Non-Destructive Estimation of Area and Greenness in Leaf and Seedling Scales: A Case Study in Cucumber
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Morphometric and Colorimetric Analysis of Individual Cotyledons and Leaves
2.3. SPAD Value of Individual Cotyledons and Leaves
2.4. Morphometric, Colorimetric Analysis and SPAD Value of Whole-Seedlings
2.5. Statistical Analysis and Model Development
3. Results
3.1. Employed Range of Variation in Morphological Traits
3.2. Calculation of Area of Single Cotyledons or Leaves Based on Dimensions
3.3. Calculation of Whole-Seedling Area Based on Image-Derived Projected Area
3.4. Calculation of SPAD Value at Either Organ or Whole-Seedling Level
4. Discussion
4.1. Individual Cotyledon/Leaf Area Estimation by Employing Linear Dimensions
4.2. Whole-Seedling LA Estimation by Employing the Projected Area
4.3. Cotyledon/Leaf, and Whole-Seedling SPAD Value Estimation by Employing Colorimetric Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Gallegos-Cedillo, V.M.; Nájera, C.; Gruda, N.S.; Signore, A.; Gallegos, J.; Rodríguez, R.; Ochoa, J.; Egea-Gilabert, C.; Fernández, J.A. An In-Depth Analysis of Sustainable Practices in Vegetable Seedlings Nurseries: A Review. Sci. Hortic. 2024, 334, 113342. [Google Scholar] [CrossRef]
- Gallegos-Cedillo, V.M.; Nájera, C.; Signore, A.; Ochoa, J.; Gallegos, J.; Egea-Gilabert, C.; Gruda, N.S.; Fernández, J.A. Analysis of Global Research on Vegetable Seedlings and Transplants and Their Impacts on Product Quality. J. Sci. Food Agric. 2024, 104, 4950–4965. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Ning, K.; Naznin, A.; Wang, Y.; Chen, C.; Zuo, Y.; Zhou, M.; Li, C.; Varshney, R.; Chen, Z.-H. Technological Advances in Imaging and Modelling of Leaf Structural Traits: A Review of Heat Stress in Wheat. J. Exp. Bot. 2025, eraf070. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Jin, X.; Bian, L.; Ge, Y. High-Throughput Phenotyping of Plant Leaf Morphological, Physiological, and Biochemical Traits on Multiple Scales Using Optical Sensing. Crop J. 2023, 11, 1303–1318. [Google Scholar] [CrossRef]
- Moosavi-Nezhad, M.; Salehi, R.; Aliniaeifard, S.; Tsaniklidis, G.; Woltering, E.J.; Fanourakis, D.; Żuk-Gołaszewska, K.; Kalaji, H.M. Blue Light Improves Photosynthetic Performance during Healing and Acclimatization of Grafted Watermelon Seedlings. Int. J. Mol. Sci. 2021, 22, 8043. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Adhikari, R.; Yao, Y.; Miller, A.G.; Kalbaugh, K.; Li, D.; Nemali, K. Measuring Plant Growth Characteristics Using Smartphone Based Image Analysis Technique in Controlled Environment Agriculture. Comput. Electron. Agric. 2020, 168, 105123. [Google Scholar] [CrossRef]
- Fanourakis, D.; Kazakos, F.; Nektarios, P.A. Allometric Individual Leaf Area Estimation in Chrysanthemum. Agronomy 2021, 11, 795. [Google Scholar] [CrossRef]
- Cho, Y.Y.; Oh, S.; Oh, M.M.; Son, J.E. Estimation of Individual Leaf Area, Fresh Weight, and Dry Weight of Hydroponically Grown Cucumbers (Cucumis sativus L.) Using Leaf Length, Width, and SPAD Value. Sci. Hortic. 2007, 111, 330–334. [Google Scholar] [CrossRef]
- Blanco, F.F.; Folegatti, M.V. Estimation of Leaf Area for Greenhouse Cucumber by Linear Measurements under Salinity and Grafting. Sci. Agric. 2005, 62, 305–309. [Google Scholar] [CrossRef]
- Huaccha-Castillo, A.E.; Fernandez-Zarate, F.H.; Pérez-Delgado, L.J.; Tantalean-Osores, K.S.; Vaca-Marquina, S.P.; Sanchez-Santillan, T.; Morales-Rojas, E.; Seminario-Cunya, A.; Quiñones-Huatangari, L. Non-Destructive Estimation of Leaf Area and Leaf Weight of Cinchona officinalis L. (Rubiaceae) Based on Linear Models. For. Sci. Technol. 2023, 19, 59–67. [Google Scholar] [CrossRef]
- Gao, M.; Van der Heijden, G.W.A.M.; Vos, J.; Eveleens, B.A.; Marcelis, L.F.M. Estimation of Leaf Area for Large Scale Phenotyping and Modeling of Rose Genotypes. Sci. Hortic. 2012, 138, 227–234. [Google Scholar] [CrossRef]
- Koubouris, G.; Bouranis, D.; Vogiatzis, E.; Nejad, A.R.; Giday, H.; Tsaniklidis, G.; Ligoxigakis, E.K.; Blazakis, K.; Kalaitzis, P.; Fanourakis, D. Leaf Area Estimation by Considering Leaf Dimensions in Olive Tree. Sci. Hortic. 2018, 240, 440–445. [Google Scholar] [CrossRef]
- Hayashi, E.; Amagai, Y.; Kozai, T.; Maruo, T.; Tsukagoshi, S.; Nakano, A.; Johkan, M. Variations in the Growth of Cotyledons and Initial True Leaves as Affected by Photosynthetic Photon Flux Density at Individual Seedlings and Nutrients. Agronomy 2022, 12, 194. [Google Scholar] [CrossRef]
- Poorter, H.; Hummel, G.M.; Nagel, K.A.; Fiorani, F.; von Gillhaussen, P.; Virnich, O.; Schurr, U.; Postma, J.A.; van de Zedde, R.; Wiese-Klinkenberg, A. Pitfalls and Potential of High-Throughput Plant Phenotyping Platforms. Front. Plant Sci. 2023, 14, 1233794. [Google Scholar] [CrossRef]
- Li, D.; Li, C.; Yao, Y.; Li, M.; Liu, L. Modern Imaging Techniques in Plant Nutrition Analysis: A Review. Comput. Electron. Agric. 2020, 174, 105459. [Google Scholar] [CrossRef]
- Xiong, D.; Chen, J.; Yu, T.; Gao, W.; Ling, X.; Li, Y.; Peng, S.; Huang, J. SPAD-Based Leaf Nitrogen Estimation Is Impacted by Environmental Factors and Crop Leaf Characteristics. Sci. Rep. 2015, 5, 13389. [Google Scholar] [CrossRef]
- Padilla, F.M.; Peña-Fleitas, M.T.; Gallardo, M.; Giménez, C.; Thompson, R.B. Derivation of Sufficiency Values of a Chlorophyll Meter to Estimate Cucumber Nitrogen Status and Yield. Comput. Electron. Agric. 2017, 141, 54–64. [Google Scholar] [CrossRef]
- Hlavinka, J.; Nauš, J.; Špundová, M. Anthocyanin Contribution to Chlorophyll Meter Readings and Its Correction. Photosynth. Res. 2013, 118, 277–295. [Google Scholar] [CrossRef]
- Yang, H.; Li, J.; Yang, J.; Wang, H.; Zou, J.; He, J. Effects of Nitrogen Application Rate and Leaf Age on the Distribution Pattern of Leaf SPAD Readings in the Rice Canopy. PLoS ONE 2014, 9, e88421. [Google Scholar] [CrossRef]
- Rigon, J.P.G.; Capuani, S.; Fernandes, D.M.; Guimarães, T.M. A Novel Method for the Estimation of Soybean Chlorophyll Content Using a Smartphone and Image Analysis. Photosynthetica 2016, 54, 559–566. [Google Scholar] [CrossRef]
- Mohan, P.J.; Gupta, S.D. Intelligent Image Analysis for Retrieval of Leaf Chlorophyll Content of Rice from Digital Images of Smartphone under Natural Light. Photosynthetica 2019, 57, 388–398. [Google Scholar] [CrossRef]
- Peters, R.D.; Noble, S.D. Characterization of Leaf Surface Phenotypes Based on Light Interaction. Plant Methods 2023, 19, 26. [Google Scholar] [CrossRef] [PubMed]
- Taheri-Garavand, A.; Mumivand, H.; Fanourakis, D.; Fatahi, S.; Taghipour, S. An Artificial Neural Network Approach for Non-Invasive Estimation of Essential Oil Content and Composition through Considering Drying Processing Factors: A Case Study in Mentha aquatica. Ind. Crops Prod. 2021, 171, 113985. [Google Scholar] [CrossRef]
- Rahimikhoob, H.; Delshad, M.; Habibi, R. Leaf Area Estimation in Lettuce: Comparison of Artificial Intelligence-Based Methods with Image Analysis Technique. Measurement 2023, 222, 113636. [Google Scholar] [CrossRef]
- Nasiri, A.; Taheri-Garavand, A.; Fanourakis, D.; Zhang, Y.-D.; Nikoloudakis, N. Automated Grapevine Cultivar Identification via Leaf Imaging and Deep Convolutional Neural Networks: A Proof-of-Concept Study Employing Primary Iranian Varieties. Plants 2021, 10, 1628. [Google Scholar] [CrossRef]
- Fanourakis, D.; Papadakis, V.M.; Machado, M.; Psyllakis, E.; Nektarios, P.A. Non-invasive Leaf Hydration Status Determination through Convolutional Neural Networks Based on Multispectral Images in Chrysanthemum. Plant Growth Regul. 2024, 102, 485–496. [Google Scholar] [CrossRef]
- Taheri-Garavand, A.; Rezaei Nejad, A.; Fanourakis, D.; Fatahi, S.; Ahmadi Majd, M. Employment of Artificial Neural Networks for Non-Invasive Estimation of Leaf Water Status Using Color Features: A Case Study in Spathiphyllum wallisii. Acta Physiol. Plant 2021, 43, 78. [Google Scholar] [CrossRef]
- Gupta, S.; Rosenthal, D.M.; Stinchcombe, J.R.; Baucom, R.S. The Remarkable Morphological Diversity of Leaf Shape in Sweet Potato (Ipomoea batatas): The Influence of Genetics, Environment, and G × E. New Phytol. 2020, 225, 2183–2195. [Google Scholar] [CrossRef]
- Tjandra, A.D.; Heywood, T.; Chandrawati, R. Trigit: A Free Web Application for Rapid Colorimetric Analysis of Images. Biosens. Bioelectron. X 2023, 14, 100361. [Google Scholar] [CrossRef]
- Lhotáková, Z.; Neuwirthová, E.; Potůčková, M.; Červená, L.; Hunt, L.; Kupková, L.; Lukeš, P.; Campbell, P.; Albrechtová, J. Mind the Leaf Anatomy While Taking Ground Truth with Portable Chlorophyll Meters. Sci. Rep. 2025, 15, 1855. [Google Scholar] [CrossRef]
- Dai, W.; Hamasaki, T. Statistics in Medicine, 4th ed.; Riffenburgh, R.H., Gillen, D.L., Eds.; Academic Press: Oxford, UK, 2020; Volume 76. [Google Scholar]
- Hagan, M.T.; Demuth, H.B.; Beale, M. Neural Network Design; PWS Publishing Co.: Boston, MA, USA, 1996. [Google Scholar]
- Koyama, K.; Smith, D.D. Scaling the Leaf Length-Times-Width Equation to Predict Total Leaf Area of Shoots. Ann. Bot. 2022, 130, 215–230. [Google Scholar] [CrossRef]
- Qian, T.; Zheng, X.; Guo, X.; Wen, W.; Yang, J.; Lu, S. Influence of Temperature and Light Gradient on Leaf Arrangement and Geometry in Cucumber Canopies: Structural Phenotyping Analysis and Modelling. Inf. Process. Agric. 2019, 6, 224–232. [Google Scholar] [CrossRef]
- Huang, Y.; Lechowicz, M.J.; Price, C.A.; Li, L.; Wang, Y.; Zhou, D. The Underlying Basis for the Trade-off between Leaf Size and Leafing Intensity. Funct. Ecol. 2016, 30, 199–205. [Google Scholar] [CrossRef]
- Schwarz, D.; Kläring, H.-P. Allometry to Estimate Leaf Area of Tomato. J. Plant Nutr. 2001, 24, 1291–1309. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, L. Leaf Shape Alters the Coefficients of Leaf Area Estimation Models for Saussurea stoliczkai in Central Tibet. Photosynthetica 2012, 50, 337–342. [Google Scholar] [CrossRef]
- Kumar, M.K.; Kumar, R.S.; Sankar, V.; Sakthivel, T.; Karunakaran, G.; Tripathi, P. Non-Destructive Estimation of Leaf Area of Durian (Durio zibethinus)—An Artificial Neural Network Approach. Sci. Hortic. 2017, 219, 319–325. [Google Scholar]
- Cemek, B.; Ünlükara, A.; Kurunç, A.; Küçüktopcu, E. Leaf Area Modeling of Bell Pepper (Capsicum annuum L.) Grown under Different Stress Conditions by Soft Computing Approaches. Comput. Electron. Agric. 2020, 174, 105514. [Google Scholar] [CrossRef]
- Li, G.; Yang, D.; Sun, S. Allometric Relationships between Lamina Area, Lamina Mass and Petiole Mass of 93 Temperate Woody Species Vary with Leaf Habit, Leaf Form and Altitude. Funct. Ecol. 2008, 22, 557–564. [Google Scholar] [CrossRef]
- Montero, F.J.; de Juan, J.A.; Cuesta, A.; Brasa, A. Nondestructive Methods to Estimate Leaf Area in Vitis vinifera L. HortScience 2000, 35, 696–698. [Google Scholar] [CrossRef]
- Fallovo, C.; Cristofori, V.; Mendoza de-Gyves, E.; Rivera, C.M.; Rea, R.; Fanasca, S.; Bignami, C.; Sassine, Y.; Rouphael, Y. Leaf Area Estimation Model for Small Fruits from Linear Measurements. HortScience 2008, 43, 2263–2267. [Google Scholar] [CrossRef]
- Sala, F.; Arsene, G.-G.; Iordănescu, O.; Boldea, M. Leaf Area Constant Model in Optimizing Foliar Area Measurement in Plants: A Case Study in Apple Tree. Sci. Hortic. 2015, 193, 218–224. [Google Scholar] [CrossRef]
- Jiang, N.; Yang, W.; Duan, L.; Chen, G.; Fang, W.; Xiong, L.; Liu, Q. A Nondestructive Method for Estimating the Total Green Leaf Area of Individual Rice Plants Using Multi-Angle Color Images. J. Innov. Opt. Health Sci. 2015, 8, 1550002. [Google Scholar] [CrossRef]
- Nagel, K.A.; Putz, A.; Gilmer, F.; Heinz, K.; Fischbach, A.; Pfeifer, J.; Faget, M.; Blossfeld, S.; Ernst, M.; Dimaki, C.; et al. Growscreen-Rhizo Is a Novel Phenotyping Robot Enabling Simultaneous Measurements of Root and Shoot Growth for Plants Grown in Soil-Filled Rhizotrons. Funct. Plant Biol. 2012, 39, 891. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Wang, X.; Sun, Y.; Ding, Y.; Lu, W. Measurement Method Based on Multispectral Three-Dimensional Imaging for the Chlorophyll Contents of Greenhouse Tomato Plants. Sensors 2019, 19, 3345. [Google Scholar] [CrossRef] [PubMed]
- Sunoj, S.; Igathinathane, C.; Saliendra, N.; Hendrickson, J.; Archer, D. Color Calibration of Digital Images for Agriculture and Other Applications. ISPRS J. Photogramm. Remote Sens. 2018, 146, 221–234. [Google Scholar] [CrossRef]
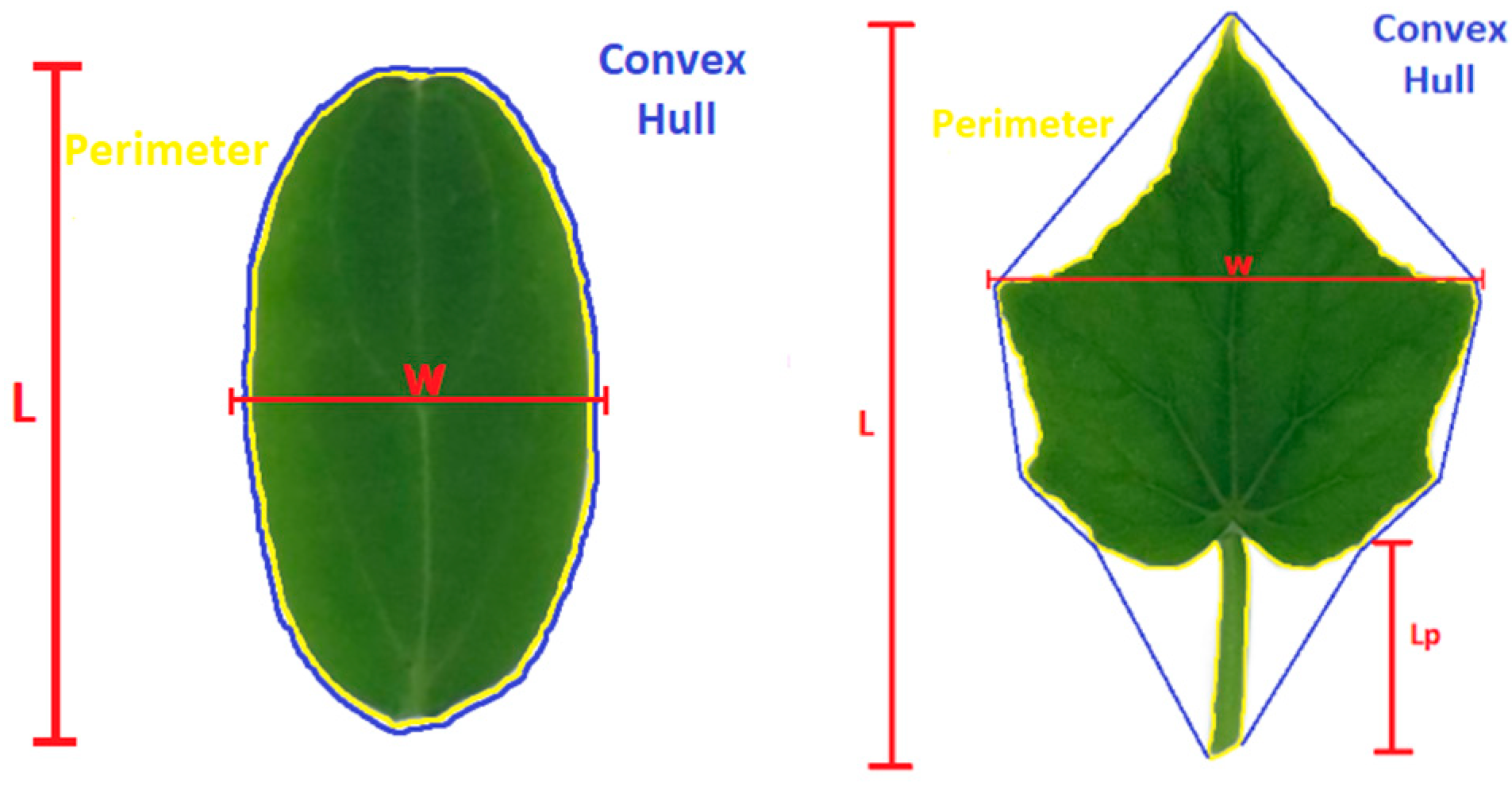


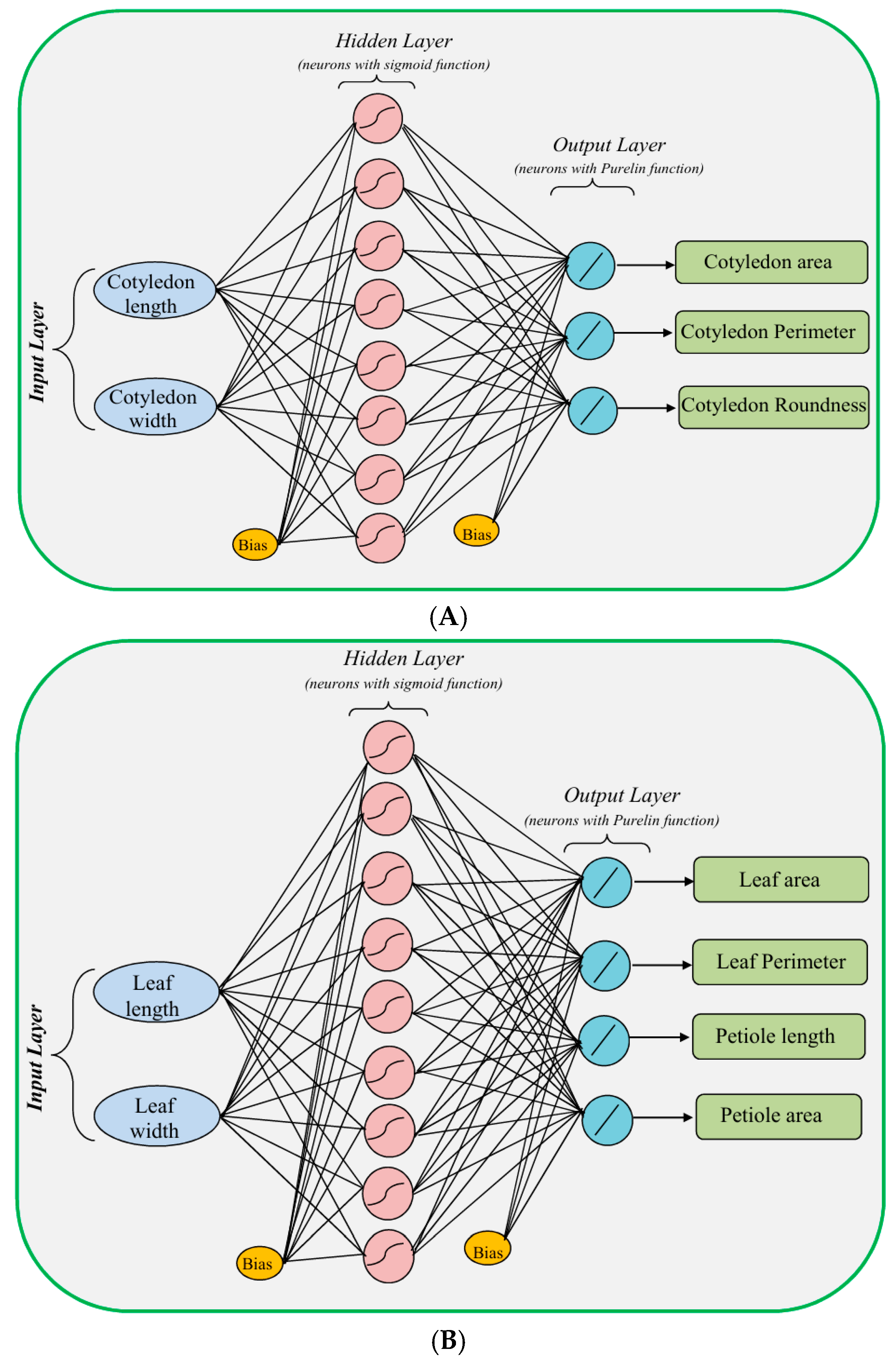
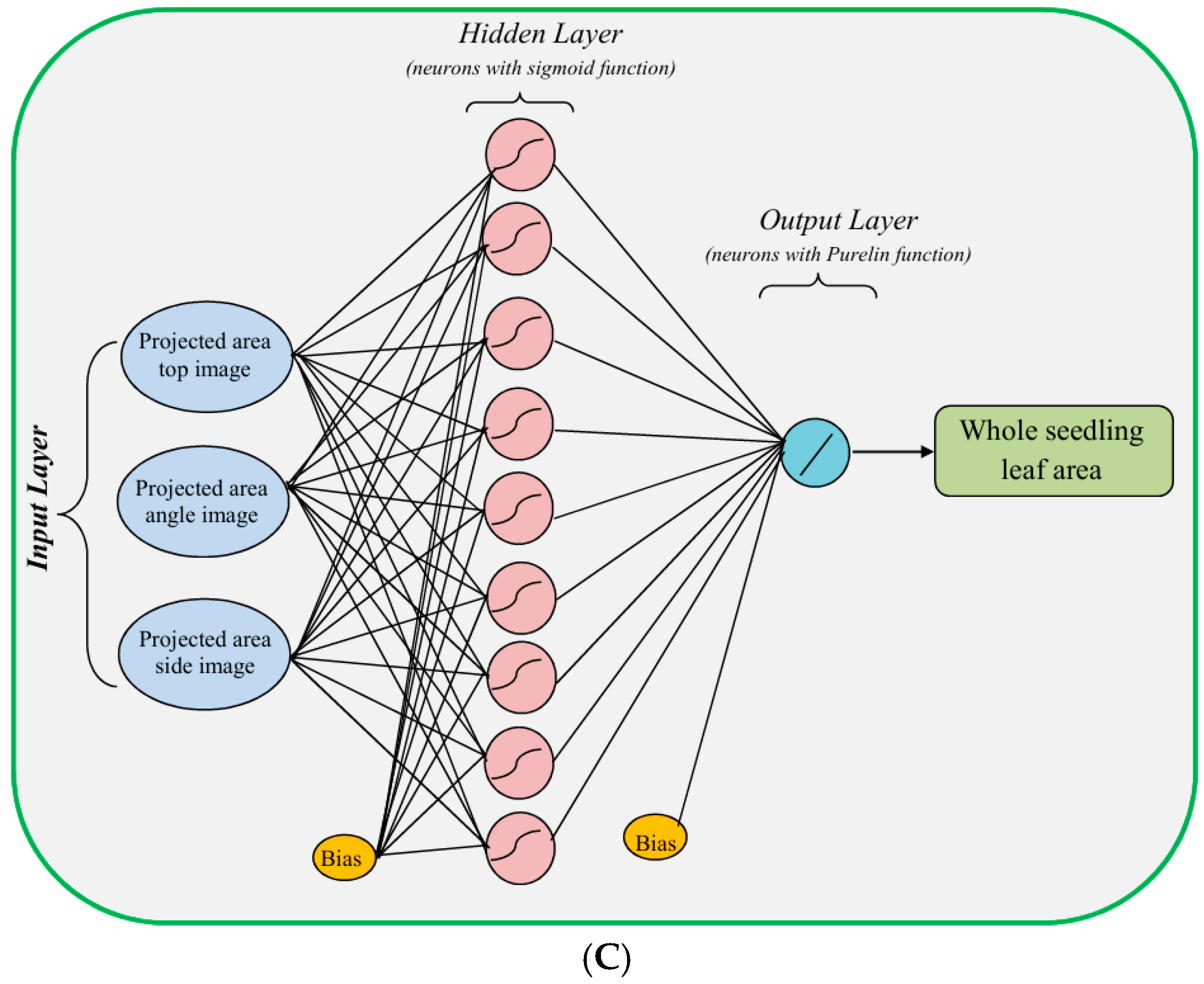

| Description | Level | Parameter | Formula | Range (unit) |
|---|---|---|---|---|
| Dimension | Cotyledon | Length (L) | From base to top along middle vein (major axis) | (cm) |
| Leaf | Petiole (stalk) L (Lp) | From petiole base to leaf base | ||
| Lamina (blade) L | From base to top along middle vein | |||
| Leaf L | Lp + lamina length (major axis) | |||
| Cotyledon/leaf | Width (W) | Widest point along the transverse axis perpendicular to the longitudinal one (minor axis) | ||
| Perimeter | Length of the path enclosing the outline along the margins (edges) | |||
| Area | Cotyledon | Cotyledon area | One-sided planar surface area | (cm2) |
| Leaf | Petiole area | |||
| Lamina area | ||||
| Leaf area (LA) | Petiole area + lamina area | |||
| Seedling 1 | Projected area (PA) | Area of green pixels obtained by seedling image | ||
| Shape | Cotyledon/leaf | Aspect ratio | 1–∞ (−) | |
| Circularity | 0–1 (−) | |||
| Roundness | 0–1 (−) | |||
| Solidity | 0–1 (−) | |||
| Leaf | Lp to leaf L ratio | 0≤ <100 (%) | ||
| Petiole area to LA ratio | ||||
| Color | Leaf/seedling 2 | Red (R) 4 | amount of red | 0–255 (−) |
| Green (G) 4 | amount of green | |||
| Blue (B) 4 | amount of blue | |||
| %R | 0≤ ≤100 (%) | |||
| %G | ||||
| %B | ||||
| Cyan (C) 5 | 0–100 (%) | |||
| Magenta (M) 5 | ||||
| Yellow (Y) 5 | ||||
| Black (K) 5 | ||||
| Lightness (L*) 6 | 0 specifies black, 100 specifies white | |||
| Red/green (a*) 6 | Amount of red or green tones | −110–110 (−) | ||
| Yellow/blue (b*) 6 | Amount of yellow or blue tones | |||
| Hue (H) 7 | Location on the color wheel | 0–360 (°) | ||
| Saturation (S) 7 | Vividness or dullness | 0–255 (−) | ||
| Value (V) 7 | Amount of white | |||
| Content | Leaf/seedling 3 | SPAD value | Non-invasive optical measurement of total chlorophyll content | 0–100 (−) |
| Level | Model | b (Fitted Coefficient) | a (Constant) | R2 | MSE | BC Lower | BC Upper | |
|---|---|---|---|---|---|---|---|---|
| Cotyledon | 1 | Ac = a + b · L | 2.99 | −5.32 | 0.833 | 0.34 | 0.809 | 0.858 |
| 2 | Ac = a + b · L2 | 0.29 | 2.14 | 0.840 | 0.32 | 0.814 | 0.869 | |
| 3 | Ac = a + b · L3 | 0.04 | 4.68 | 0.846 | 0.32 | 0.813 | 0.868 | |
| 4 | Ac = a + b · W | 5.78 | −4.72 | 0.720 | 0.58 | 0.676 | 0.755 | |
| 5 | Ac = a + b · W2 | 1.14 | 2.52 | 0.727 | 0.56 | 0.680 | 0.764 | |
| 6 | Ac = a + b · W3 | 0.30 | 4.99 | 0.728 | 0.56 | 0.685 | 0.769 | |
| 7 | Ac = a + b · L · W | 0.68 | 1.13 | 0.916 | 0.17 | 0.899 | 0.929 | |
| 8 | Ac = a + b (L + W) | 2.27 | −7.35 | 0.913 | 0.18 | 0.898 | 0.927 | |
| 9 | Ac = a + b (L + W)2 | 0.15 | 1.20 | 0.920 | 0.16 | 0.904 | 0.934 | |
| 10 | Ac = a (L + W)3 | 0.01 | 4.11 | 0.920 | 0.17 | 0.905 | 0.930 | |
| Leaf | 1 | LA = a + b · L | 4.23 | −11.87 | 0.936 | 5.68 | 0.930 | 0.940 |
| 2 | LA = a + b · L2 | 0.30 | 1.48 | 0.939 | 5.44 | 0.933 | 0.945 | |
| 3 | LA = a + b · L3 | 0.03 | 6.42 | 0.890 | 9.46 | 0.887 | 0.903 | |
| 4 | LA = a + b · Llamina | 6.67 | −14.09 | 0.925 | 6.67 | 0.918 | 0.933 | |
| 5 | LA = a + b · Llamina2 | 0.69 | 0.58 | 0.928 | 6.46 | 0.921 | 0.935 | |
| 6 | LA = a + b · Llamina3 | 0.09 | 6.05 | 0.880 | 11.00 | 0.873 | 0.890 | |
| 7 | LA = a + b · W | 6.44 | −14.11 | 0.906 | 8.42 | 0.899 | 0.913 | |
| 8 | LA = a + b · W2 | −0.66 | 0.70 | 0.937 | 5.59 | 0.932 | 0.943 | |
| 9 | LA = a + b · W3 | 0.09 | 4.33 | 0.920 | 6.80 | 0.916 | 0.931 | |
| 10 | LA = a + b · L · W | 0.48 | −0.12 | 0.973 | 2.37 | 0.971 | 0.976 | |
| 11 | LA = a + b (L + W) | 2.63 | −13.62 | 0.951 | 4.43 | 0.948 | 0.954 | |
| 12 | LA = a + b (L + W)2 | 0.11 | 0.08 | 0.972 | 2.52 | 0.969 | 0.975 | |
| 13 | LA = a (L + W)3 | 0.006 | 5.12 | 0.947 | 4.74 | 0.942 | 0.951 | |
| 14 | LA = a + b · Llamina · W | 0.73 | −0.79 | 0.976 | 2.15 | 0.973 | 0.979 | |
| 15 | LA = a + b (Llamina + W) | 3.40 | −15.29 | 0.949 | 4.48 | 0.946 | 0.954 | |
| 16 | LA = a + b (Llamina + W)2 | 0.30 | 1.48 | 0.939 | 2.33 | 0.934 | 0.945 | |
| 17 | LA = a (Llamina + W)3 | 0.012 | 4.55 | 0.954 | 4.11 | 0.949 | 0.959 | |
| Whole-seedling | 1 | LA = a + b · PAtop | 0.76 | 8.68 | 0.880 | 70 | 0.860 | 0.895 |
| 2 | LA = a + b · PAside | 1.66 | 15.8 | 0.730 | 157 | 0.698 | 0.765 | |
| 3 | LA = a + b · PAangle | 1.05 | 8.49 | 0.817 | 108 | 0.789 | 0.839 | |
| 4 | LA = a + b · (PAtop + PAside) | 0.58 | 6.34 | 0.920 | 46 | 0.908 | 0.933 | |
| 5 | LA = a + b · (PAtop + PAangle) | 0.46 | 6.96 | 0.880 | 68 | 0.864 | 0.899 | |
| 6 | LA = a + b · (PAangle + PAside) | 0.73 | 5.44 | 0.890 | 64 | 0.871 | 0.904 | |
| 7 | LA = a + b · (PAtop + PAside + PAangle) | 0.39 | 5.27 | 0.918 | 48 | 0.906 | 0.929 | |
| Main Output Trait | Area | SPAD Value | |||
|---|---|---|---|---|---|
| Level | Cotyledon | Leaf | Seedling | Cotyledon/Leaf | Seedling |
| ANN Feature | |||||
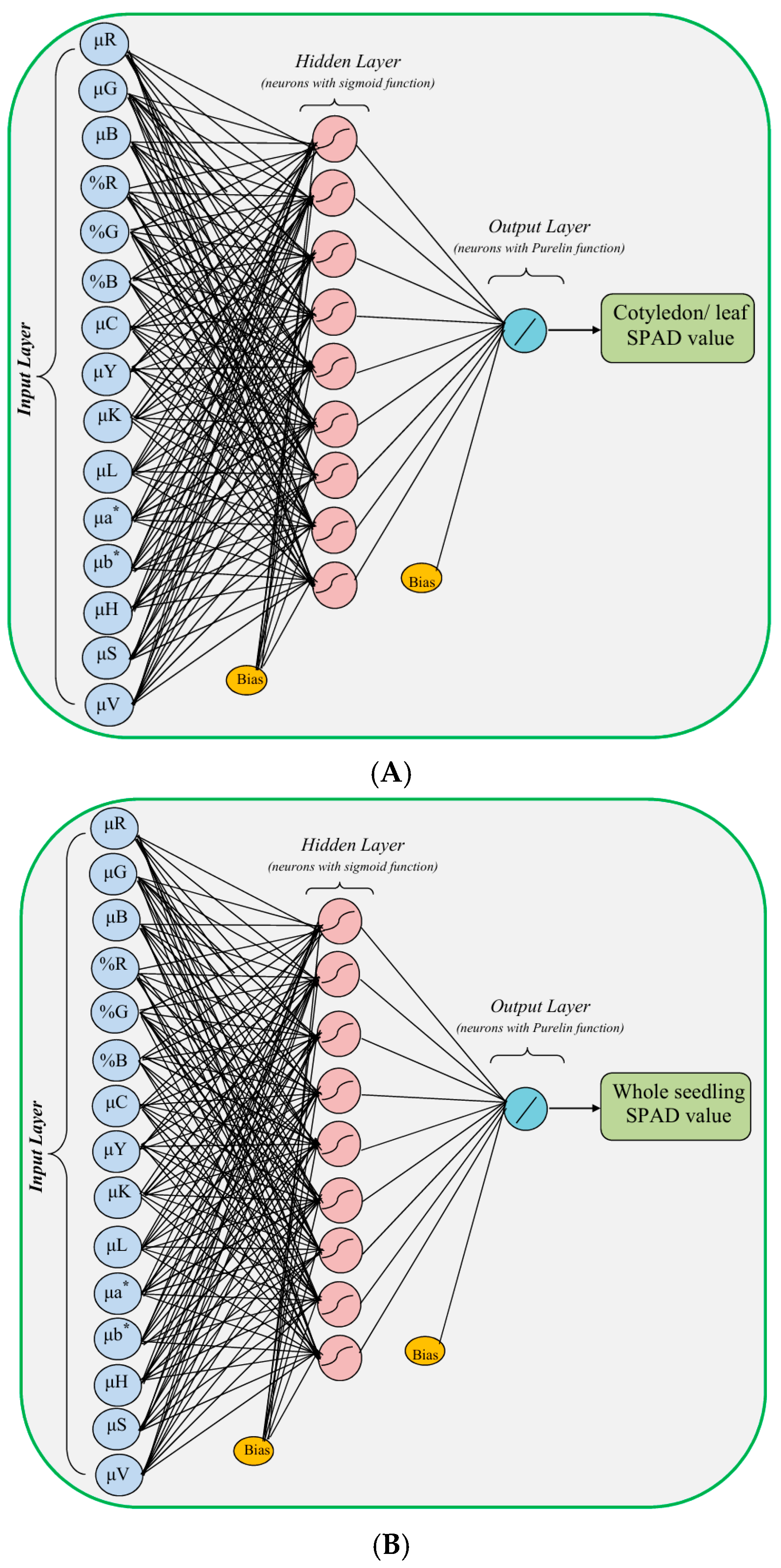
| Number of input layer units | 2 | 2 | 3 | 15 | 15 |
| Number of hidden layers | 1 | 1 | 1 | 1 | 1 |
| Number of hidden layer units | 3–10 | 4–12 | 3–10 | 3–10 | 3–10 |
| Number of output layer units | 3 | 4 | 1 | 1 | 1 |
| Transfer function in hidden layer | Sigmoid | Sigmoid | Sigmoid | Sigmoid | Sigmoid |
| Transfer function in output layer | Purelin | Purelin | Purelin | Purelin | Purelin |
| Training function | Levenberg–Marquardt | Levenberg–Marquardt | Levenberg–Marquardt | Levenberg–Marquardt | Levenberg–Marquardt |
| Learning rate | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| Performance goal | 0 | 0 | 0 | 0 | 0 |
| Maximum number of epochs | 100 | 100 | 100 | 100 | 100 |
| Level | Trait | Dataset | Statistical Index | |||||
|---|---|---|---|---|---|---|---|---|
| R | R2 | MSE | RMSE | MAE | SEP | |||
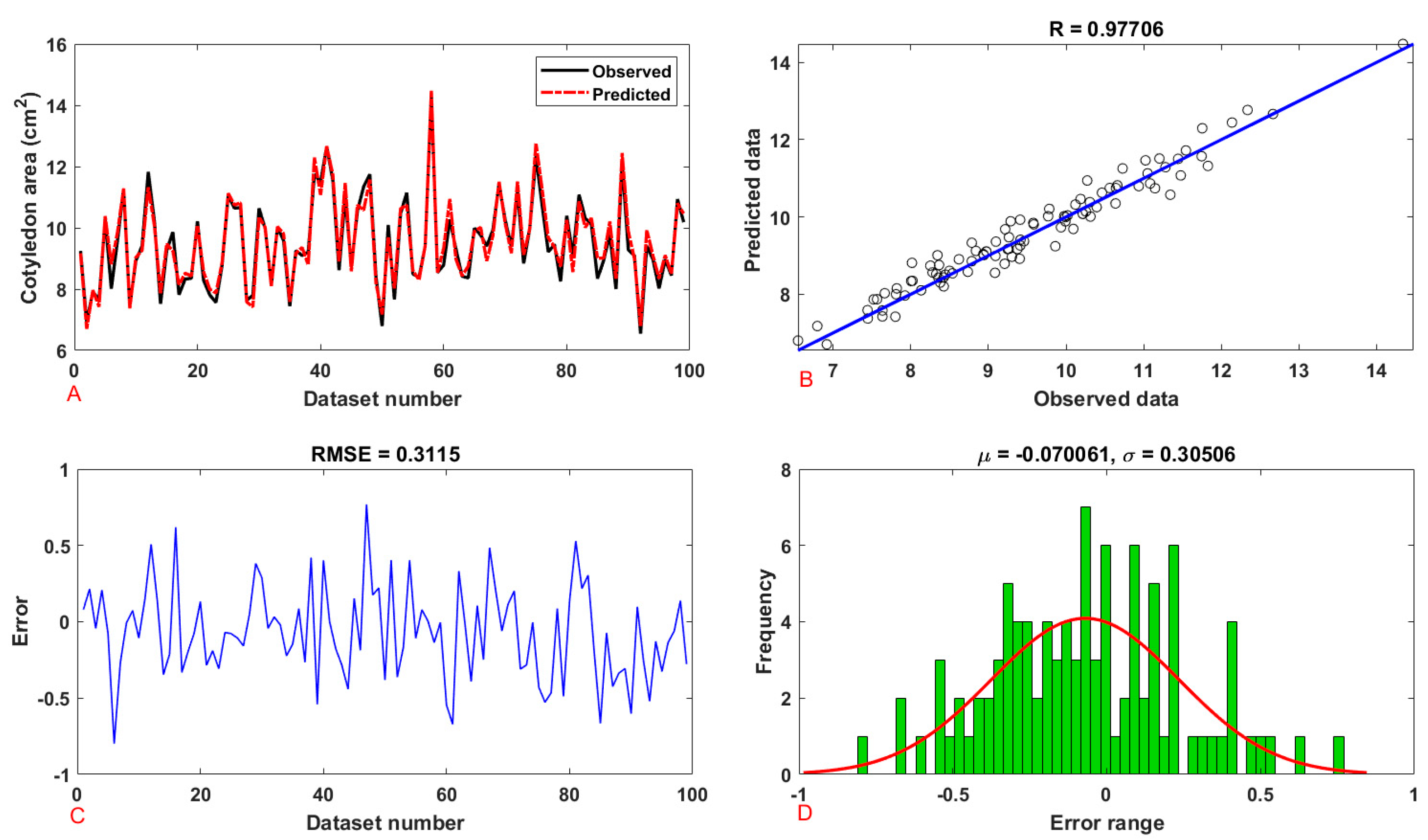
| Cotyledon | Area | training | 0.9650 | 0.9311 | 0.1515 | 0.3892 | 0.3124 | 0.3889 |
| testing | 0.9771 | 0.9546 | 0.097 | 0.3115 | 0.2752 | 0.3487 | ||
| Perimeter | training | 0.9408 | 0.8851 | 0.1246 | 0.3529 | 0.2684 | 0.3529 | |
| testing | 0.9558 | 0.9136 | 0.0988 | 0.3144 | 0.2538 | 0.3144 | ||
| Roundness | training | 0.8518 | 0.7256 | 0.0002 | 0.0154 | 0.0120 | 0.0154 | |
| testing | 0.8785 | 0.7718 | 0.0002 | 0.0145 | 0.0115 | 0.0145 | ||
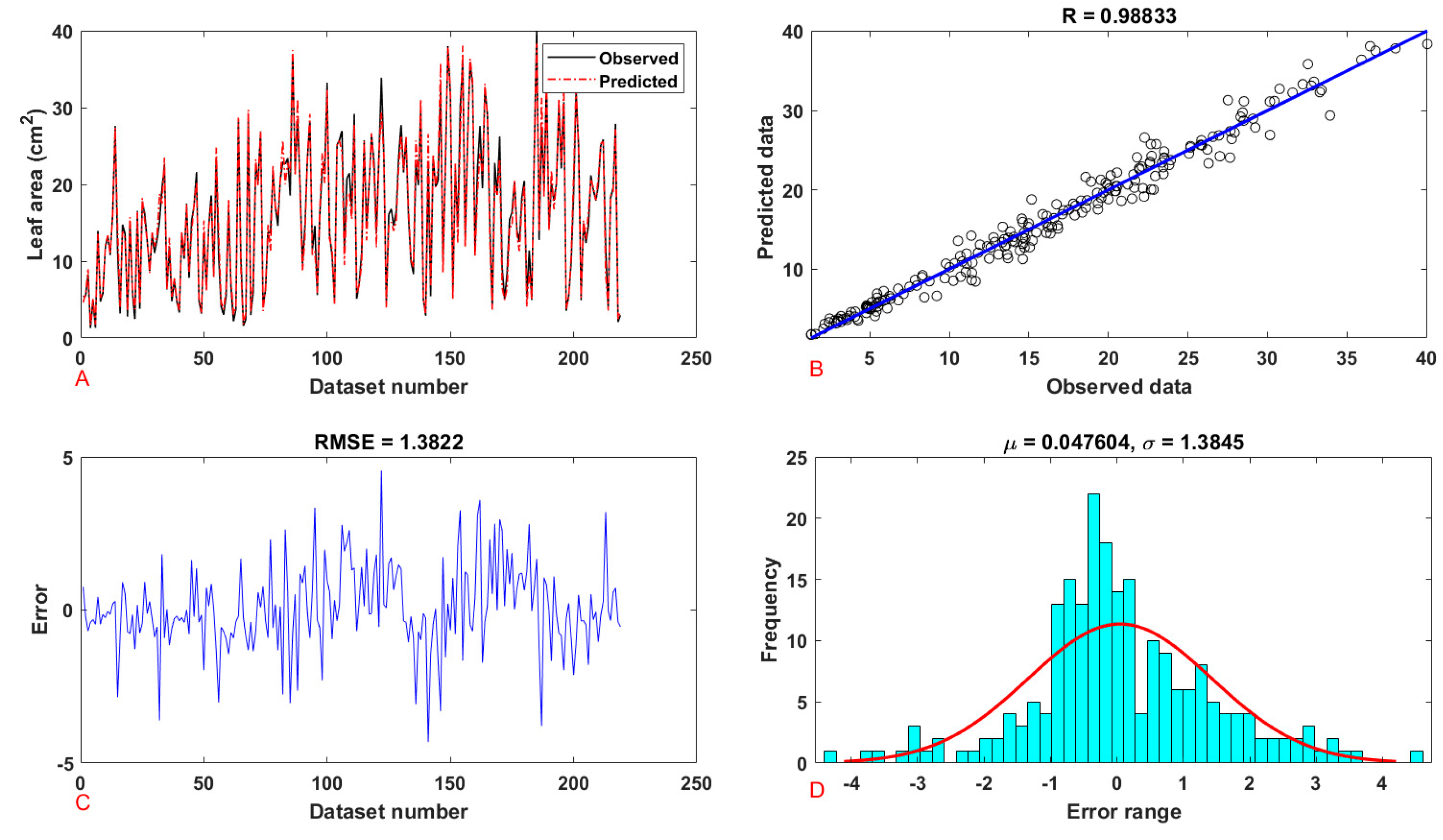
| Leaf | Area | training | 0.9867 | 0.9735 | 2.4177 | 1.5549 | 1.1520 | 1.5549 |
| testing | 0.9883 | 0.9768 | 1.9105 | 1.3822 | 1.0187 | 1.3822 | ||
| Perimeter | training | 0.9385 | 0.8808 | 4.9150 | 2.2170 | 1.7824 | 2.2170 | |
| testing | 0.9440 | 0.8912 | 4.4359 | 2.1062 | 1.6669 | 2.1062 | ||
| Petiole length | training | 0.9381 | 0.8801 | 0.0939 | 0.3065 | 0.2363 | 0.3065 | |
| testing | 0.9542 | 0.9105 | 0.0774 | 0.2781 | 0.2122 | 0.2781 | ||
| Petiole area | training | 0.9184 | 0.8435 | 0.0204 | 0.1430 | 0.1091 | 0.1430 | |
| testing | 0.9325 | 0.8696 | 0.0159 | 0.1262 | 0.0957 | 0.1262 | ||
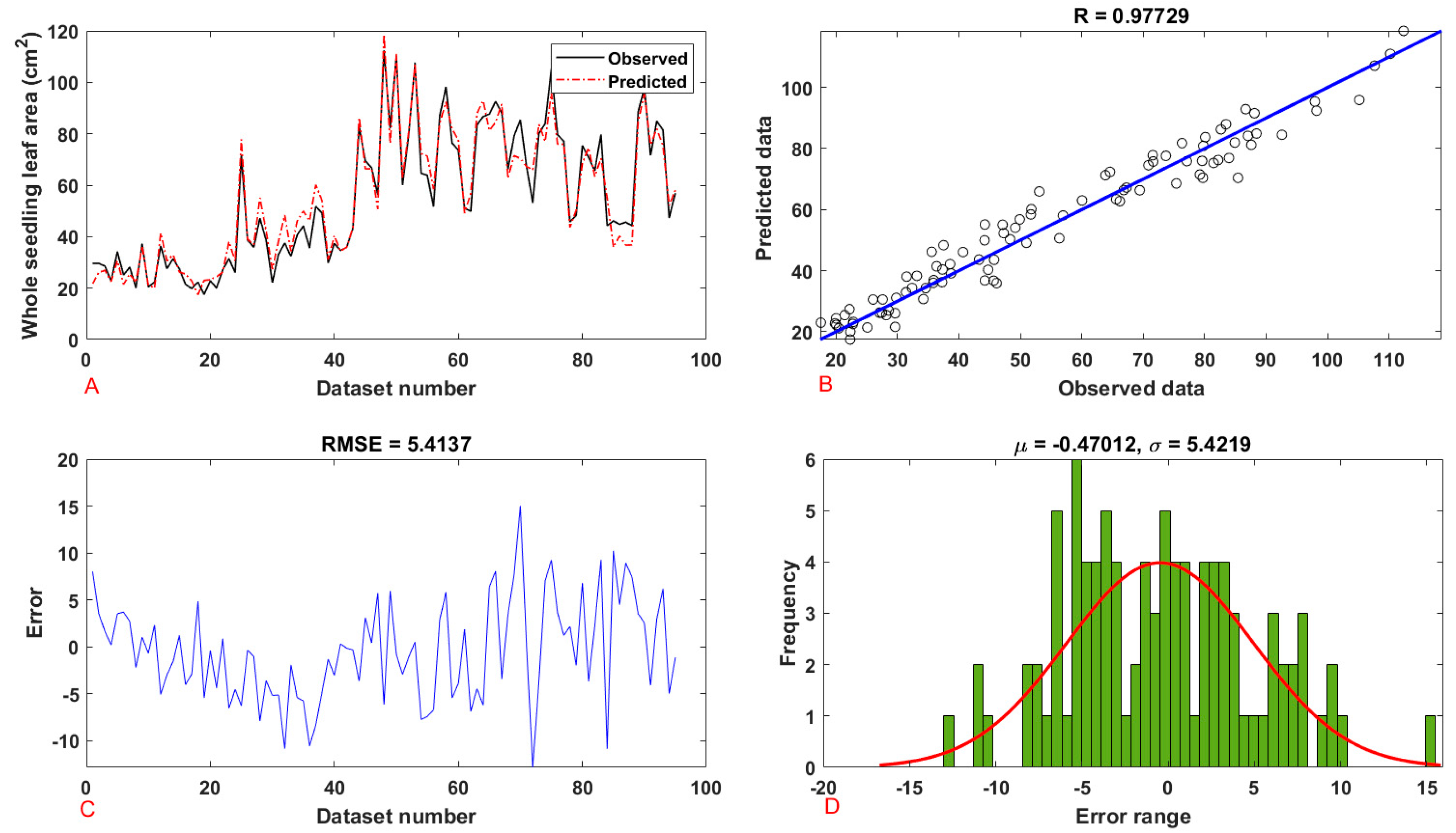
| Seedling | Leaf area | training | 0.9708 | 0.9424 | 33.8354 | 5.8168 | 4.3459 | 5.8168 |
| testing | 0.9773 | 0.9551 | 29.3080 | 5.4137 | 4.4429 | 5.4137 | ||
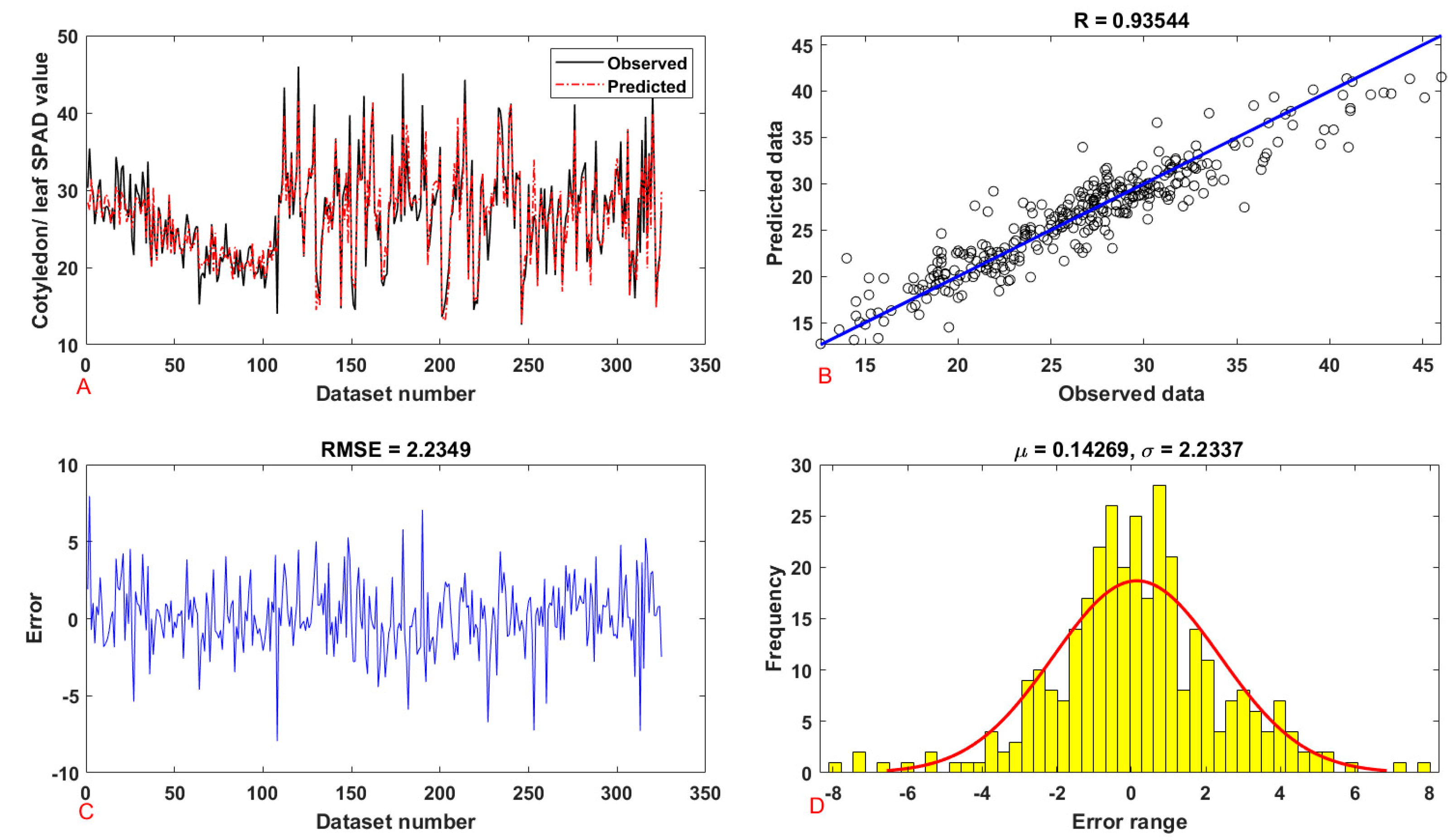
| Cotyledon/leaf | SPAD value | training | 0.9210 | 0.8482 | 5.0770 | 2.2532 | 1.7103 | 2.2532 |
| testing | 0.9354 | 0.8751 | 4.9946 | 2.2349 | 1.6678 | 2.2349 | ||
| Seedling | training | 0.6714 | 0.4507 | 9.9203 | 3.1497 | 2.6194 | 3.1497 | |
| testing | 0.7726 | 0.5969 | 8.6380 | 2.9390 | 2.4156 | 2.9390 | ||
| Level | Model | b (Fitted Coefficient) | a (Constant) | R2 | MSE | BC Lower | BC Upper | |
|---|---|---|---|---|---|---|---|---|
| Cotyledon/Leaf | 1 | SPAD value = a + b · μR | −0.25 | 42.09 | 0.605 | 13.90 | 0.578 | 0.627 |
| 2 | SPAD value = a + b · μG | −0.24 | 49.74 | 0.693 | 10.80 | 0.673 | 0.715 | |
| 3 | SPAD value = a + b · μB | −0.20 | 33.43 | 0.030 | 34.13 | 0.015 | 0.049 | |
| 4 | SPAD value = a + b · %R | −177.89 | 82.33 | 0.581 | 14.80 | 0.556 | 0.605 | |
| 5 | SPAD value = a + b · %G | −93.98 | 72.99 | 0.125 | 30.79 | 0.096 | 0.154 | |
| 6 | SPAD value = a + b · %B | 159.80 | −3.70 | 0.736 | 9.30 | 0.719 | 0.755 | |
| 7 | SPAD value = a + b · μC | 0.48 | 8.66 | 0.241 | 26.70 | 0.211 | 0.278 | |
| 8 | SPAD value = a + b · μY | −0.63 | 65.43 | 0.662 | 11.90 | 0.636 | 0.683 | |
| 9 | SPAD value = a + b · μK | 0.612 | −11.51 | 0.691 | 10.90 | 0.661 | 0.716 | |
| 10 | SPAD value = a + b · μL | −0.61 | 49.20 | 0.688 | 11.00 | 0.668 | 0.713 | |
| 11 | SPAD value = a + b · μa* | 1.27 | 53.82 | 0.488 | 18.00 | 0.455 | 0.533 | |
| 12 | SPAD value = a + b · μb* | −0.63 | 45.03 | 0.771 | 8.1 | 0.755 | 0.786 | |
| 13 | SPAD value = a + b · μH | 0.61 | −32.08 | 0.703 | 10.4 | 0.678 | 0.726 | |
| 14 | SPAD value = a + b · μS | −0.63 | 65.47 | 0.662 | 11.9 | 0.641 | 0.686 | |
| 15 | SPAD value = a + b · μV | −0.61 | 49.76 | 0.693 | 10.8 | 0.674 | 0.715 | |
| Whole seedling | 1 | SPAD value = a + b · μR | −0.21 | 41.85 | 0.439 | 10.19 | 0.407 | 0.482 |
| 2 | SPAD value = a + b · μG | −0.16 | 47.82 | 0.417 | 10.60 | 0.373 | 0.469 | |
| 3 | SPAD value = a + b · μB | −0.03 | 27.21 | 0.001 | 18.16 | 0.001 | 0.001 | |
| 4 | SPAD value = a + b · %R | −161.19 | 75.17 | 0.430 | 10.36 | 0.377 | 0.4720 | |
| 5 | SPAD value = a + b · %G | −7.55 | 30.24 | 0.001 | 18.17 | 0.001 | 0.001 | |
| 6 | SPAD value = a + b · %B | 97.21 | 12.46 | 0.267 | 13.31 | 0.227 | 0.313 | |
| 7 | SPAD value = a + b · μC | 0.79 | −9.70 | 0.373 | 11.38 | 0.331 | 0.434 | |
| 8 | SPAD value = a + b · μY | −0.44 | 59.03 | 0.223 | 14.11 | 0.178 | 0.269 | |
| 9 | SPAD value = a + b · μK | 0.40 | 7.66 | 0.415 | 10.63 | 0.374 | 0.461 | |
| 10 | SPAD value = a + b · μL | −0.43 | 48.44 | 0.419 | 10.56 | 0.363 | 0.460 | |
| 11 | SPAD value = a + b · μa* | 0.81 | 54.02 | 0.276 | 13.17 | 0.228 | 0.320 | |
| 12 | SPAD value = a + b · μb* | −0.41 | 44.62 | 0.401 | 10.89 | 0.364 | 0.453 | |
| 13 | SPAD value = a + b · μH | 0.65 | −36.30 | 0.418 | 10.58 | 0.369 | 0.455 | |
| 14 | SPAD value = a + b · μS | −0.44 | 59.25 | 0.225 | 14.10 | 0.177 | 0.264 | |
| 15 | SPAD value = a + b · μV | −0.001 | 27.46 | 0.017 | 17.87 | 0.002 | 0.039 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsaniklidis, G.; Makraki, T.; Papadimitriou, D.; Nikoloudakis, N.; Taheri-Garavand, A.; Fanourakis, D. Non-Destructive Estimation of Area and Greenness in Leaf and Seedling Scales: A Case Study in Cucumber. Agronomy 2025, 15, 2294. https://doi.org/10.3390/agronomy15102294
Tsaniklidis G, Makraki T, Papadimitriou D, Nikoloudakis N, Taheri-Garavand A, Fanourakis D. Non-Destructive Estimation of Area and Greenness in Leaf and Seedling Scales: A Case Study in Cucumber. Agronomy. 2025; 15(10):2294. https://doi.org/10.3390/agronomy15102294
Chicago/Turabian StyleTsaniklidis, Georgios, Theodora Makraki, Dimitrios Papadimitriou, Nikolaos Nikoloudakis, Amin Taheri-Garavand, and Dimitrios Fanourakis. 2025. "Non-Destructive Estimation of Area and Greenness in Leaf and Seedling Scales: A Case Study in Cucumber" Agronomy 15, no. 10: 2294. https://doi.org/10.3390/agronomy15102294
APA StyleTsaniklidis, G., Makraki, T., Papadimitriou, D., Nikoloudakis, N., Taheri-Garavand, A., & Fanourakis, D. (2025). Non-Destructive Estimation of Area and Greenness in Leaf and Seedling Scales: A Case Study in Cucumber. Agronomy, 15(10), 2294. https://doi.org/10.3390/agronomy15102294









