Genome-Wide Identification, Characterization, and Expression Profiles of the CCO Gene Family in Raphanus sativus L. Response to Fusarium oxysporum Resistance" Agronomy 15, no. 12: 2722. https://doi.org/10.3390/agronomy15122722
APA StyleMa, Y., Zuo, W., Shi, P., Cao, X., Xia, Y., Tan, R., Hao, S., Sun, H., & Hu, W. (2025). Genome-Wide Identification, Characterization, and Expression Profiles of the CCO Gene Family in Raphanus sativus L. Response to Fusarium oxysporum Resistance. Agronomy, 15(12), 2722. https://doi.org/10.3390/agronomy15122722




