Comparative Transcriptome Analysis of Leaves and Roots Revealed Organ-Specific and Cross-Stress Defense Strategies of Pearl Millet Under Different Abiotic Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Plant Growth and Stress Treatment
2.3. RNA Extraction, Library Preparation, and Sequencing
2.4. Sequencing Data Standardization and Quality Assessment
2.5. Transcriptome Data Analysis
3. Results
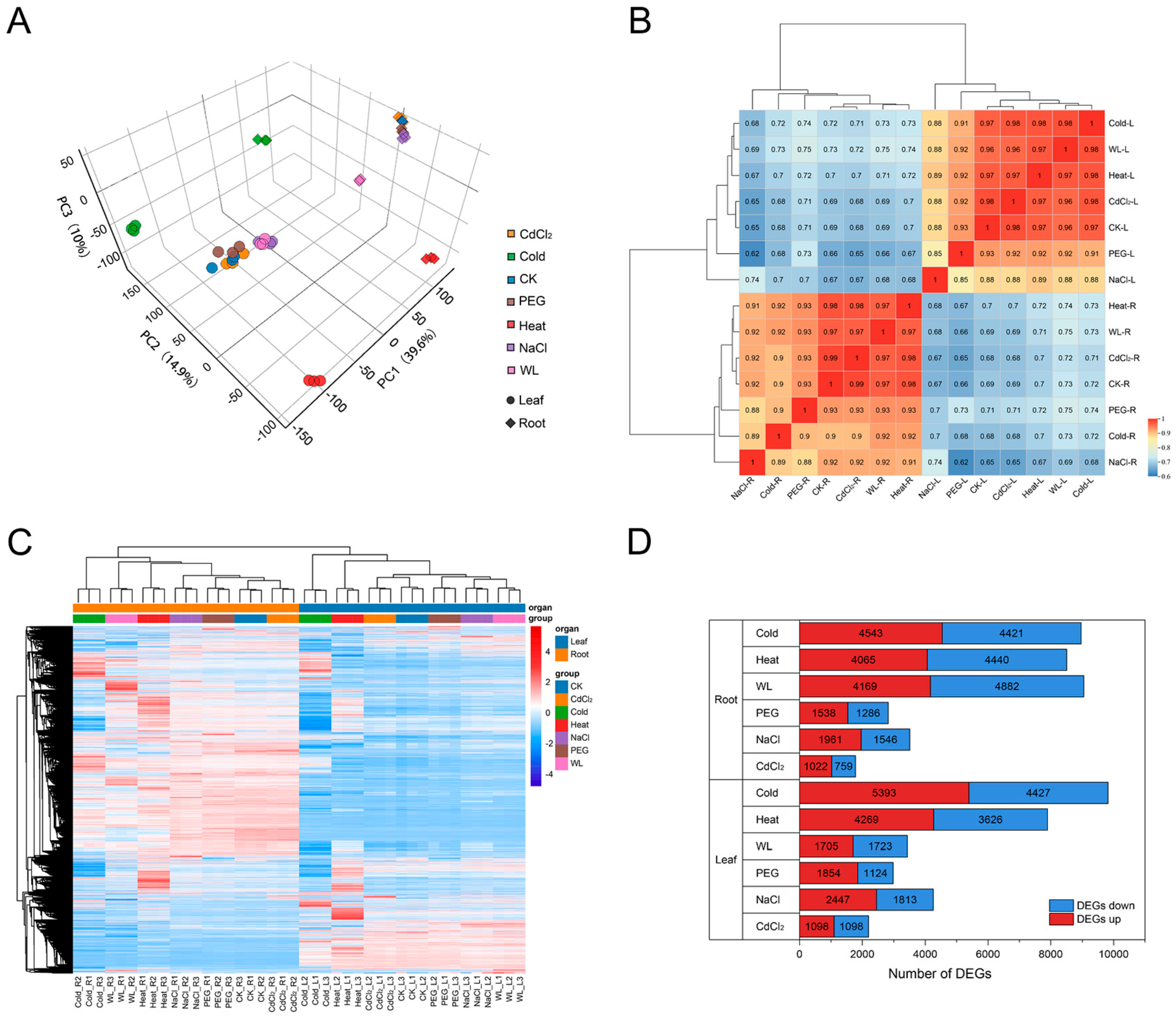
3.1. Transcriptome Sequencing Overview and Global Response Characteristics
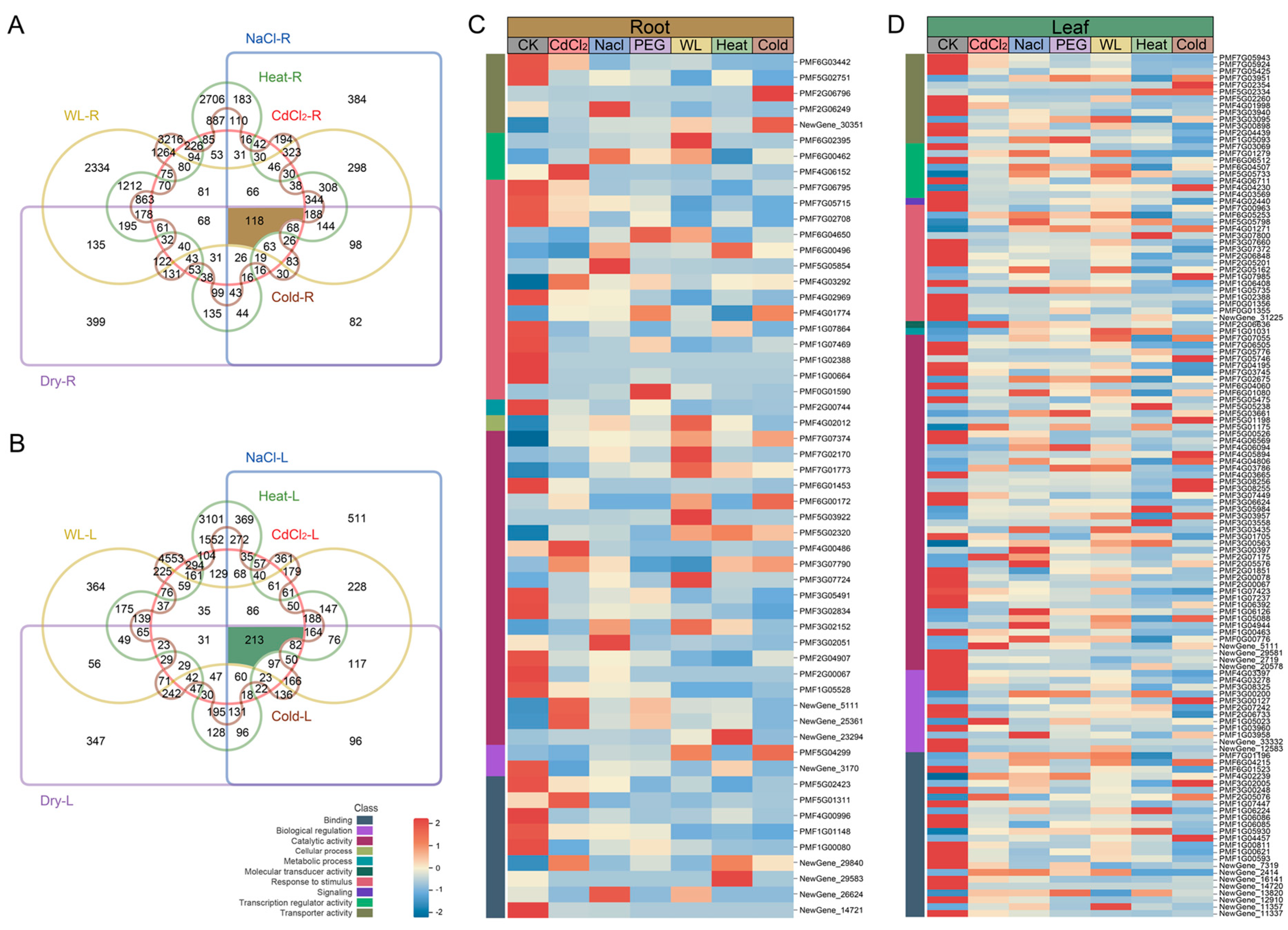
3.2. Differential Expression Gene Analysis
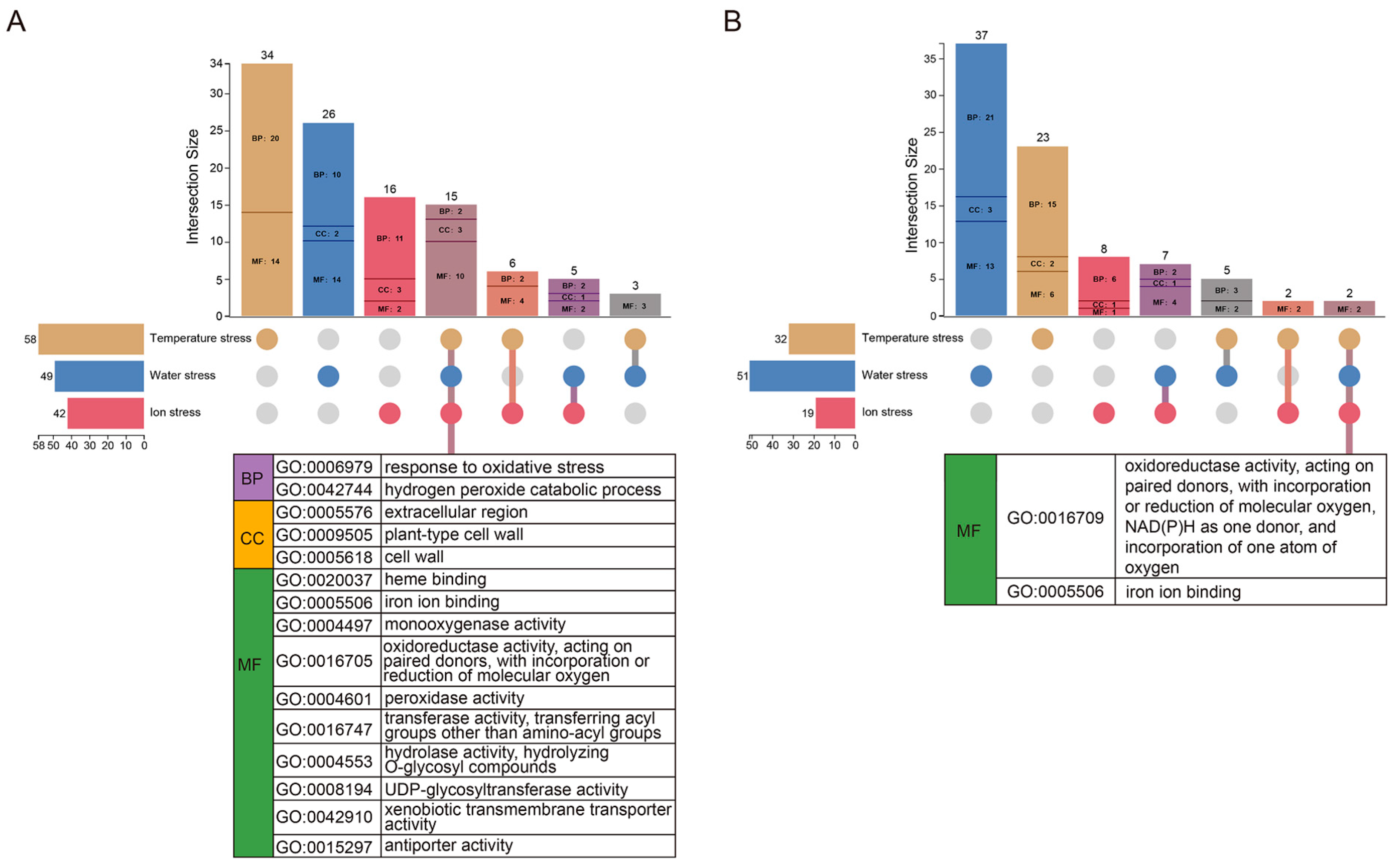
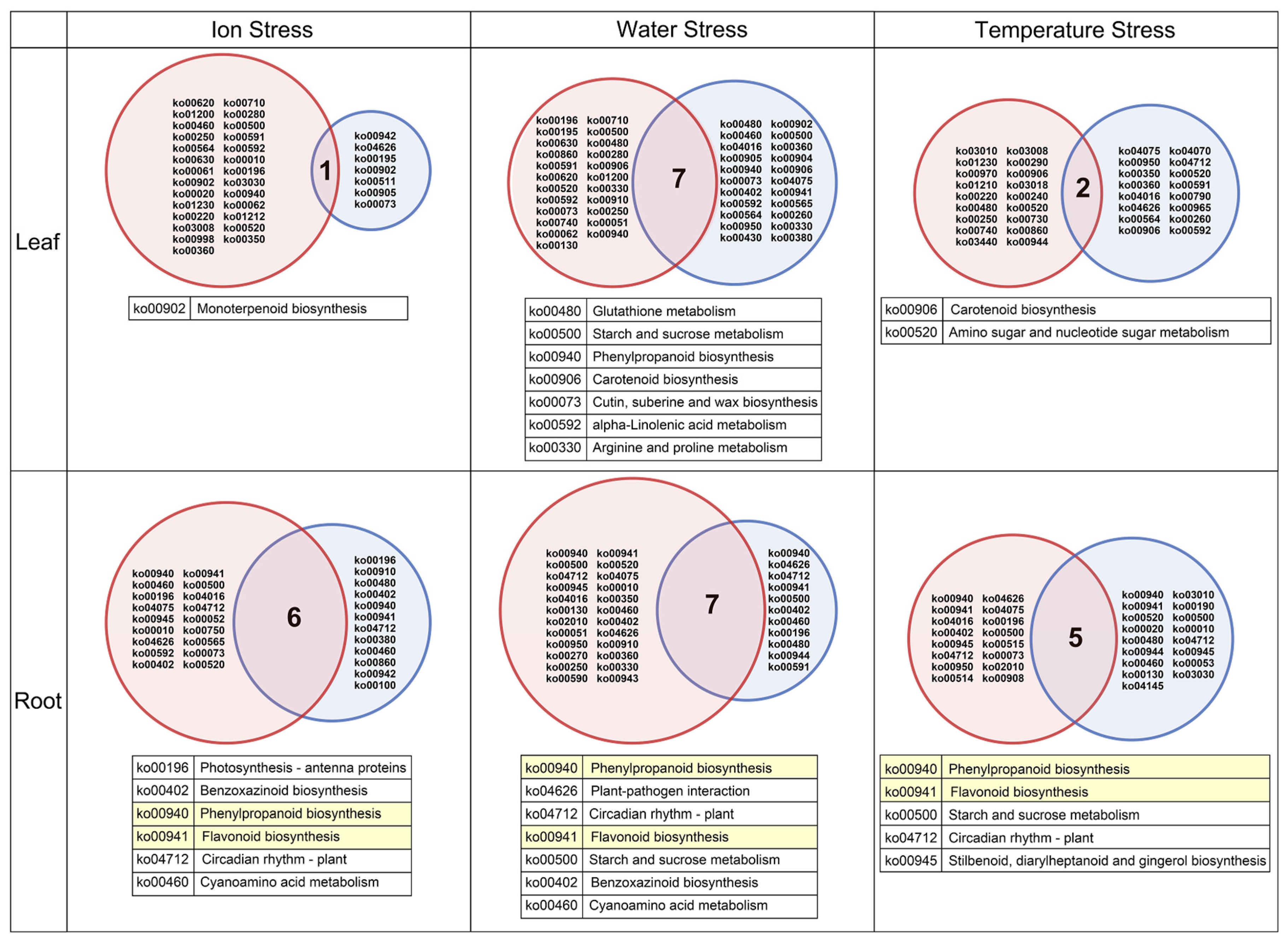
3.3. Enrichment Analysis Based on DEGs
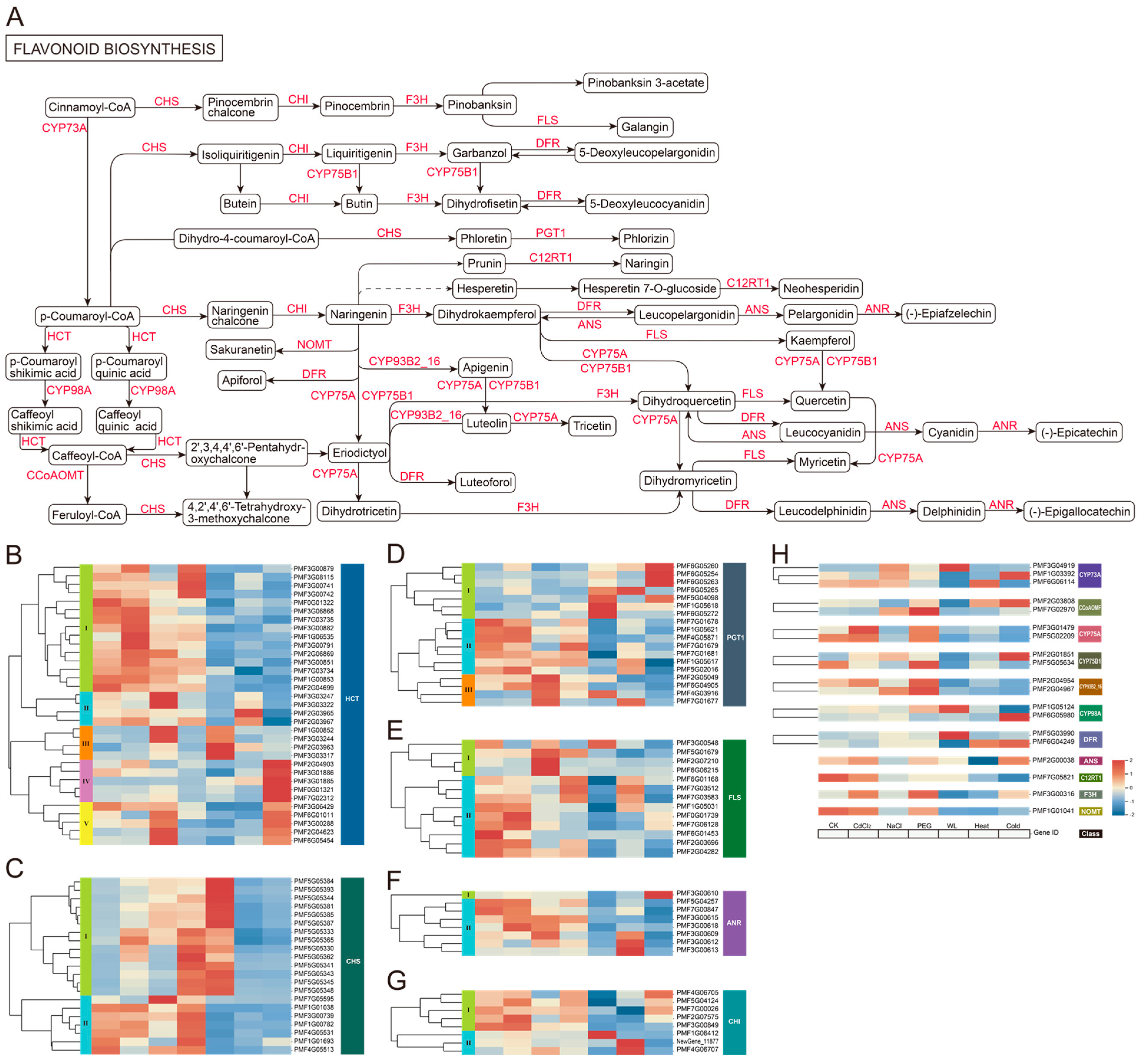
3.4. Analysis of Phenylpropanoid and Flavonoid Biosynthesis Pathways in Roots
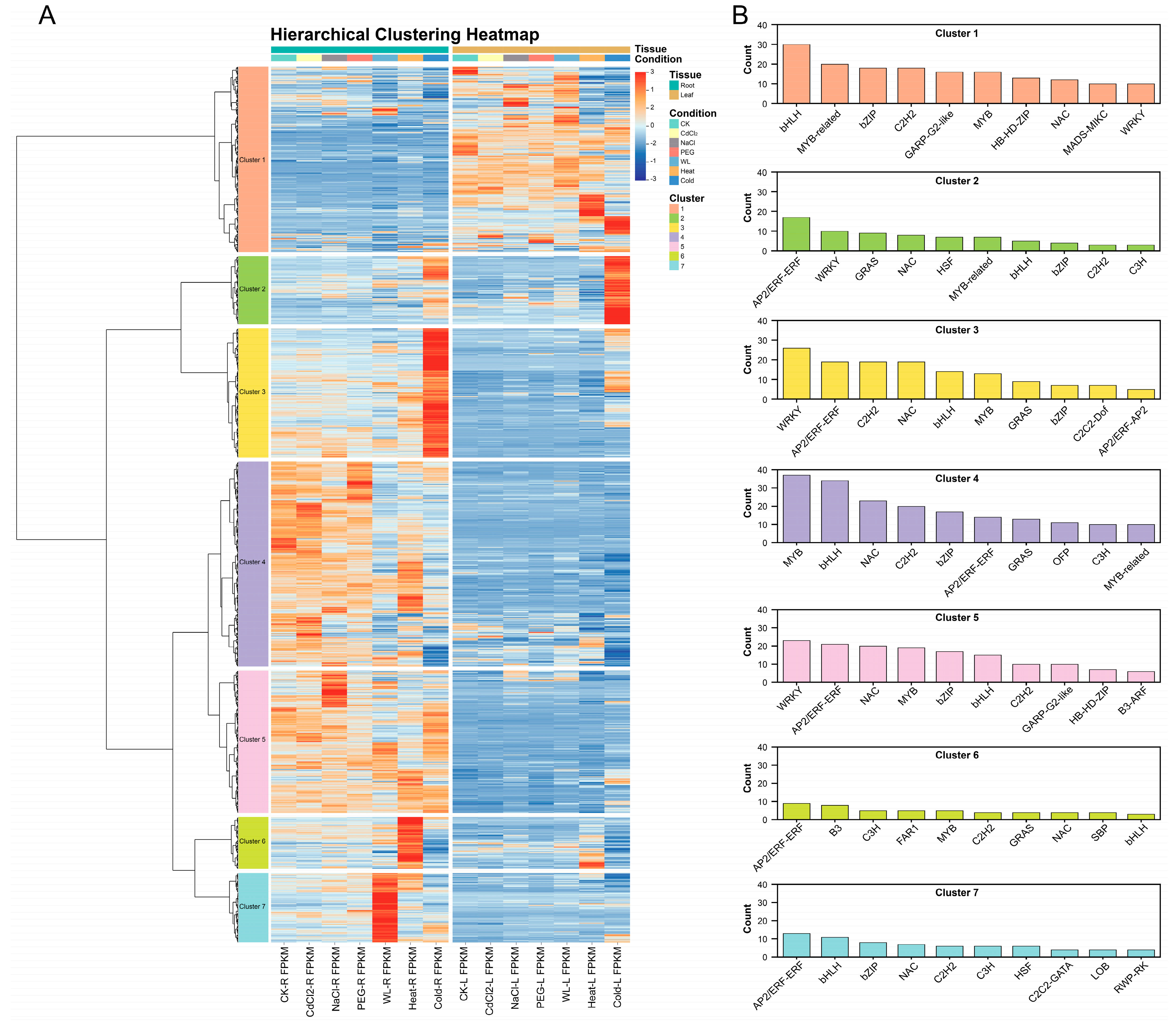
3.5. Cluster Analysis of TF Expression Profiles Under Different Stresses
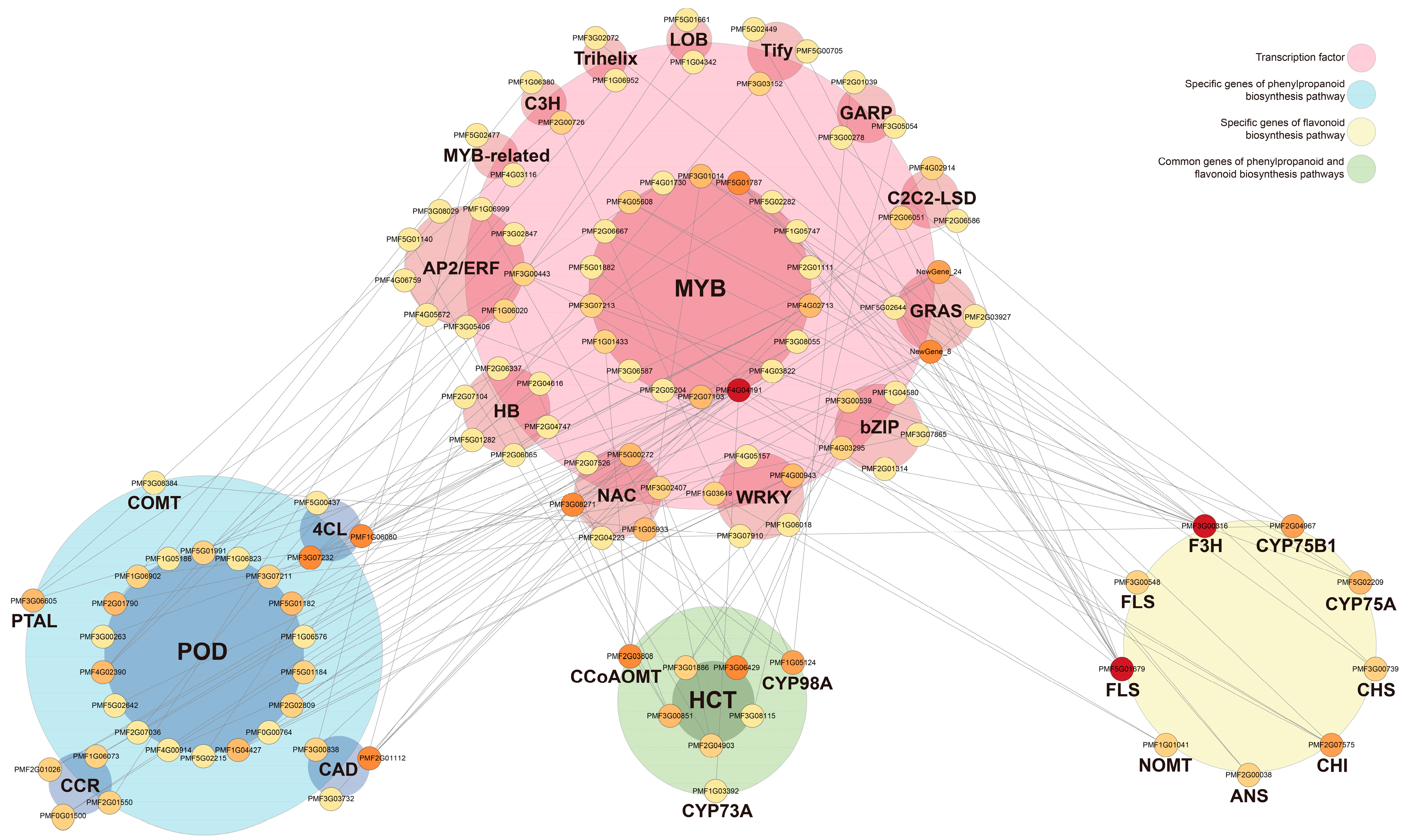
3.6. Interaction Network Analysis of TFs Regulating Phenylpropanoid/Flavonoid Biosynthesis Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zandalinas, S.I.; Fritschi, F.B.; Mittler, R. Global Warming, Climate Change, and Environmental Pollution: Recipe for a Multifactorial Stress Combination Disaster. Trends Plant Sci. 2021, 26, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Chele, K.H.; Tinte, M.M.; Piater, L.A.; Dubery, I.A.; Tugizimana, F. Soil Salinity, a Serious Environmental Issue and Plant Responses: A Metabolomics Perspective. Metabolites 2021, 11, 724. [Google Scholar] [CrossRef]
- Sperdouli, I. Heavy Metal Toxicity Effects on Plants. Toxics 2022, 10, 715. [Google Scholar] [CrossRef]
- Yadav, B.; Jogawat, A.; Rahman, M.S.; Narayan, O.P. Secondary Metabolites in the Drought Stress Tolerance of Crop Plants: A Review. Gene Rep. 2021, 23, 101040. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of Extreme Weather Disasters on Global Crop Production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Saleem, M.H.; Noreen, S.; Ishaq, I.; Saleem, A.; Khan, K.A.; Ercisli, S.; Anas, M.; Khalid, A.; Ahmed, T.; Hassan, A.; et al. Omics Technologies: Unraveling Abiotic Stress Tolerance Mechanisms for Sustainable Crop Improvement. J. Plant Growth Regul. 2025, 44, 4165–4178. [Google Scholar] [CrossRef]
- Serba, D.D.; Yadav, R.S. Genomic Tools in Pearl Millet Breeding for Drought Tolerance: Status and Prospects. Front. Plant Sci. 2016, 7, 1724. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, R. Genetic Variation and Diversity of Pearl Millet [Pennisetum glaucum (L.)] Genotypes Assessed for Millet Head Miner, Heliocheilus Albipunctella Resistance, in West Africa. Euphytica 2020, 216, 158. [Google Scholar] [CrossRef]
- Satyavathi, C.T. Pearl Millet: A Climate-Resilient Nutricereal for Mitigating Hidden Hunger and Provide Nutritional Security. Front. Plant Sci. 2021, 12, 659938. [Google Scholar] [CrossRef] [PubMed]
- Raheem, D.; Dayoub, M.; Birech, R.; Nakiyemba, A. The Contribution of Cereal Grains to Food Security and Sustainability in Africa: Potential Application of UAV in Ghana, Nigeria, Uganda, and Namibia. Urban Sci. 2021, 5, 8. [Google Scholar] [CrossRef]
- Srivastava, R.K. Genome-Wide Association Studies and Genomic Selection in Pearl Millet: Advances and Prospects. Front. Plant Sci. 2020, 10, 1389. [Google Scholar] [CrossRef]
- Zhang, A.; Ji, Y.; Sun, M.; Lin, C.; Zhou, P.; Ren, J.; Luo, D.; Wang, X.; Ma, C.; Zhang, X.; et al. Research on the Drought Tolerance Mechanism of Pennisetum glaucum (L.) in the Root during the Seedling Stage. BMC Genom. 2021, 22, 568. [Google Scholar] [CrossRef]
- Jacob, J.; Sanjana, P.; Visarada, K.B.R.S.; Shobha, E.; Ratnavathi, C.V.; Sooganna, D. Seedling Stage Heat Tolerance Mechanisms in Pearl Millet (Pennisetum glaucum (L.) R. Br.). Russ. J. Plant Physiol. 2022, 69, 128. [Google Scholar] [CrossRef]
- Sayed, M.R.I.; Zayed, E.M.; Morsi, N.A.A. Identification of Molecular Genetic Markers Associated with Salt Tolerance in PearlMillet (Pennisetum glaucum L.). Middle East J. Agric. Res. 2022, 11, 451–465. [Google Scholar] [CrossRef]
- Silva, Q.M.; Andrade-Vieria, L.F. Is Pearl Millet (Pennisetum glaucum) a Good Plant Species for Ecotoxicological Tests? Environ. Sci. Pollut. Res. 2024, 31, 41953–41963. [Google Scholar] [CrossRef]
- Merga, D. Pearl Millet (Pennisetum glaucum L.) Breeding for Adaptation and Performance under Drought Condition: Review. J. Environ. Earth Sci. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Nambiar, V.S.; Dhaduk, J.J.; Sareen, N.; Shahu, T.; Desai, R. Potential Functional Implications of Pearl millet (Pennisetum glaucum) in Health and Disease. J. Appl. Pharm. Sci. 2011, 1, 62–67. [Google Scholar]
- Ashraf, M.; Hafeez, M. Thermotolerance of Pearl Millet and Maize at Early Growth Stages: Growth and Nutrient Relations. Biol. Plant. 2004, 48, 81–86. [Google Scholar] [CrossRef]
- Huang, D.; Sun, M.; Zhang, A.; Chen, J.; Zhang, J.; Lin, C.; Zhang, H.; Lu, X.; Wang, X.; Yan, H.; et al. Transcriptional Changes in Pearl Millet Leaves under Heat Stress. Genes 2021, 12, 1716. [Google Scholar] [CrossRef]
- Sun, M.; Huang, D.; Zhang, A.; Khan, I.; Yan, H.; Wang, X.; Zhang, X.; Zhang, J.; Huang, L. Transcriptome Analysis of Heat Stress and Drought Stress in Pearl Millet Based on Pacbio Full-Length Transcriptome Sequencing. BMC Plant Biol. 2020, 20, 323. [Google Scholar] [CrossRef] [PubMed]
- Imadi, S.R.; Kazi, A.G.; Ahanger, M.A.; Gucel, S.; Ahmad, P. Plant Transcriptomics and Responses to Environmental Stress: An Overview. J. Genet. 2015, 94, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Soniya, E.V.; Srinivasan, A.; Menon, A.; Kattupalli, D. Transcriptomics in Response of Biotic Stress in Plants. In Transcriptome Profiling, 1st ed.; Ali, M.A., Lee, J., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 285–303. [Google Scholar]
- Zhou, Z.; Fan, J.; Zhang, J.; Yang, Y.; Zhang, Y.; Zan, X.; Li, X.; Wan, J.; Gao, X.; Chen, R.; et al. OsMLP423 Is a Positive Regulator of Tolerance to Drought and Salt Stresses in Rice. Plants 2022, 11, 1653. [Google Scholar] [CrossRef]
- Almira-Casellas, M.; Busoms, S.; Pérez-Martín, L.; Escolà, G.; López-Valiñas, Á.; Garcia-Molina, A.; Llugany, M.; Poschenrieder, C. At the Core of Salinity: Divergent Transcriptomic Responses to Neutral and Alkaline Salinity in Arabidopsis Thaliana. Environ. Exp. Bot. 2024, 228, 105982. [Google Scholar] [CrossRef]
- Nazari, L.; Zinati, Z. Transcriptional Survey of Abiotic Stress Response in Maize (Zea mays) in the Level of Gene Co-Expression Network and Differential Gene Correlation Analysis. AoB Plants 2024, 16, plad087. [Google Scholar] [CrossRef]
- Liu, X.; Lan, S.; Sha, J.; Zhang, Z.; Chang, Y.; Chen, Y.; Yang, S. Transcriptome Analysis Reveals the Molecular Mechanisms through Which Exogenous Methyl Jasmonate Enhances Mercury Stress Tolerance in Maize Seedlings. Agronomy 2025, 15, 1369. [Google Scholar] [CrossRef]
- Kim, Y.-C.; Chen, M.; Kim, G.W.; Lee, H.; Lee, J.H. Comparative Transcriptomic Analysis in Roots of a Domestic Wheat Cultivar (Triticum aestivum L. Cv. Keumkang) Seedlings under Waterlogging Stress. J. Plant Biol. 2024, 67, 35–48. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Turgut Yiğit, A.; Yilmaz, O.; Uzi̇Lday, B.; Özgür Uzi̇Lday, R.; Türkan, İ. Plant Response to Salinity: An Analysis of ROS Formation, Signaling, and Antioxidant Defense. Turk. J. Bot. 2020, 44, 1–13. [Google Scholar] [CrossRef]
- Zhang, Q.; Teng, R.; Yuan, Z.; Sheng, S.; Xiao, Y.; Deng, H.; Tang, W.; Wang, F. Integrative Transcriptomic Analysis Deciphering the Role of Rice bHLH Transcription Factor Os04g0301500 in Mediating Responses to Biotic and Abiotic Stresses. Front. Plant Sci. 2023, 14, 1266242. [Google Scholar] [CrossRef]
- Ji, Y.; Lu, X.; Zhang, H.; Luo, D.; Zhang, A.; Sun, M.; Wu, Q.; Wang, X.; Huang, L. Transcriptome Reveals the Dynamic Response Mechanism of Pearl Millet Roots under Drought Stress. Genes 2021, 12, 1988. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Awan, S.A.; Rizwan, M.; Akram, M.A.; Zia-ur-Rehman, M.; Wang, X.; Zhang, X.; Huang, L. Physiological and Transcriptome Analyses Demonstrate the Silver Nanoparticles Mediated Alleviation of Salt Stress in Pearl Millet (Pennisetum glaucum L). Environ. Pollut. 2023, 318, 120863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Choudhary, M.; Jayanand; Padaria, J.C. Transcriptional Profiling in Pearl Millet (Pennisetum glaucum L.R. Br.) for Identification of Differentially Expressed Drought Responsive Genes. Physiol. Mol. Biol. Plants 2015, 21, 187–196. [Google Scholar] [CrossRef]
- Chen, S.; Xu, X.; Ma, Z.; Liu, J.; Zhang, B. Organ-Specific Transcriptome Analysis Identifies Candidate Genes Involved in the Stem Specialization of Bermudagrass (Cynodon dactylon L.). Front. Genet. 2021, 12, 678673. [Google Scholar] [CrossRef]
- Booth, M.W.; Breed, M.F.; Kendrick, G.A.; Bayer, P.E.; Severn-Ellis, A.A.; Sinclair, E.A. Tissue-Specific Transcriptome Profiles Identify Functional Differences Key to Understanding Whole Plant Response to Life in Variable Salinity. Biol. Open 2022, 11, bio059147. [Google Scholar] [CrossRef]
- Aher, R.R.; Palakolanu, S.R. Advancements in Genetic Enhancement Addressing Key Challenges in Pearl Millet (Pennisetum glaucum (L.) R. Br.). J. Plant Biochem. Biotechnol. 2025, 34, 1078. [Google Scholar] [CrossRef]
- Awan, S.A.; Khan, I.; Rizwan, M.; Irshad, M.A.; Xiaosan, W.; Zhang, X.; Huang, L. Reduction in the Cadmium (Cd) Accumulation and Toxicity in Pearl Millet (Pennisetum glaucum L.) by Regulating Physio-Biochemical and Antioxidant Defense System via Soil and Foliar Application of Melatonin. Environ. Pollut. 2023, 328, 121658. [Google Scholar] [CrossRef]
- Iwuala, E.; Olajide, O.; Abiodun, I.; Odjegba, V.; Utoblo, O.; Ajewole, T.; Oluwajobi, A.; Uzochukwu, S. Silicon Ameliorates Cadmium (Cd) Toxicity in Pearl Millet by Inducing Antioxidant Defense System. Heliyon 2024, 10, e25514. [Google Scholar] [CrossRef]
- Divya, K.; Palakolanu, S.R.; Kavi Kishor, P.; Rajesh, A.S.; Vadez, V.; Sharma, K.K.; Mathur, P.B. Functional Characterization of Late Embryogenesis Abundant Genes and Promoters in Pearl Millet (Pennisetum glaucum L.) for Abiotic Stress Tolerance. Physiol. Plant. 2021, 173, 1616–1628. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Abundance Estimation from RNA-Seq Reveals Thousands of New Transcripts and Switching among Isoforms. Nat Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Guo, A.-Y.; Chen, X.; Gao, G.; Zhang, H.; Zhu, Q.-H.; Liu, X.-C.; Zhong, Y.-F.; Gu, X.; He, K.; Luo, J. PlantTFDB: A Comprehensive Plant Transcription Factor Database. Nucleic Acids Res. 2007, 36, D966–D969. [Google Scholar] [CrossRef]
- Altschul, S. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-Protein Interaction Networks, with Increased Coverage and Integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into Plant Salt Stress Signaling and Tolerance. J. Genet. Genom. 2024, 51, 16–34. [Google Scholar] [CrossRef]
- Mondal, S.; Karmakar, S.; Panda, D.; Pramanik, K.; Bose, B.; Singhal, R.K. Crucial Plant Processes under Heat Stress and Tolerance through Heat Shock Proteins. Plant Stress 2023, 10, 100227. [Google Scholar] [CrossRef]
- Azad, M.; Tohidfar, M.; Ghanbari Moheb Seraj, R.; Mehralian, M.; Esmaeilzadeh-Salestani, K. Identification of Responsive Genes to Multiple Abiotic Stresses in Rice (Oryza sativa): A Meta-Analysis of Transcriptomics Data. Sci. Rep. 2024, 14, 5463. [Google Scholar] [CrossRef]
- Sun, M.; Yan, H.; Zhang, A.; Jin, Y.; Lin, C.; Luo, J.; Zhou, P.; Wang, X.; Zhang, X.; Huang, L. The Transcriptional Dynamic Landscape Map of Pearl Millet in Response to Abiotic Stress Provides New Insights into the Responses of Plants to Stress. Trop. Plants 2025, 4, e024. [Google Scholar] [CrossRef]
- Shi, C.; Guo, F.; Sun, Y.; Han, J.; Zheng, X.; Zhang, J.; Qin, C.; Tan, Z.; Lin, J.; Wang, J. Physiological and Transcriptomic Analysis of Hordeum Jubatum Seedlings in Response to Salt, Alkali and Drought Stresses under Uniform Water Potential. Environ. Exp. Bot. 2024, 220, 105677. [Google Scholar] [CrossRef]
- Tenorio Berrío, R.; Verstaen, K.; Vandamme, N.; Pevernagie, J.; Achon, I.; Van Duyse, J.; Van Isterdael, G.; Saeys, Y.; De Veylder, L.; Inzé, D.; et al. Single-Cell Transcriptomics Sheds Light on the Identity and Metabolism of Developing Leaf Cells. Plant Physiol. 2022, 188, 898–918. [Google Scholar] [CrossRef]
- Nelissen, H.; Sun, X.; Rymen, B.; Jikumaru, Y.; Kojima, M.; Takebayashi, Y.; Abbeloos, R.; Demuynck, K.; Storme, V.; Vuylsteke, M.; et al. The Reduction in Maize Leaf Growth under Mild Drought Affects the Transition between Cell Division and Cell Expansion and Cannot Be Restored by Elevated Gibberellic Acid Levels. Plant Biotechnol. J. 2018, 16, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Ke, Q.; Kang, L.; Kim, H.S.; Xie, T.; Liu, C.; Ji, C.Y.; Kim, S.H.; Park, W.S.; Ahn, M.-J.; Wang, S.; et al. Down-Regulation of Lycopene ε-Cyclase Expression in Transgenic Sweetpotato Plants Increases the Carotenoid Content and Tolerance to Abiotic Stress. Plant Sci. 2019, 281, 52–60. [Google Scholar] [CrossRef]
- Mi, J.; Vallarino, J.G.; Petřík, I.; Novák, O.; Correa, S.M.; Chodasiewicz, M.; Havaux, M.; Rodriguez-Concepcion, M.; Al-Babili, S.; Fernie, A.R.; et al. A Manipulation of Carotenoid Metabolism Influence Biomass Partitioning and Fitness in Tomato. Metab. Eng. 2022, 70, 166–180. [Google Scholar] [CrossRef]
- Song, J.; Sun, B.; Chen, C.; Ning, Z.; Zhang, S.; Cai, Y.; Zheng, X.; Cao, B.; Chen, G.; Jin, D.; et al. An R-R-type MYB transcription factor promotes non-climacteric pepper fruit carotenoid pigment biosynthesis. Plant J. 2023, 115, 724–741. [Google Scholar] [CrossRef]
- Lunn, J.E.; Hatch, M.D. Primary Partitioning and Storage of Photosynthate in Sucrose and Starch in Leaves of C4 Plants. Planta 1995, 197, 385–391. [Google Scholar] [CrossRef]
- Dong, N.; Lin, H. Contribution of Phenylpropanoid Metabolism to Plant Development and Plant–Environment Interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Zhao, T.; Wu, T.; Pei, T.; Wang, Z.; Yang, H.; Jiang, J.; Zhang, H.; Chen, X.; Li, J.; Xu, X. Overexpression of SlGATA17 Promotes Drought Tolerance in Transgenic Tomato Plants by Enhancing Activation of the Phenylpropanoid Biosynthetic Pathway. Front. Plant Sci. 2021, 12, 634888. [Google Scholar] [CrossRef]
- Jia, C.; Guo, B.; Wang, B.; Li, X.; Yang, T.; Li, N.; Wang, J.; Yu, Q. Integrated Metabolomic and Transcriptomic Analysis Reveals the Role of Phenylpropanoid Biosynthesis Pathway in Tomato Roots during Salt Stress. Front. Plant Sci. 2022, 13, 1023696. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, P.; Lu, Y.; Bai, Y.; Wei, Y.; Liu, G.; Shi, H. MeRAV5 Promotes Drought Stress Resistance in Cassava by Modulating Hydrogen Peroxide and Lignin Accumulation. Plant J. 2021, 107, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dou, Y.; Geng, H.; Fu, J.; Dan, Z.; Liang, T.; Cheng, M.; Zhao, W.; Zeng, Y.; Hu, Z.; et al. OsGRP3 Enhances Drought Resistance by Altering Phenylpropanoid Biosynthesis Pathway in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 7045. [Google Scholar] [CrossRef]
- Gao, H.; Huang, X.; Lin, P.; Hu, Y.; Zheng, Z.; Yang, Q. Transcriptome-Associated Metabolomics Reveals the Molecular Mechanism of Flavonoid Biosynthesis in Desmodium styracifolium (Osbeck.) Merr under Abiotic Stress. Front. Plant Sci. 2024, 15, 1431148. [Google Scholar] [CrossRef]
- Zhao, P.; Yan, X.; Qian, C.; Ma, G.; Fan, X.; Yin, X.; Liao, Y.; Fang, T.; Zhou, S.; Awuku, I.; et al. Flavonoid Synthesis Pathway Response to Low-Temperature Stress in a Desert Medicinal Plant, Agriophyllum squarrosum (Sandrice). Genes 2024, 15, 1228. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Xu, Z.; Cong, K.; Zhu, X.; Zhang, L.; Wang, J.; Wei, J.; Ji, P. Transcriptomic Responses to Drought Stress in Polygonatum Kingianum Tuber. BMC Plant Biol. 2021, 21, 537. [Google Scholar] [CrossRef]
- Schulz, E.; Tohge, T.; Winkler, J.B.; Albert, A.; Schäffner, A.R.; Fernie, A.R.; Zuther, E.; Hincha, D.K. Natural Variation among Arabidopsis Accessions in the Regulation of Flavonoid Metabolism and Stress Gene Expression by Combined UV Radiation and Cold. Plant Cell Physiol. 2021, 62, 502–514. [Google Scholar] [CrossRef]
- Dai, Y.; Zhang, L.; Sun, X.; Li, F.; Zhang, S.; Zhang, H.; Li, G.; Fang, Z.; Sun, R.; Hou, X.; et al. Transcriptome Analysis Reveals Anthocyanin Regulation in Chinese Cabbage (Brassica rapa L.) at Low Temperatures. Sci. Rep. 2022, 12, 6308. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
- Kang, L.; Teng, Y.; Cen, Q.; Fang, Y.; Tian, Q.; Zhang, X.; Wang, H.; Zhang, X.; Xue, D. Genome-Wide Identification of R2R3-MYB Transcription Factor and Expression Analysis under Abiotic Stress in Rice. Plants 2022, 11, 1928. [Google Scholar] [CrossRef]
- Guo, H.; Wang, Y.; Wang, L.; Hu, P.; Wang, Y.; Jia, Y.; Zhang, C.; Zhang, Y.; Zhang, Y.; Wang, C.; et al. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2017, 15, 107–121. [Google Scholar] [CrossRef]
- Geng, P.; Zhang, S.; Liu, J.; Zhao, C.; Wu, J.; Cao, Y.; Fu, C.; Han, X.; He, H.; Zhao, Q. MYB20, MYB42, MYB43, and MYB85 Regulate Phenylalanine and Lignin Biosynthesis during Secondary Cell Wall Formation. Plant Physiol. 2020, 182, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Fornalé, S.; Shi, X.; Chai, C.; Encina, A.; Irar, S.; Capellades, M.; Fuguet, E.; Torres, J.-L.; Rovira, P.; Puigdomènech, P.; et al. ZmMYB31 Directly Represses Maize Lignin Genes and Redirects the Phenylpropanoid Metabolic Flux. Plant J. 2010, 64, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Chanwala, J.; Khadanga, B.; Jha, D.K.; Sandeep, I.S.; Dey, N. MYB Transcription Factor Family in Pearl Millet: Genome-Wide Identification, Evolutionary Progression and Expression Analysis under Abiotic Stress and Phytohormone Treatments. Plants 2023, 12, 355. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Dong, Z.; Zhou, H.; Wu, G.; Xu, L.; Ying, S.; Chen, M. Genome-Wide Identification and Transcriptional Analysis of the MYB Gene Family in Pearl Millet (Pennisetum glaucum). Int. J. Mol. Sci. 2023, 24, 2484. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Luo, L.; Zhou, T.; Gan, J.; Liu, N.; Lu, R.; Xu, Q.; Hu, L.; Chen, G. Comparative Transcriptome Analysis of Leaves and Roots Revealed Organ-Specific and Cross-Stress Defense Strategies of Pearl Millet Under Different Abiotic Stresses. Agronomy 2025, 15, 2707. https://doi.org/10.3390/agronomy15122707
Chen Q, Luo L, Zhou T, Gan J, Liu N, Lu R, Xu Q, Hu L, Chen G. Comparative Transcriptome Analysis of Leaves and Roots Revealed Organ-Specific and Cross-Stress Defense Strategies of Pearl Millet Under Different Abiotic Stresses. Agronomy. 2025; 15(12):2707. https://doi.org/10.3390/agronomy15122707
Chicago/Turabian StyleChen, Qi, Lixuan Luo, Tao Zhou, Jinxin Gan, Ningfang Liu, Rui Lu, Qian Xu, Longxing Hu, and Guihua Chen. 2025. "Comparative Transcriptome Analysis of Leaves and Roots Revealed Organ-Specific and Cross-Stress Defense Strategies of Pearl Millet Under Different Abiotic Stresses" Agronomy 15, no. 12: 2707. https://doi.org/10.3390/agronomy15122707
APA StyleChen, Q., Luo, L., Zhou, T., Gan, J., Liu, N., Lu, R., Xu, Q., Hu, L., & Chen, G. (2025). Comparative Transcriptome Analysis of Leaves and Roots Revealed Organ-Specific and Cross-Stress Defense Strategies of Pearl Millet Under Different Abiotic Stresses. Agronomy, 15(12), 2707. https://doi.org/10.3390/agronomy15122707






