Proteomic Insights into Edible Nut Seeds: Nutritional Value, Allergenicity, Stress Responses, and Processing Effects
Abstract
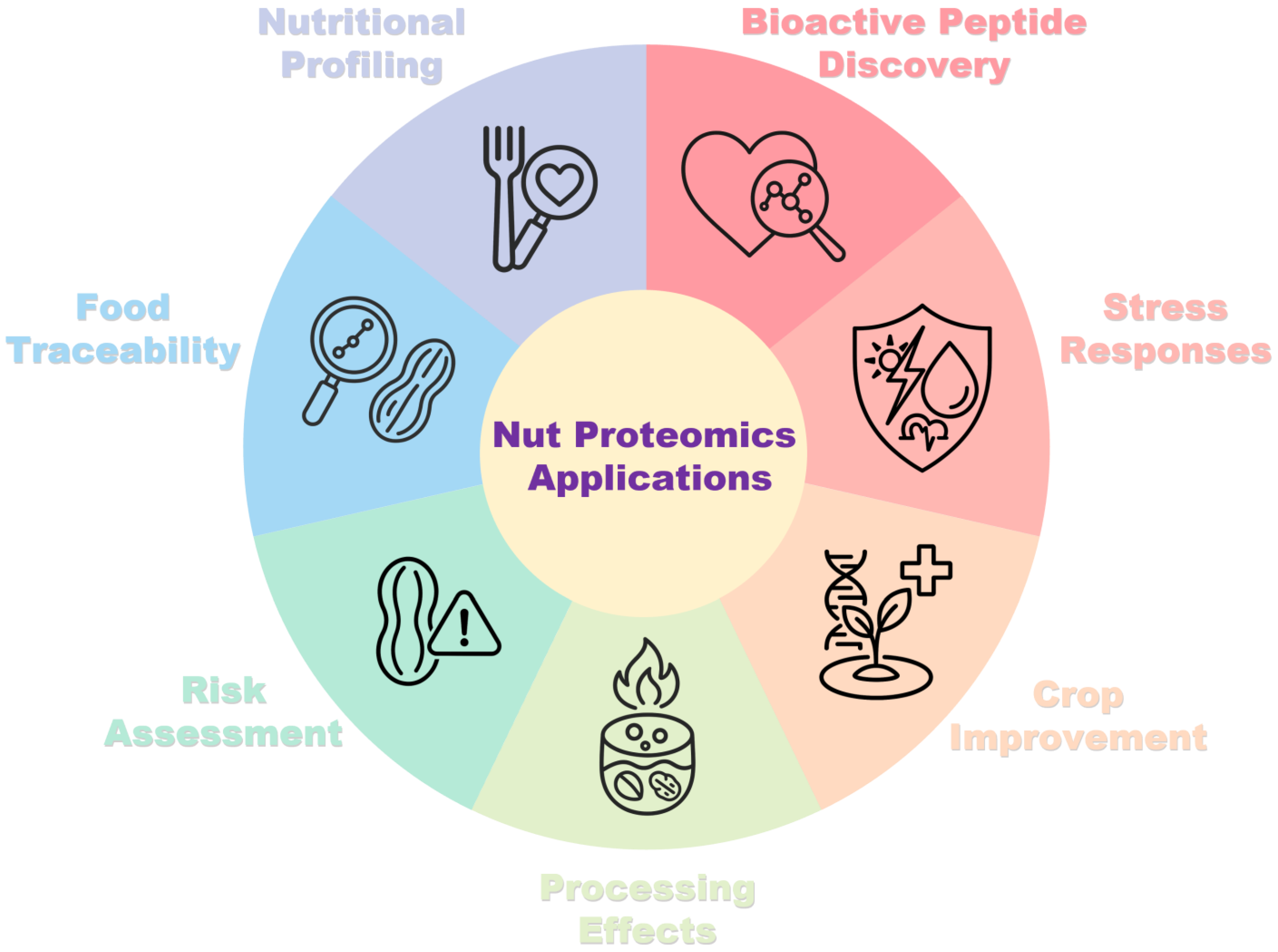
1. Introduction
2. Status and Utility of Proteomic Research in Nut Species
| Nut Species —Sample Tissue | Study Purpose | Protein Extraction Methods | Research Methods and Instruments | Search Database (If Applicable) | Key Findings | Application/Significance | Ref. |
|---|---|---|---|---|---|---|---|
| Macadamia (Macadamia integrifolia) - Nuts | Characterize proteomic diversity; assess parental genetic contributions to nut quality and allergenicity | Methanol homogenization + TCA precipitation; SDS-PAGE separation; in-gel digestion | SDS-PAGE densitometry (Bio-Rad); Nano-HPLC-MS/MS (Eksigent LC400 + TripleTOF 6600+); ProteinPilot; Scaffold; NSAF quantification | M. integrifolia genome database for ProteinPilot/Scaffold; functional annotation via BLASTP to UniProtKB/SwissProt Arabidopsis thaliana | 431 proteins identified; ~50% were vicilin/legumin SSPs; paternal genotype strongly influenced SSP profiles; PTM and sequence coverage of SSPs varied across genotypes | Informs allergenicity risk assessment, highlights natural variation in SSP abundance, supports breeding strategies for hypoallergenic and nutritionally improved cultivars | [32] |
| Various nuts - Raw/baked nut powders; incurred bakery food samples | Develop HRMS-based method for simultaneous detection of multiple nut allergens in bakery foods | Buffers with urea/thiourea/CHAPS for raw nuts; Tris–HCl buffer for bakery food | LC-Q-TOF MS (Agilent 6545B) in EDR and auto MS/MS mode; HPLC-QQQ-MS (Sciex 6500) in MRM; PEAKS Xpro bioinformatics | NCBI Protein Database (taxon-specific entries for almond, cashew, peanut, walnut) | Identified 16 exclusive heat-stable marker peptides; LODs 0.10–0.31 mg/kg; recoveries 72.5–92.1%; detection of undeclared allergens in commercial bakery foods | Provides robust, sensitive, label-free quantification of nut allergens; supports allergen labeling regulation and risk management | [30] |
| Macadamia (Macadamia integrifolia) - Nuts | Identify new allergens and assess cross-reactivity with other tree nuts in Spanish cohort | Ground nuts in liquid N2; homogenized in PBS; centrifugation and dialysis; Bradford quantification | SDS-PAGE (reducing and non-reducing); Western blot with allergic patients’ sera; Inhibition assays (hazelnut, walnut extracts); MALDI-TOF/TOF MS for IgE-binding protein identification | Non-redundant protein sequence databases (MASCOT/NCBI search for peptide fingerprint) | Three new allergens identified: oleosin, pectin acetylesterase, aspartyl protease; confirmed Mac i 1 (vicilin) and antimicrobial peptide 1; Oleosin cross-reactive with hazelnut but not walnut | Expands macadamia allergen profile; reveals regional sensitization differences (Spanish vs. Australian/Japanese); supports improved allergy diagnosis and risk management | [27] |
| Walnut (Juglans regia) - Nuts (raw, boiled, roasted); oil body (oleosome) fraction | Characterize walnut oleosome proteome; assess processing effects and IgE recognition | Oil-body enrichment via sucrose/Tween/NaCl/urea buffers; methanol–chloroform–water precipitation | Off-gel label free shotgun proteomics (Orbitrap Q Exactive Plus + UHPLC); In-gel LDS-PAGE + MS/MS; Immunoblotting | UniProt J. regia database + mature protein sequences (signal peptide/propeptide removed forms) | Identified all 8 oleosins, 1 caleosin, 1 steroleosin detected; boiling enhanced protein solubility and recovery; phylogenetic alignment showed walnut oleosins similar to hazelnut Cor a 13 and peanut Ara h 15 | First evidence of walnut oleosin allergenicity; shows thermal processing modulates allergen solubility and IgE reactivity; supports improved diagnosis and allergen risk assessment | [33] |
| Various nuts - (raw and roasted) | Develop a single LC-HRMS method to simultaneously detect/quantify nut allergens in processed bakery matrices | Tris-HCl buffer + urea; reduction (DTT), alkylation (IAA); SPE purification (Strata-X, SepPak C18); SEC cleanup for cookie extracts | LC-ESI-Q-Orbitrap MS (Q-Exactive Plus, Thermo Fisher); Full-MS/dd-MS2 & AIF modes; isotopically labelled AQUA peptides as internal standards; Proteome Discoverer | Customized database from UniProtKB entries: almond, hazelnut, peanut, walnut, cashew, pistachio | Identified reliable marker peptides common to raw and roasted nuts; 3 high-response peptides per nut selected; LOD 2.4–8.1 mg/kg, LOQ 8–27 mg/kg; roasting reduced peptide detection up to ~50% | Provides robust, multi-allergen detection at ppm levels in bakery foods; aligns with VITAL 3.0 thresholds; supports rational PAL use and consumer safety | [31] |
| Pecan (Carya illinoinensis) - Nuts from two cultivars at 6 developmental stages | Characterize proteomic changes during nut development; identify allergen accumulation timing; compare cultivars | Borate buffer; homogenization; centrifugation; SDS-PAGE quantification | SDS-PAGE + immunoblot with anti-pecan sera; LC-QTOF-MS/MS (Agilent 6520 Chip Cube); Mascot Distiller/Daemon; 2D-GE (isoelectric focusing pH 3–10, silver staining); image analysis | In-house protein libraries from Sumner (NCBI) and Pawnee transcriptomes; annotated with GO terms (InterProScan) | protein accumulation peaked at dough stage; allergens Car i 1 & Car i 2 first detected at dough stage; histones decreased as development progressed; cultivar differences | Provides molecular insight into timing of allergen accumulation and developmental regulation; informs breeding for reduced allergenicity, stress tolerance, improved seed quality | [28] |
| Almond (Prunus dulcis) - Commercial almond flour | Compare aqueous vs. protease-assisted aqueous extraction (AEP vs. EAEP) on protein profile, digestibility and antigenicity | AEP (alkaline, pH 9) vs. EAEP (AEP + neutral protease); centrifugation to separate protein/oil | SDS-PAGE; LC–MS/MS (Orbitrap); PEAKS Studio; simulated digestion and Degree of Hydrolysis (OPA); ELISA and Western blot (IgE/IgG sera) | UniProt P. dulcis (Swiss-Prot + TrEMBL) | EAEP mainly hydrolyzed prunin α-chains, improved digestibility (79→89%), increased peptide yield, and reduced immunoreactivity (~75%) with degradation of multiple allergens. | Demonstrates protease-assisted extraction improves almond protein digestibility and reduces allergenicity, supporting development of safer hypoallergenic almond-based foods | [34] |
| Chinese hickory (Carya cathayensis) - Embryos at 3 developmental stages | Characterize lipid droplet (LD) proteome during embryo development and identify key LD proteins linked to oil accumulation | Sucrose gradient centrifugation for LD isolation; protein precipitation (chloroform/acetone); lysis buffer (urea, Tris, NP40, NaDOC, protease inhibitors); reduction (DTT), alkylation (IAM) | LC–MS/MS (Q Exactive HF-X Orbitrap + Ultimate 3000 nano-UPLC); label-free quantification (LFQ, iBAQ, riBAQ); qRT-PCR validation; confocal microscopy (LD localization, GFP fusion); TEM ultrastructure | C. cathayensis genome searched with MaxQuant; Arabidopsis Plant Proteome Database for subcellular localization | Identified 6574 proteins, incl. 38 LD-associated; 62 DEPs linked to LD biogenesis; oleosins/CLO1/HSD5 increased during S2→S3, declined at S5; subcellular localization confirmed proteins at LDs | Provides first comprehensive LD proteome of Chinese hickory; reveals dynamic regulation of LD proteins during oil accumulation; supports breeding strategies for high-oil-yield nut cultivars | [35] |
| Hazelnut (Corylus avellana) - Nuts (raw and autoclaved variants) | Assess whether autoclaving (with/without hydration/drying) reduces allergenicity | Milling; protein extraction; Bradford assay for quantification; filtration | SDS-PAGE; In-gel digestion; LC–MS/MS (Q Exactive Orbitrap, Thermo) with UHPLC; Proteome Discoverer/SequestHT; Immunoblotting (IgE, IgG from 14 allergic patients) | UniProt C. avellana protein database | Autoclaving (especially with hydration/drying) reduced protein solubility (40–70%), degraded major allergens (Cor a 9, 11, 14), and markedly lowered patient skin test reactivity and IgE binding. | Demonstrates autoclaving, especially with hydration/drying, markedly reduces hazelnut allergenicity; provides proteomic and clinical evidence supporting hypoallergenic hazelnut product development | [36] |
| Pistachio (Pistacia vera) - Defatted flour from 4 cultivars | Characterize pistachio allergen proteome and compare cultivar-specific expression | Defatting with diethyl ether; extraction with urea + TBS buffer; Bradford assay; reduction (DTT), alkylation (IAA) | SDS-PAGE, 2-DE (IEF + SDS-PAGE); LC–MS/MS (Q Exactive Orbitrap + UltiMate 3000 RSLC nano-UPLC) | UniProtKB P. vera | Identified major allergens Pis v 1 (2S albumin), Pis v 2 & Pis v 5 (11S globulins), Pis v 3 (7S vicilin); multiple isoforms of 11S and 7S revealed; Pis v 5 differentially expressed in cultivars | First comprehensive pistachio allergen proteome; confirms dominance of cupin family allergens; highlights cultivar variation; supports allergen risk assessment and breeding for hypoallergenic cultivars | [26] |
| Macadamia (Macadamia integrifolia and hybrids with M. tetraphylla) - Nuts | Provide proteomic overview of macadamia nut; identify functional protein categories; assess potential allergens and cross-reactivity | Defatting (n-hexane/ethanol); protein extraction (NaCl buffer pH 8.4); acetone precipitation; SDS-PAGE; in-gel trypsin digestion | SDS-PAGE; LC–MS/MS (LTQ-FT Ultra Orbitrap + Ultimate 3000 nano-LC); Mascot and X!Tandem; Scaffold validation; spectral counting; GO/KEGG annotation | NCBI non-redundant Viridiplantae protein database; AllergenOnline (FARRP v16); IEDB-AR (linear epitopes) | Identified 1079 proteins; proteins classified into 46 categories; defense proteins most abundant; identified seed storage proteins; in silico matched macadamia proteins with allergens; cross-reactivity with peanut, walnut, lupin, sesame, birch pollen, fungi, dust mite indicated | First comprehensive macadamia nut proteome; highlights abundant defense and storage proteins; identifies potential allergenic proteins and cross-reactivity risks; provides resource for allergy assessment and breeding | [37] |
| Peanut (Arachis hypogaea) - Developing seeds at 7 underground stages (R1–R7) | Investigate peanut proteins at different development stages; analyze allergen accumulation and protein interaction networks | TCA/acetone precipitation; phenol extraction; MeOH/NH4OAc precipitation; guanidine-HCl solubilization; reduction (TBP); alkylation (2-VP); urea/thiourea/CHAPS lysis | 2-DE (IEF pH 3–10, SDS-PAGE); colloidal CBB staining; densitometry (GS-800); MALDI-TOF/TOF-MS (Sciex 5800); Mascot search; Mfuzz clustering; qRT-PCR validation | Peanut A-genome (A. duranensis) and B-genome (A. ipaensis); EggNOG for functional annotation; STRING v9.1 for PPI network | Identified 264 proteins; clustered into 8 groups; 5 major functional categories; 14 allergen-like proteins identified, expression low at R1–R4, increased sharply from R5–R7; storage proteins mainly in cluster 7 | Provides first proteome map of peanut underground seed development; reveals allergen accumulation timing; suggests harvesting immature seeds may lower allergenicity risk | [23] |
| Cashew (Anacardium occidentale) - Blanched raw nuts | Develop customized protein database; analyze effects of oil-roasting on allergen stability; improve MS peptide targets | Tris-HCl buffer + urea; reduction (DTT), alkylation (IAA); in-solution trypsin/chymotrypsin digestion; C18 cleanup | SDS-PAGE; LC–MS/MS (Q Exactive HF and Plus Orbitrap + UltiMate 3000 UHPLC); DDA and PRM targeted analysis | Customized cashew protein database from UniProt, Phytozome v12.1 genome (primary + selected alternative transcripts), and AUGUSTUS predictions | Identified 6595 peptides; revealed 17 new sequences for major allergens; Ana o 3 (2S) most heat-stable; roasting reduced solubility and abundance of Ana o 1; Maillard adducts identified on 11S peptides; | Provides optimized cashew protein database; reveals isoforms and processing impact on allergenicity; enables high-quality MS-based allergen detection; supports food safety regulation and risk assessment | [38] |
| Peanut (Arachis hypogaea) - Seeds from high and low-oleate cultivar at 6 developmental stages | Identify proteins linked to oleic acid accumulation during seed development | TCA/acetone precipitation; methanol/ammonium acetate wash; urea/Tris lysis; reduction (DTT), alkylation (IAM); iTRAQ labeling | iTRAQ-based LC–ESI–MS/MS (TripleTOF 5600, SCIEX); Mascot and ProteinPilot search; GO and KEGG annotation; STRING PPI analysis; qRT-PCR validation | UniprotKB peanut protein database; functional annotation with Arabidopsis TAIR and GO annotation; KEGG pathways | 7666 proteins identified; 389 DEPs associated with FA metabolism; FAB2 upregulated early in high-oleate cultivar, downregulated later; KAS I/II upregulated early; multiple lipid oxidation enzymes identified; OA biosynthesis enriched at stage 3 | Provides molecular insight into oleic acid accumulation dynamics; identifies FAB2 as key regulator; supports breeding strategies for high-oleate peanut varieties with improved oil stability and nutritional quality | [39] |
| Cashew (Anacardium occidentale) - Cashew nut protein fractions and protein isolate | Characterize molecular and functional properties of cashew protein fractions vs. isolate | Osborne fractionation (water, NaCl, ethanol, NaOH) for fractions; alkali extraction + isoelectric precipitation for protein isolate; petroleum ether defatting | SDS-PAGE (reducing/non-reducing), Circular dichroism spectroscopy, SEM, solubility tests, water/Oil holding capacity, foaming and emulsifying assays, amino acid analyzer (Hitachi L-8800) | N/A | Cashew proteins showed varied functional properties | Demonstrates functional specialization of fractions: albumin/globulin for beverages (solubility), glutelin for bakery/meat (foaming and water/oil binding), protein isolate for emulsified foods; supports cashew as valuable protein source for food industry | [40] |
| Hazelnut (Corylus avellana) - Nuts | Characterize storage, allergenic, and defense proteins; improve allergen detection | PBS extraction, defatting (hexane), TCA/acetone precipitation, buffer optimization (temp, ratio) | SDS-PAGE, 2-DE, MALDI-TOF, LC–ESI–MS/MS; RP–HPLC; Western blot; ELISA; MS-based targeted detection (SRM/PRM) | NCBI and UniProtKB databases (SwissProt entries for Cor a proteins); Mascot search; AllergenOnline cross-references | Identified major allergens; multiple isoforms and proteoforms; MS marker peptides developed for food allergen detection; roasting altered extractability but not IgE binding significantly | First comprehensive hazelnut proteome; provides allergen catalog and biomarker peptides; supports allergy diagnosis, risk assessment, and food authentication | [41] |
| Almond (Prunus dulcis) - Nuts | Characterize almond nut proteome; compare with American almonds; identify allergens and stress-related proteins | Phenol extraction with sucrose/KCl/EDTA Tris buffer; methanol–ammonium acetate precipitation; acetone wash; SDS-PAGE and in-gel trypsin digestion | 2-DE + SDS-PAGE; LC–MS/MS (Q Exactive Orbitrap); Mascot search; GO annotation (Blast2GO); ImageMaster 2D Elite analysis | NCBI protein database (taxonomy: P. dulcis); UniProt for GO annotation; SwissProt cross-references | Identified 434 proteins (259 novel/hypothetical); 2104 protein spots in 2-DE; allergens Pru du 1, 2, 3, 4, 6 detected; 22 hypothetical proteins with glycosylation motifs; abundant metabolic/catalytic proteins; stress-related proteins | Provides reference almond proteome map; improves understanding of allergenicity, stress response proteins, and nutritional value; supports development of almond protein-based foods | [29] |
| Walnut (Juglans regia) - Defatted walnut flour (raw and roasted) | Assess effects of roasting on solubility and detectability of walnut allergens | Defatting (hexane); sequential vs. simultaneous extraction/trypsin digestion; DTT, IAA | SDS-PAGE; LC–MS/MS (Orbitrap Elite, UPLC C18); Label-free quantification (Progenesis LC-MS); Mascot search; Maillard adduct screening | UniProt Juglandaceae + UniProt A. hypogaea + AllergenOnline v12 + contaminants | 2S albumin (Jug r 1), nsLTP (Jug r 3), 7S N-terminal showed minor roasting effects; Mature 7S (Jug r 2) and 11S (Jug r 4) increased detectability after roasting, likely due to improved digestibility; Maillard adducts mainly on 7S and 11S globulins | Reveals allergen-specific responses to roasting; highlights MS as powerful tool for monitoring allergen modifications; informs allergen risk assessment and MS-based detection in processed foods | [42] |
| Hazelnut (Corylus avellana) - Defatted flour; water-soluble protein extracts | Evaluate effect of high hydrostatic pressure (HHP) on protein solubility and immunoreactivity | Defatting (n-hexane); soaking in distilled water; extraction with borate saline buffer + PVP; centrifugation; dialysis; freeze-drying | SDS-PAGE; Western blot (IgE sera from 15 allergic patients); 2D-LC fractionation (ProteomeLab PF-2D: chromatofocusing + RP-HPLC); MALDI-TOF/TOF-MS; LC–ESI–MS/MS | SwissProt/NCBI protein databases (taxonomy: C. avellana); Mascot for peptide mass fingerprinting | HHP reduced solubility of major allergens Cor a 9 (11S globulin) and Cor a 11 (7S vicilin); SDS-PAGE and immunoblot showed similar IgE reactivity between control and HHP samples; PF-2D revealed fewer protein peaks in HHP extracts; Cor a 9 and Cor a 11 remained identifiable but less soluble | Demonstrates HHP alters solubility but not IgE-binding of hazelnut allergens; provides proteomic evidence for limited effect of HHP alone on allergenicity; suggests combining HHP with protease digestion for hypoallergenic food development | [43] |
| Hazelnut (Corylus avellana) - Nuts | Identify and characterize novel IgE-binding proteins in hazelnut | Defatting (diethyl ether); PBS extraction; reduction (DTT), alkylation (IAA); dialysis + lyophilization | SDS-PAGE (reducing and non-reducing), 2-DE; MALDI-TOF-MS; nano-LC–ESI-Q-TOF MS/MS (Mascot, Batch-Tag, Protein Prospector); de novo sequencing; Immunoblotting and inhibition assays; RP-HPLC purification | NCBI and SwissProt databases (green plants); homology search via BLAST/MS-Pattern; incomplete hazelnut genome noted | Identified a novel alkaline subunit (homologous to 11S globulin Cor a 9) as predominant IgE-binding protein; minor reactivity also to Cor a 11, Cor a 9, Cor a 8; de novo peptides showed homology with 11S isoforms from pistachio, cashew, soybean | First evidence of new 11S globulin isoallergen in hazelnut; explains complex IgE-reactivity patterns; provides platform for component-resolved diagnostics and new therapeutic strategies | [44] |
| Peanut (Arachis hypogaea) - Nuts | Compare proteomes of allergenic vs. hypoallergenic peanut cultivars | Defatting (hexane); extraction with Tris-HCl + urea + thiourea + CHAPS; reduction (DTT), alkylation (IAA); trypsin digestion | 2-DE; MALDI-TOF/TOF-MS (AB Sciex 5800); LC–MS/MS; Progenesis PG240 software; Mascot and X!Tandem search; Scaffold validation | NCBI A. hypogaea database + expressed sequence tags | 162 protein spots identified; 40 differential spots between allergenic and hypoallergenic lines; allergens Ara h 1, 2, 3, 6 more abundant in allergenic varieties; hypoallergenic types showed lower SSPs and higher stress-response proteins (LEA, HSPs, PR proteins) | Provides proteomic evidence for reduced allergenicity in selected peanut cultivars; supports breeding of hypoallergenic peanuts | [45] |
| Brazil nut (Bertholletia excelsa) - Nuts | Purify and characterize major 2S albumin isoforms; assess heterogeneity and structural features | Defatting (petroleum ether); water extraction; dialysis; gel filtration (Superdex 75); chromatofocusing | SDS-PAGE, IEF, MALDI-TOF-MS (Bruker Reflex III), RP-HPLC–ESI-MS (Micromass Quattro II), Circular dichroism spectroscopy, Fourier transform-infrared spectroscopy | NCBI protein database (Viridiplantae) searched with Mascot peptide mass fingerprinting | Identified ~10 2S albumin isoforms; heterogeneity due to post-translational processing; all predominantly α-helical; no glycosylation detected | Provides first detailed proteomic and structural characterization of Brazil nut 2S albumin isoforms; explains allergen heterogeneity (Ber e 1); supports allergen risk assessment and structure–function studies | [46] |
| Pili nut (Canarium ovatum) - Nuts | Characterize storage protein composition and physicochemical properties | Modified Osborne fractionation: water (albumins), NaCl (globulins), ethanol (prolamins), NaOH (glutelins); defatting with hexane | SDS-PAGE (reducing and non-reducing); Native-PAGE; Amino acid analysis (Beckman Analyzer); DSC (MC-2D calorimeter); proximate and mineral analysis | N/A | Protein content ~11.9% of nuts; Globulins major fraction (60.3% of total protein), mainly 11S globulin with 22.6 and 31.6 kDa subunits; Albumins minor (2.9%); No prolamins; Glutelins ~36.8% | First detailed characterization of pili nut proteins; shows similarity to other oilseed 11S globulins (soy, peanut, hemp); supports potential use of pili nut proteins as isolates in food applications | [47] |
| Peanut (Arachis hypogaea) - Nuts (raw and roasted, aqueous extracts) | Assess roasting effects on major allergens and PTM profiles | Defatting (n-hexane, tetrachloroethylene); aqueous PBS extraction under mild conditions | 1D/2D SDS-PAGE; In-gel digestion; nLC–MS/MS (LTQ Orbitrap XL); PTM profiling via PEAKS XPro; Western blot with PTM-specific Abs; ELISA (IgE binding, patient sera) | UniProtKB A. hypogaea + contaminants; PTM search included 313 mods from Unimod | Roasting reduced protein solubility (~4 × lower); Ara h 3 enriched in raw, Ara h 6 in roasted; >40 PTMs; oxidation (Met), deamidation (Asn/Gln), hydroxylation (Pro), carbamoylation (Lys/Arg), and methylation more frequent in roasted; Ara h 1 most extensively modified; PTMs mapped to IgE-binding epitopes. | Provides first semi-quantitative PTM profiling of peanut allergens; shows roasting alters PTM patterns without markedly changing IgE binding; supports understanding of allergen proteoforms in food processing and allergy risk assessment | [48] |
3. Functional Proteomic Landscape of Nut Seeds
3.1. Storage Proteins and Allergens in Nuts
| Nut Species | 2S Albumin (Family) | 7S Vicilin (Family) | 11S Legumin (Family) | Other Allergenic Proteins (Family) | Note | Ref. |
|---|---|---|---|---|---|---|
| Almond (Prunus dulcis) | Pru du 1 (AMP or 2S albumin) | Pru du 2 (7S conglutin), Pru du 5 (amposin; 7S vicilin), Pru du 8 (antimicrobial protein, related to 7S storage globulin) | Pru du 6 (amandin; 11S legumin) | Pru du 3 (nsLTP), Pru du 4 (profilin), Pru du 7 (Bet v 1-like PR-10 protein), Pru du 10 (mandelonitrile lyase) | Amandin (Pru du 6) is the major almond allergen and can account for ~65% of soluble protein in almonds. | [74] |
| Brazil Nut (Bertholletia excelsa) | Ber e 1 (2S albumin) | (7S vicilin not well-characterized) | Ber e 2 (11S globulin) | - | Ber e 1 heat-/digestion-stable, key diagnostic marker for true Brazil nut allergy. Ber e 2 also IgE-reactive. Cross-reactivity with other tree nut and seed 2S/11S proteins. | [75] |
| Cashew (Anacardium occidentale) | Ana o 3 (2S albumin) | Ana o 1 (7S vicilin) | Ana o 2 (11S legumin) | - | 2S/7S/11S allergens are all IgE-reactive; contribute to severe reactions and cross-reactivity with pistachio, hazelnut, etc. | [76] |
| Hazelnut (Corylus avellana) | Cor a 14 (2S albumin) | Cor a 11 (7S vicilin) | Cor a 9 (11S legumin) | Cor a 8 (nsLTP), Cor a 1 (PR-10, Bet v 1 homolog), Cor a 12, 13, 15 (Oleosins) | Cor a 9, 11, 14 are major allergens. Oleosins (Cor a 12, 13, 15) are minor allergens. | [71] |
| Peanut (Arachis hypogaea) | Ara h 2,6, 7 (2S albumins) | Ara h 1 (7S vicilin) | Ara h 3 (11S legumin; Ara h 4 isoform) | Ara h 8 (Bet v 1–like PR-10 protein), Ara h 9 (nsLTP), Ara h 5 (profilin), Ara h 10/11 (oleosins), Ara h 12/13 (defensins) | Ara h 2 and Ara h 6 are the most potent peanut allergens. Major allergens include Ara h 1 (7S), Ara h 3/4 (11S), Ara h 2/6/7 (2S). At least 17 allergens identified | [77] |
| Pecan (Carya illinoinensis) | Car i 1 (2S albumin) | Car i 2 (7S vicilin) | Car i 4 (11S legumin) | - | Car i 1, 2 are major allergens, accumulate in late seed filling; show IgE cross-reactivity with walnut, hazelnut. | [78] |
| Pine Nut (Pinus pinea) | Pin p 1 (2S albumin) | (Globulins present, not allergenic) | (Globulins present, not allergenic) | - | Pin p 1 is the major pine nut allergen. Though less common as an allergen, pine nut allergy can be severe and may cross-react with peanut or tree nut allergens. | [79] |
| Pistachio (Pistacia vera) | Pis v 1 (2S albumin) | Pis v 3 (7S vicilin) | Pis v 2, 5 (11S globulin) | Pis v 4 (superoxide dismutase) | Pistachio shares major allergens (2S, 7S, 11S) with cashew, causing frequent cross-reactivity. Interestingly, a defense enzyme (Pis v 4) is also an allergen in pistachio. | [80] |
| Walnut (Juglans regia) | Jug r 1 (2S albumin) | Jug r 2 (7S vicilin) | Jug r 4 (11S legumin-like) | Jug r 3 (nsLTP), Jug r 5 (profilin) | Jug r 4 (11S globulin) is the major walnut allergen and shares homology with 11S proteins from hazelnut and cashew, explaining cross-reactivity. | [81] |
| Macadamia (Macadamia integrifolia) | (Not yet identified) | Mac i 1 (7S vicilin) | Mac i 2 (11S globulin) | Putative: oleosin, pectin acetylesterase, aspartyl protease | Putative allergens identified by IgE-reactive proteomics; oleosin cross-reacts with hazelnut IgE. Allergy rare but may be severe. | [27] |
3.2. Metabolism-Related Proteins in Nut Tissues
3.3. Abiotic Stress Response Proteins
3.4. Disease and Pest Defense Proteins (Pathogenesis-Related)
4. Proteomic Insights into Processing-Induced Changes in Nut Proteins
5. Challenges and Technical Advances in Nut Proteomics
6. Future Prospects in Nut Proteomics
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Thomas, E.; Alcázar Caicedo, C.; McMichael, C.H.; Corvera, R.; Loo, J. Uncovering spatial patterns in the natural and human history of Brazil nut (Bertholletia excelsa) across the Amazon Basin. J. Biogeogr. 2015, 42, 1367–1382. [Google Scholar] [CrossRef]
- Pollegioni, P.; Woeste, K.; Chiocchini, F.; Del Lungo, S.; Ciolfi, M.; Olimpieri, I.; Tortolano, V.; Clark, J.; Hemery, G.E.; Mapelli, S.; et al. Rethinking the history of common walnut (Juglans regia L.) in Europe: Its origins and human interactions. PLoS ONE 2017, 12, e0172541. [Google Scholar] [CrossRef] [PubMed]
- Hammons, R.O.; Herman, D.; Stalker, H.T. Chapter 1—Origin and Early History of the Peanut. In Peanuts; Stalker, H.T., Wilson, R.F., Eds.; AOCS Press: Champaign, IL, USA, 2016; pp. 1–26. [Google Scholar]
- Nuts and Dried Fruits Statistical Yearbook; INC International Nut and Dried Fruit Council: Reus, Spain, 2024.
- Tu, X.H.; Wu, B.F.; Xie, Y.; Xu, S.L.; Wu, Z.Y.; Lv, X.; Wei, F.; Du, L.Q.; Chen, H. A comprehensive study of raw and roasted macadamia nuts: Lipid profile, physicochemical, nutritional, and sensory properties. Food Sci. Nutr. 2021, 9, 1688–1697. [Google Scholar] [CrossRef] [PubMed]
- Devadoss, S. Trends and Challenges in Fruit and Tree Nut Sectors. Choices 2021, 36, 1–3. [Google Scholar]
- Becerra-Tomas, N.; Paz-Graniel, I.; C, W.C.K.; Kahleova, H.; Rahelic, D.; Sievenpiper, J.L.; Salas-Salvado, J. Nut consumption and incidence of cardiovascular diseases and cardiovascular disease mortality: A meta-analysis of prospective cohort studies. Nutr. Rev. 2019, 77, 691–709. [Google Scholar] [CrossRef]
- Aune, D.; Keum, N.; Giovannucci, E.; Fadnes, L.T.; Boffetta, P.; Greenwood, D.C.; Tonstad, S.; Vatten, L.J.; Riboli, E.; Norat, T. Nut consumption and risk of cardiovascular disease, total cancer, all-cause and cause-specific mortality: A systematic review and dose-response meta-analysis of prospective studies. BMC Med. 2016, 14, 207. [Google Scholar] [CrossRef]
- Fernandez-Montero, A.; Bes-Rastrollo, M.; Beunza, J.J.; Barrio-Lopez, M.T.; de la Fuente-Arrillaga, C.; Moreno-Galarraga, L.; Martinez-Gonzalez, M.A. Nut consumption and incidence of metabolic syndrome after 6-year follow-up: The SUN (Seguimiento Universidad de Navarra, University of Navarra Follow-up) cohort. Public. Health Nutr. 2013, 16, 2064–2072. [Google Scholar] [CrossRef]
- Gorji, N.; Moeini, R.; Memariani, Z. Almond, hazelnut and walnut, three nuts for neuroprotection in Alzheimer’s disease: A neuropharmacological review of their bioactive constituents. Pharmacol. Res. 2018, 129, 115–127. [Google Scholar] [CrossRef]
- Ahola, A.J.; Forsblom, C.M.; Harjutsalo, V.; Groop, P.H. Nut consumption is associated with lower risk of metabolic syndrome and its components in type 1 diabetes. Nutrients 2021, 13, 3909. [Google Scholar] [CrossRef]
- Shataer, D.; Li, J.; Duan, X.M.; Liu, L.; Xin, X.L.; Aisa, H.A. Chemical composition of the hazelnut kernel (Corylus avellana L.) and its anti-inflammatory, antimicrobial, and antioxidant activities. J. Agric. Food Chem. 2021, 69, 4111–4119. [Google Scholar] [CrossRef]
- Beltran Sanahuja, A.; Maestre Perez, S.E.; Grane Teruel, N.; Valdes Garcia, A.; Prats Moya, M.S. Variability of chemical profile in almonds (Prunus dulcis) of different cultivars and origins. Foods 2021, 10, 153. [Google Scholar] [CrossRef]
- Reis Ribeiro, S.; Klein, B.; Machado Ribeiro, Q.; Duarte Dos Santos, I.; Gomes Genro, A.L.; de Freitas Ferreira, D.; Janner Hamann, J.; Smanioto Barin, J.; Cichoski, A.J.; Fronza, D.; et al. Chemical composition and oxidative stability of eleven pecan cultivars produced in southern Brazil. Food Res. Int. 2020, 136, 109596. [Google Scholar] [CrossRef]
- Simsek, M. Chemical, mineral, and fatty acid compositions of various types of walnut (Juglans regia L.) in Turkey. Bulg. Chem. Commun. 2016, 48, 66–70. [Google Scholar]
- Amini-Noori, F.; Ziarati, P. Chemical composition of native hazelnut (Corylus avellana L.) barieties in Iran, association with ecological conditions. Biosci. Biotechnol. Res. Asia 2015, 12, 2053–2060. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, S.; Wang, R.; Wu, Y.; Zhang, W.; Han, Y.; Tang, F.; Shen, D.; Liu, Y. Analysis and correlationship of chemical components of various walnut (Juglans regia L.) cultivars. J. Food Meas. Charact. 2020, 14, 3605–3614. [Google Scholar] [CrossRef]
- Queirós, C.S.G.P.; Cardoso, S.; Lourenço, A.; Ferreira, J.; Miranda, I.; Lourenço, M.J.V.; Pereira, H. Characterization of walnut, almond, and pine nut shells regarding chemical composition and extract composition. Biomass Convers. Biorefin 2019, 10, 175–188. [Google Scholar] [CrossRef]
- Venkatachalam, M.; Kshirsagar, H.H.; Seeram, N.P.; Heber, D.; Thompson, T.E.; Roux, K.H.; Sathe, S.K. Biochemical composition and immunological comparison of select pecan [Carya illinoinensis (Wangenh.) K. Koch] cultivars. J. Agric. Food Chem. 2007, 55, 9899–9907. [Google Scholar] [CrossRef]
- Benzitoune, N.; Kadri, N.; Adouane, M.; Berkani, F.; Abbou, A.; Dahmoune, F.; Remini, H.; Bensmail, S. Pine nuts (Pinus pinea L.) as a potential novel plant-based source of functional protein isolates: Optimization of alkali extraction conditions, evaluation of functional properties, and biochemical characterization. J. Food Process Preserv. 2022, 46, e16471. [Google Scholar] [CrossRef]
- Zhao, F.; Liu, C.; Bordoni, L.; Petracci, I.; Wu, D.; Fang, L.; Wang, J.; Wang, X.; Gabbianelli, R.; Min, W. Advances on the Antioxidant Peptides from Nuts: A Narrow Review. Antioxidants 2022, 11, 2020. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Zhao, Y.; Xu, Y.; Gong, C.; Jiao, S. Effects of hot air-assisted radio frequency roasting on nutritional quality and aroma composition of cashew nut kernels. LWT 2019, 116, 108551. [Google Scholar] [CrossRef]
- Li, H.; Liang, X.; Zhou, B.; Chen, X.; Hong, Y.; Zhou, R.; Li, S.; Liu, H.; Lu, Q.; Liu, H.; et al. A proteomic analysis of peanut seed at different stages of underground development to understand the changes of seed proteins. PLoS ONE 2020, 15, e0243132. [Google Scholar] [CrossRef]
- Ribeiro, M.; Freitas, M.; Domínguez-Perles, R.; Barros, A.I.R.N.A.; Ferreira-Cardoso, J.; Igrejas, G. Nutriproteomics survey of sweet chestnut (Castanea sativa Miller) genetic resources in Portugal. Food Biosci. 2020, 36, 100622. [Google Scholar] [CrossRef]
- Xiong, W.; McFarland, M.A.; Pirone, C.; Parker, C.H. Selection of Tree Nut Allergen Peptide Markers: A Need for Improved Protein Sequence Databases. J. AOAC Int. 2019, 102, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Di Stasio, L.; Sciammaro, L.; De Caro, S.; Salinas, M.V.; Puppo, M.C.; Mamone, G. Proteomic characterization of pistachio nut allergen proteins. J. Food Compos. Anal. 2022, 106, 104337. [Google Scholar] [CrossRef]
- Gutierrez-Diaz, G.; Betancor, D.; Parron-Ballesteros, J.; Gordo, R.G.; Castromil-Benito, E.S.; Haroun, E.; Vazquez de la Torre, M.; Turnay, J.; Villalba, M.; Cuesta-Herranz, J.; et al. Identification of new allergens in macadamia nut and cross-reactivity with other tree nuts in a Spanish cohort. Nutrients 2024, 16, 947. [Google Scholar] [CrossRef]
- Clermont, K.; Graham, C.J.; Lloyd, S.W.; Grimm, C.C.; Randall, J.J.; Mattison, C.P. Proteomic analysis of pecan (Carya illinoinensis) nut development. Foods 2023, 12, 866. [Google Scholar] [CrossRef]
- Li, S.; Geng, F.; Wang, P.; Lu, J.; Ma, M. Proteome analysis of the almond kernel (Prunus dulcis). J. Sci. Food Agric. 2016, 96, 3351–3357. [Google Scholar] [CrossRef]
- Xu, D.; Huang, H.; Liu, Z.; Wang, Y.; Liu, Q.; Jiang, X.; Yang, J.; Ling, R. Comprehensive characterization and detection of nut allergens in bakery foods using Q-TOF mass spectrometry and bioinformatics. Food Qual. Saf. 2024, 8, fyad061. [Google Scholar] [CrossRef]
- Luparelli, A.; Losito, I.; De Angelis, E.; Pilolli, R.; Monaci, L. Multi-Target Detection of Nuts and Peanuts as Hidden Allergens in Bakery Products through Bottom-Up Proteomics and High-Resolution Mass Spectrometry. Foods 2023, 12, 726. [Google Scholar] [CrossRef]
- Guo, Q.; Barkla, B.; Barker, R.; Liu, L. Genotype-driven proteomic diversity in macadamia nuts: Implications for allergenicity, nutritional quality, and breeding strategies. J. Agric. Food Chem. 2025, 73, 22272–22282. [Google Scholar] [CrossRef]
- Cirrincione, S.; Aiuto, B.; Gosso, E.; Schiavone, C.; Portesi, C.; Rossi, A.M.; Monti, G.; Cavallarin, L.; Lamberti, C.; Giuffrida, G.M. Proteomic study of walnut oleosome and first evidence on oleosin sensitization in allergic patients. J. Food Compos. Anal. 2023, 121, 105386. [Google Scholar] [CrossRef]
- Furlan Goncalves Dias, F.; Huang, Y.P.; Schauer, J.; Barile, D.; Van de Water, J.; Leite Nobrega de Moura Bell, J.M. Effects of protease-assisted aqueous extraction on almond protein profile, digestibility, and antigenicity. Curr. Res. Food Sci. 2023, 6, 100488. [Google Scholar] [CrossRef]
- Chen, A.; Hu, S.; Zhu, D.; Zhao, R.; Huang, C.; Gao, Y. Lipid droplets proteome reveals dynamic changes of lipid droplets protein during embryonic development of Carya cathayensis nuts. Plant Sci. 2023, 334, 111753. [Google Scholar] [CrossRef]
- De Angelis, E.; Di Bona, D.; Pilolli, R.; Loiodice, R.; Luparelli, A.; Giliberti, L.; D’Uggento, A.M.; Rossi, M.P.; Macchia, L.; Monaci, L. In Vivo and In Vitro Assessment and Proteomic Analysis of the Effectiveness of Physical Treatments in Reducing Allergenicity of Hazelnut Proteins. Nutrients 2022, 14, 874. [Google Scholar] [CrossRef] [PubMed]
- Rost, J.; Muralidharan, S.; Lee, N.A. A label-free shotgun proteomics analysis of macadamia nut. Food Res. Int. 2020, 129, 108838. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Downs, M.L. Proteomic Analysis of Oil-Roasted Cashews Using a Customized Allergen-Focused Protein Database. J. Proteome Res. 2022, 21, 1694–1706. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, H.; Gu, J.; Deng, L.; Ren, L.; Hong, Y.; Lu, Q.; Chen, X.; Liang, X. Identification of the candidate proteins related to oleic acid accumulation during peanut (Arachis hypogaea L.) seed development through comparative proteome analysis. Int. J. Mol. Sci. 2018, 19, 1235. [Google Scholar] [CrossRef]
- Liu, C.M.; Peng, Q.; Zhong, J.Z.; Liu, W.; Zhong, Y.J.; Wang, F. Molecular and Functional Properties of Protein Fractions and Isolate from Cashew Nut (Anacardium occidentale L.). Molecules 2018, 23, 393. [Google Scholar] [CrossRef]
- Nitride, C.; Picariello, G.; Mamone, G.; Ferranti, P. Proteomics of Hazelnut (Corylus avellana). In Proteomics in Food Science; Academic Press: Cambridge, MA, USA, 2017; pp. 107–125. [Google Scholar] [CrossRef]
- Downs, M.L.; Baumert, J.L.; Taylor, S.L.; Mills, E.N. Mass spectrometric analysis of allergens in roasted walnuts. J. Proteom. 2016, 142, 62–69. [Google Scholar] [CrossRef]
- Prieto, N.; Burbano, C.; Iniesto, E.; Rodriguez, J.; Cabanillas, B.; Crespo, J.F.; Pedrosa, M.M.; Muzquiz, M.; Del Pozo, J.C.; Linacero, R.; et al. A Novel Proteomic Analysis of the Modifications Induced by High Hydrostatic Pressure on Hazelnut Water-Soluble Proteins. Foods 2014, 3, 279–289. [Google Scholar] [CrossRef]
- Nitride, C.; Mamone, G.; Picariello, G.; Mills, C.; Nocerino, R.; Berni Canani, R.; Ferranti, P. Proteomic and immunological characterization of a new food allergen from hazelnut (Corylus avellana). J. Proteom. 2013, 86, 16–26. [Google Scholar] [CrossRef]
- White, B.L.; Gokce, E.; Nepomuceno, A.I.; Muddiman, D.C.; Sanders, T.H.; Davis, J.P. Comparative proteomic analysis and IgE binding properties of peanut seed and testa (skin). J. Agric. Food Chem. 2013, 61, 3957–3968. [Google Scholar] [CrossRef]
- Moreno, F.J.; Jenkins, J.A.; Mellon, F.A.; Rigby, N.M.; Robertson, J.A.; Wellner, N.; Clare Mills, E.N. Mass spectrometry and structural characterization of 2S albumin isoforms from Brazil nuts (Bertholletia excelsa). Biochim. Biophys. Acta 2004, 1698, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Marcone, M.R.; Kakuda, Y.; Jahaniaval, F.; Yada, R.Y.; Montevirgen, L.S. Characterization of the proteins of Pili nut (Canarium ovatum, Engl.). Plant Foods Hum. Nutr. 2002, 57, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Dukic, T.; Smiljanic, K.; Mihailovic, J.; Prodic, I.; Apostolovic, D.; Liu, S.H.; Epstein, M.M.; van Hage, M.; Stanic-Vucinic, D.; Cirkovic Velickovic, T. Proteomic Profiling of Major Peanut Allergens and Their Post-Translational Modifications Affected by Roasting. Foods 2022, 11, 3993. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Huang, Y.; Sun, Z.; Huang, J.; Wang, Z. Transcriptome analysis of genes involved in lipid biosynthesis in the developing embryo of pecan (Carya illinoinensis). J. Agric. Food Chem. 2017, 65, 4223–4236. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, T.; Zhang, Q.; Chen, M.; Wang, Z.; Zheng, B.; Xia, G.; Yang, X.; Huang, C.; Huang, Y. The mechanism of high contents of oil and oleic acid revealed by transcriptomic and lipidomic analysis during embryogenesis in Carya cathayensis Sarg. BMC Genomics 2016, 17, 113. [Google Scholar] [CrossRef]
- Jin, F.; Zhou, Y.; Zhang, P.; Huang, R.; Fan, W.; Li, B.; Li, G.; Song, X.; Pei, D. Identification of Key Lipogenesis Stages and Proteins Involved in Walnut Kernel Development. J. Agric. Food Chem. 2023, 71, 4306–4318. [Google Scholar] [CrossRef]
- Ghorab, H.; Lammi, C.; Arnoldi, A.; Kabouche, Z.; Aiello, G. Proteomic analysis of sweet algerian apricot kernels (Prunus armeniaca L.) by combinatorial peptide ligand libraries and LC-MS/MS. Food Chem. 2018, 239, 935–945. [Google Scholar] [CrossRef]
- Cai, L.; Xiao, L.; Liu, C.; Ying, T. Functional properties and bioactivities of pine nut (Pinus gerardiana) protein isolates and its enzymatic hydrolysates. Food Bioprocess. Technol. 2012, 6, 2109–2117. [Google Scholar] [CrossRef]
- Lin, S.; Liang, R.; Xue, P.; Zhang, S.; Liu, Z.; Dong, X. Antioxidant activity improvement of identified pine nut peptides by pulsed electric field (PEF) and the mechanism exploration. LWT 2017, 75, 366–372. [Google Scholar] [CrossRef]
- Katam, R.; Basha, S.M.; Suravajhala, P.; Pechan, T. Analysis of peanut leaf proteome. J. Proteome Res. 2010, 9, 2236–2254. [Google Scholar] [CrossRef]
- Cipriano, A.K.; Gondim, D.M.; Vasconcelos, I.M.; Martins, J.A.; Moura, A.A.; Moreno, F.B.; Monteiro-Moreira, A.C.; Melo, J.G.; Cardoso, J.E.; Paiva, A.L.; et al. Proteomic analysis of responsive stem proteins of resistant and susceptible cashew plants after Lasiodiplodia theobromae infection. J. Proteom. 2015, 113, 90–109. [Google Scholar] [CrossRef] [PubMed]
- Hafizi, A.; Fatehi, F.; Pakzad, R. SWATH-based quantitative proteomics gives insight into a significant proteome shift in UCB-1 pistachio rootstock under salt stress. Sci. Hortic. 2023, 311, 111822. [Google Scholar] [CrossRef]
- Martínez-García, P.J.; Gãmez, E.M.; Casado-Vela, J.; Elortza, F.; Dicenta, F.; Ortega, E. Differential protein expression in compatible and incompatible pollen-pistil interactions in almond [Prunus dulcis (Miller) D. A. Webb] by 2D-DIGE and HPLC-MS/MS. J. Hortic. Sci. Biotechnol. 2015, 90, 71–77. [Google Scholar] [CrossRef]
- Zaini, P.A.; Feinberg, N.G.; Grilo, F.S.; Saxe, H.J.; Salemi, M.R.; Phinney, B.S.; Crisosto, C.H.; Dandekar, A.M. Comparative proteomic analysis of walnut (Juglans regia L.) pellicle tissues reveals the regulation of nut quality attributes. Life 2020, 10, 314. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, Q.; Zhang, X.; Wang, J.; Ayup, M.; Yang, B.; Guo, C.; Gong, P.; Dong, W. The proteome reveals the involvement of serine/threonine kinase in the recognition of self- incompatibility in almond. J. Proteom. 2022, 256, 104505. [Google Scholar] [CrossRef]
- Larkins, B. Seed storage proteins: Characterization and biosynthesis. In Proteins and Nucleic Acids; Elsevier: Amsterdam, The Netherlands, 1981; pp. 449–489. [Google Scholar]
- Shewry, P.R.; Napier, J.A.; Tatham, A.S. Seed storage proteins: Structures and biosynthesis. Plant Cell 1995, 7, 945. [Google Scholar]
- Wolf, W.J.; Sathe, S.K. Ultracentrifugal and polyacrylamide gel electrophoretic studies of extractability and stability of almond meal proteins. J. Sci. Food Agric. 1998, 78, 511–521. [Google Scholar] [CrossRef]
- Sebei, K.; Gnouma, A.; Herchi, W.; Sakouhi, F.; Boukhchina, S. Lipids, proteins, phenolic composition, antioxidant and antibacterial activities of seeds of peanuts (Arachis hypogaea L.) cultivated in Tunisia. Biol. Res. 2013, 46, 257–263. [Google Scholar] [CrossRef]
- Prodic, I.; Krstic Ristivojevic, M.; Smiljanic, K. Antioxidant Properties of Protein-Rich Plant Foods in Gastrointestinal Digestion-Peanuts as Our Antioxidant Friend or Foe in Allergies. Antioxidants 2023, 12, 886. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.S.; Altenbach, S.B.; Leung, F.W. Properties, biosynthesis and processing of a sulfur-rich protein in Brazil nut (Bertholletia excelsa H.B.K.). Eur. J. Biochem. 1987, 162, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Koppelman, S.J.; Hefle, S.L.; Taylor, S.L.; de Jong, G.A. Digestion of peanut allergens Ara h 1, Ara h 2, Ara h 3, and Ara h 6: A comparative in vitro study and partial characterization of digestion-resistant peptides. Mol. Nutr. Food Res. 2010, 54, 1711–1721. [Google Scholar] [CrossRef] [PubMed]
- Gipson, S.A.Y.; Nesbit, J.B.; Swientoniewski, L.T.; Rogers, S.I.; Mustafa, S.S.; Dreskin, S.C.; Teuber, S.S.; Cheng, H.; Maleki, S.J. Purification and Epitope Mapping of Jug r 4, a Major Walnut Allergen. Allergies 2025, 5, 8. [Google Scholar] [CrossRef]
- Bezerra, M.; Ribeiro, M.; Igrejas, G. An Updated Overview of Almond Allergens. Nutrients 2021, 13, 2578. [Google Scholar] [CrossRef]
- Weinberger, T.; Sicherer, S. Current perspectives on tree nut allergy: A review. J. Asthma Allergy 2018, 11, 41–51. [Google Scholar] [CrossRef]
- Costa, J.; Mafra, I.; Carrapatoso, I.; Oliveira, M.B. Hazelnut Allergens: Molecular Characterization, Detection, and Clinical Relevance. Crit. Rev. Food Sci. Nutr. 2016, 56, 2579–2605. [Google Scholar] [CrossRef]
- Breiteneder, H.; Ebner, C. Molecular and biochemical classification of plant-derived food allergens. J. Allergy Clin. Immunol. 2000, 106, 27–36. [Google Scholar] [CrossRef]
- Cuadrado, C.; Sanchiz, Á.; Linacero, R. Nut allergenicity: Effect of food processing. Allergies 2021, 1, 150–162. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, T. Almond allergens: Update and perspective on identification and characterization. J. Sci. Food Agric. 2020, 100, 4657–4663. [Google Scholar] [CrossRef]
- Kluczkovski, A.M.; Scussel, V.M. Brazil nut allergy: A review. Afr. J. Pharm. Pharmacol. 2015, 9, 633–644. [Google Scholar] [CrossRef]
- Mendes, C.; Costa, J.; Vicente, A.A.; Oliveira, M.; Mafra, I. Cashew Nut Allergy: Clinical Relevance and Allergen Characterisation. Clin. Rev. Allergy Immunol. 2019, 57, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Palladino, C.; Breiteneder, H. Peanut allergens. Mol. Immunol. 2018, 100, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lee, B.; Du, W.X.; Lyu, S.C.; Nadeau, K.C.; Grauke, L.J.; Zhang, Y.; Wang, S.; Fan, Y.; Yi, J.; et al. Identification and Characterization of a New Pecan [Carya illinoinensis (Wangenh.) K. Koch] Allergen, Car i 2. J. Agric. Food Chem. 2016, 64, 4146–4151. [Google Scholar] [CrossRef]
- Cabanillas, B.; Crespo, J.F.; Maleki, S.J.; Rodriguez, J.; Novak, N. Pin p 1 is a major allergen in pine nut and the first food allergen described in the plant group of gymnosperms. Food Chem. 2016, 210, 70–77. [Google Scholar] [CrossRef]
- Costa, J.; Silva, I.; Vicente, A.A.; Oliveira, M.; Mafra, I. Pistachio nut allergy: An updated overview. Crit. Rev. Food Sci. Nutr. 2019, 59, 546–562. [Google Scholar] [CrossRef]
- Costa, J.; Carrapatoso, I.; Oliveira, M.B.; Mafra, I. Walnut allergens: Molecular characterization, detection and clinical relevance. Clin. Exp. Allergy 2014, 44, 319–341. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, X.; Zhang, X.; He, X.; Li, H.; Cui, D.; Yin, D. ITRAQ-Based Proteomic Analysis of the Metabolic Mechanisms Behind Lipid Accumulation and Degradation during Peanut Seed Development and Postgermination. J. Proteome Res. 2016, 15, 4277–4289. [Google Scholar] [CrossRef]
- Liu, H.; Hong, Y.; Lu, Q.; Li, H.; Gu, J.; Ren, L.; Deng, L.; Zhou, B.; Chen, X.; Liang, X. Integrated Analysis of Comparative Lipidomics and Proteomics Reveals the Dynamic Changes of Lipid Molecular Species in High-Oleic Acid Peanut Seed. J. Agric. Food Chem. 2020, 68, 426–438. [Google Scholar] [CrossRef]
- Frandsen, G.I.; Mundy, J.; Tzen, J.T. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant 2001, 112, 301–307. [Google Scholar] [CrossRef]
- Nebbia, S.; Lamberti, C.; Cirrincione, S.; Acquadro, A.; Abba, S.; Ciuffo, M.; Torello Marinoni, D.; Manfredi, M.; Marengo, E.; Calzedda, R.; et al. Oleosin Cor a 15 is a novel allergen for Italian hazelnut allergic children. Pediatr. Allergy Immunol. 2021, 32, 1743–1755. [Google Scholar] [CrossRef]
- Johnson, P.E.; Sayers, R.L.; Gethings, L.A.; Balasundaram, A.; Marsh, J.T.; Langridge, J.I.; Mills, E.N. Quantitative Proteomic Profiling of Peanut Allergens in Food Ingredients Used for Oral Food Challenges. Anal. Chem. 2016, 88, 5689–5695. [Google Scholar] [CrossRef]
- Li, W.; Huang, L.; Liu, N.; Pandey, M.K.; Chen, Y.; Cheng, L.; Guo, J.; Yu, B.; Luo, H.; Zhou, X.; et al. Key Regulators of Sucrose Metabolism Identified through Comprehensive Comparative Transcriptome Analysis in Peanuts. Int. J. Mol. Sci. 2021, 22, 7266. [Google Scholar] [CrossRef]
- Salas-Salvado, J.; Casas-Agustench, P.; Salas-Huetos, A. Cultural and historical aspects of Mediterranean nuts with emphasis on their attributed healthy and nutritional properties. Nutr. Metab. Cardiovasc. Dis. 2011, 21, S1–S6. [Google Scholar] [CrossRef]
- Casas-Agustench, P.; Salas-Huetos, A.; Salas-Salvado, J. Mediterranean nuts: Origins, ancient medicinal benefits and symbolism. Public. Health Nutr. 2011, 14, 2296–2301. [Google Scholar] [CrossRef]
- Kottapalli, K.R.; Zabet-Moghaddam, M.; Rowland, D.; Faircloth, W.; Mirzaei, M.; Haynes, P.A.; Payton, P. Shotgun label-free quantitative proteomics of water-deficit-stressed midmature peanut (Arachis hypogaea L.) seed. J. Proteome Res. 2013, 12, 5048–5057. [Google Scholar] [CrossRef] [PubMed]
- Puppala, N.; Nayak, S.N.; Sanz-Saez, A.; Chen, C.; Devi, M.J.; Nivedita, N.; Bao, Y.; He, G.; Traore, S.M.; Wright, D.A.; et al. Sustaining yield and nutritional quality of peanuts in harsh environments: Physiological and molecular basis of drought and heat stress tolerance. Front. Genet. 2023, 14, 1121462. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Group II late embryogenesis abundant (LEA) proteins: Structural and functional aspects in plant abiotic stress. Plant Growth Regul. 2015, 79, 1–17. [Google Scholar] [CrossRef]
- ZamaniBahramabadi, E.; Jonoubi, P.; Rezanejad, F. Dehydrin content in fresh and desiccated pistachio (Pistacia vera L.) seeds. Iran. J. Sci. Technol. Trans. A Sci. 2019, 43, 2099–2105. [Google Scholar] [CrossRef]
- Yakubov, B.; Barazani, O.; Shachack, A.; Rowland, L.J.; Shoseyov, O.; Golan-Goldhirsh, A. Cloning and expression of a dehydrin-like protein from Pistacia vera L. Trees 2004, 19, 224–230. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Zhu, Y.; Jones, A.; Rose, R.J.; Song, Y. Heat Stress in Legume Seed Setting: Effects, Causes, and Future Prospects. Front. Plant Sci. 2019, 10, 938. [Google Scholar] [CrossRef]
- Chen, H.; Liu, N.; Xu, R.; Chen, X.; Zhang, Y.; Hu, R.; Lan, X.; Tang, Z.; Lin, G. Quantitative proteomics analysis reveals the response mechanism of peanut (Arachis hypogaea L.) to imbibitional chilling stress. Plant Biol. 2021, 23, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Lotfi, N.; Soleimani, A.; Vahdati, K.; Çakmakçı, R. Comprehensive biochemical insights into the seed germination of walnut under drought stress. Sci. Hortic. 2019, 250, 329–343. [Google Scholar] [CrossRef]
- Chen, Y.; Pei, H.; Dai, Q.; Zhang, C.; Kong, X.; Hua, Y. Raw walnut kernel: A natural source for dietary proteases and bioactive proteins. Food Chem. 2022, 369, 130961. [Google Scholar] [CrossRef] [PubMed]
- Hubert, B.; Leprince, O.; Buitink, J. Sleeping but not defenceless: Seed dormancy and protection. J. Exp. Bot. 2024, 75, 6110–6124. [Google Scholar] [CrossRef]
- Sudisha, J.; Sharathchandra, R.G.; Amruthesh, K.N.; Kumar, A.; Shetty, H.S. Pathogenesis related proteins in plant defense response. In Plant Defence: Biological Control; Springer: Berlin/Heidelberg, Germany, 2012; pp. 379–403. [Google Scholar]
- Liang, X.Q.; Holbrook, C.C.; Lynch, R.E.; Guo, B.Z. Beta-1,3-glucanase activity in peanut seed (Arachis hypogaea) is induced by inoculation with Aspergillus flavus and copurifies with a conglutin-like protein. Phytopathology 2005, 95, 506–511. [Google Scholar] [CrossRef]
- Muller, V.; Bonacci, G.; Batthyany, C.; Ame, M.V.; Carrari, F.; Gieco, J.; Asis, R. Peanut seed cultivars with contrasting resistance to Aspergillus parasiticus colonization display differential temporal response of protease inhibitors. Phytopathology 2017, 107, 474–482. [Google Scholar] [CrossRef]
- Sagawa, C.H.D.; Assis, R.d.A.B.; Zaini, P.A.; Wilmarth, P.A.; Phinney, B.S.; Moreira, L.M.; Dandekar, A.M. Proteome Analysis of Walnut Bacterial Blight Disease. Int. J. Mol. Sci. 2020, 21, 7453. [Google Scholar] [CrossRef]
- Wang, T.; Xie, M.; Hou, S.; Ma, J.; Lin, Y.; Chen, S.; Li, D.; Yang, G. Walnut PR10/Bet v1-like proteins interact with chitinase in response to anthracnose stress. J. Evol. Biol. 2025, 38, 391–403. [Google Scholar] [CrossRef]
- Zeng, L.; Tu, X.L.; Dai, H.; Han, F.M.; Lu, B.-S.; Wang, M.S.; Nanaei, H.A.; Tajabadipour, A.; Mansouri, M.; Li, X.L.; et al. Whole genomes and transcriptomes reveal adaptation and domestication of pistachio. Genome Biol. 2019, 20, 79. [Google Scholar] [CrossRef]
- Smolikova, G.; Gorbach, D.; Lukasheva, E.; Mavropolo-Stolyarenko, G.; Bilova, T.; Soboleva, A.; Tsarev, A.; Romanovskaya, E.; Podolskaya, E.; Zhukov, V.; et al. Bringing new methods to the seed proteomics platform: Challenges and perspectives. Int. J. Mol. Sci. 2020, 21, 9162. [Google Scholar] [CrossRef]
- Yan, X.; Wang, Y.; Chen, Y.; Xie, J.; Yu, Q. Effect of roasting duration on the solubility, structure, and IgE-binding capacity of cashew nut proteins. Innov. Food Sci. Emerg. Technol. 2021, 68, 102635. [Google Scholar] [CrossRef]
- Sealey-Voyksner, J.; Zweigenbaum, J.; Voyksner, R. Discovery of highly conserved unique peanut and tree nut peptides by LC-MS/MS for multi-allergen detection. Food Chem. 2016, 194, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, S.S.; Krishnan, H.B.; Lakshman, S.; Garrett, W.M. An efficient extraction method to enhance analysis of low abundant proteins from soybean seed. Anal. Biochem. 2009, 394, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Wang, Y.; Gupta, R.; Kim, S.W.; Min, C.W.; Kim, Y.C.; Park, K.H.; Agrawal, G.K.; Rakwal, R.; Choung, M.G.; et al. Protamine sulfate precipitation method depletes abundant plant seed-storage proteins: A case study on legume plants. Proteomics 2015, 15, 1760–1764. [Google Scholar] [CrossRef]
- Krishnan, H.B.; Oehrle, N.W.; Natarajan, S.S. A rapid and simple procedure for the depletion of abundant storage proteins from legume seeds to advance proteome analysis: A case study using Glycine max. Proteomics 2009, 9, 3174–3188. [Google Scholar] [CrossRef]
- Righetti, P.G.; Boschetti, E. Low-abundance plant protein enrichment with peptide libraries to enlarge proteome coverage and related applications. Plant Sci. 2020, 290, 110302. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, L.; Yim, W.C.; Cushman, J.C.; Barkla, B.J. Membrane profiling by free flow electrophoresis and SWATH-MS to characterize subcellular compartment proteomes in Mesembryanthemum crystallinum. Int. J. Mol. Sci. 2021, 22, 5020. [Google Scholar] [CrossRef]
- Pirone-Davies, C.; McFarland, M.A.; Parker, C.H.; Adachi, Y.; Croley, T.R. The Utility of Genomic and Transcriptomic Data in the Construction of Proxy Protein Sequence Databases for Unsequenced Tree Nuts. Biology 2020, 9, 104. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Nock, C.J.; Baten, A.; Mauleon, R.; Langdon, K.S.; Topp, B.; Hardner, C.; Furtado, A.; Henry, R.J.; King, G.J. Chromosome-scale assembly and annotation of the macadamia genome (Macadamia integrifolia HAES 741). Genes Genomes Genet. 2020, 10, 3497–3504. [Google Scholar] [CrossRef]
- Marrano, A.; Britton, M.; Zaini, P.A.; Zimin, A.V.; Workman, R.E.; Puiu, D.; Bianco, L.; Pierro, E.A.D.; Allen, B.J.; Chakraborty, S.; et al. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. Gigascience 2020, 9, giaa050. [Google Scholar] [CrossRef]
- Adaskaveg, J.A.; Lee, C.; Wei, Y.; Wang, F.; Grilo, F.S.; Mesquida-Pesci, S.D.; Davis, M.; Wang, S.C.; Marino, G.; Ferguson, L.; et al. In a nutshell: Pistachio genome and kernel development. New Phytol. 2025, 246, 1032–1048. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.J.; Kahraman, K.; Avsar, B.; Buggs, R.J.A.; Bilge, I. A chromosome-scale genome assembly of European hazel (Corylus avellana L.) reveals targets for crop improvement. Plant J. 2021, 105, 1413–1430. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Orsburn, B.C. Proteome Discoverer-A Community Enhanced Data Processing Suite for Protein Informatics. Proteomes 2021, 9, 15. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]
- Baker, C.P.; Bruderer, R.; Abbott, J.; Arthur, J.S.C.; Brenes, A.J. Optimizing Spectronaut search parameters to improve data quality with minimal proteome coverage reductions in DIA analyses of heterogeneous samples. J. Proteome Res. 2024, 23, 1926–1936. [Google Scholar] [CrossRef]
- Zubarev, R.A.; Makarov, A. Orbitrap mass spectrometry. Anal. Chem. 2013, 85, 5288–5296. [Google Scholar] [CrossRef]
- Meier, F.; Brunner, A.-D.; Frank, M.; Ha, A.; Bludau, I.; Voytik, E.; Kaspar-Schoenefeld, S.; Lubeck, M.; Raether, O.; Bache, N.; et al. diaPASEF: Parallel accumulation–serial fragmentation combined with data-independent acquisition. Nat. Methods 2020, 17, 1229–1236. [Google Scholar] [CrossRef]
- Zhao, R.; Zhou, Y.; Shen, H.; Guan, L.; Wang, Y.; Shen, X.; Wang, F.; Yao, X. Preparation and Encapsulation of DPP-IV Inhibitory Peptides: Challenges and Strategies for Functional Food Development. Foods 2025, 14, 1479. [Google Scholar] [CrossRef]
- Thangella, P.A.V.; Pasumarti, S.; Pullakhandam, R.; Geereddy, B.R.; Daggu, M.R. Differential expression of leaf proteins in four cultivars of peanut (Arachis hypogaea L.) under water stress. 3 Biotech 2018, 8, 157. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Q.; Barkla, B.J. Proteomic Insights into Edible Nut Seeds: Nutritional Value, Allergenicity, Stress Responses, and Processing Effects. Agronomy 2025, 15, 2353. https://doi.org/10.3390/agronomy15102353
Guo Q, Barkla BJ. Proteomic Insights into Edible Nut Seeds: Nutritional Value, Allergenicity, Stress Responses, and Processing Effects. Agronomy. 2025; 15(10):2353. https://doi.org/10.3390/agronomy15102353
Chicago/Turabian StyleGuo, Qi, and Bronwyn J. Barkla. 2025. "Proteomic Insights into Edible Nut Seeds: Nutritional Value, Allergenicity, Stress Responses, and Processing Effects" Agronomy 15, no. 10: 2353. https://doi.org/10.3390/agronomy15102353
APA StyleGuo, Q., & Barkla, B. J. (2025). Proteomic Insights into Edible Nut Seeds: Nutritional Value, Allergenicity, Stress Responses, and Processing Effects. Agronomy, 15(10), 2353. https://doi.org/10.3390/agronomy15102353






