Genome-Wide Identification of MADS-box Family Genes and Analysis of Their Expression Patterns in the Common Oat (Avena sativa L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth and Treatments
2.2. Retrieval of Genome Sequences
2.3. Identification of AsMADS-box Genes in the Common Oat and Analysis of Their Chromosomal Location
2.4. Phylogenetic Analysis of MADS-box Proteins
2.5. Analysis of the Physicochemical Properties, Gene Structure, and Conserved Motif and Domains of the MADS-box TFs
2.6. Prediction of Gene Duplication and Cis-Acting Regulatory Elements
2.7. Prediction of Putative microRNAs Targeting AsMADS-box Genes
2.8. Analysis of AsMADS-box Expression Profiles Based on Transcriptome Sequencing
2.9. Overexpression Plasmid Construction
2.10. Validation of AsMADS-box Gene Expression by qRT-PCR
3. Results
3.1. Identification and Chromosomal Location of AsMADS-box Genes
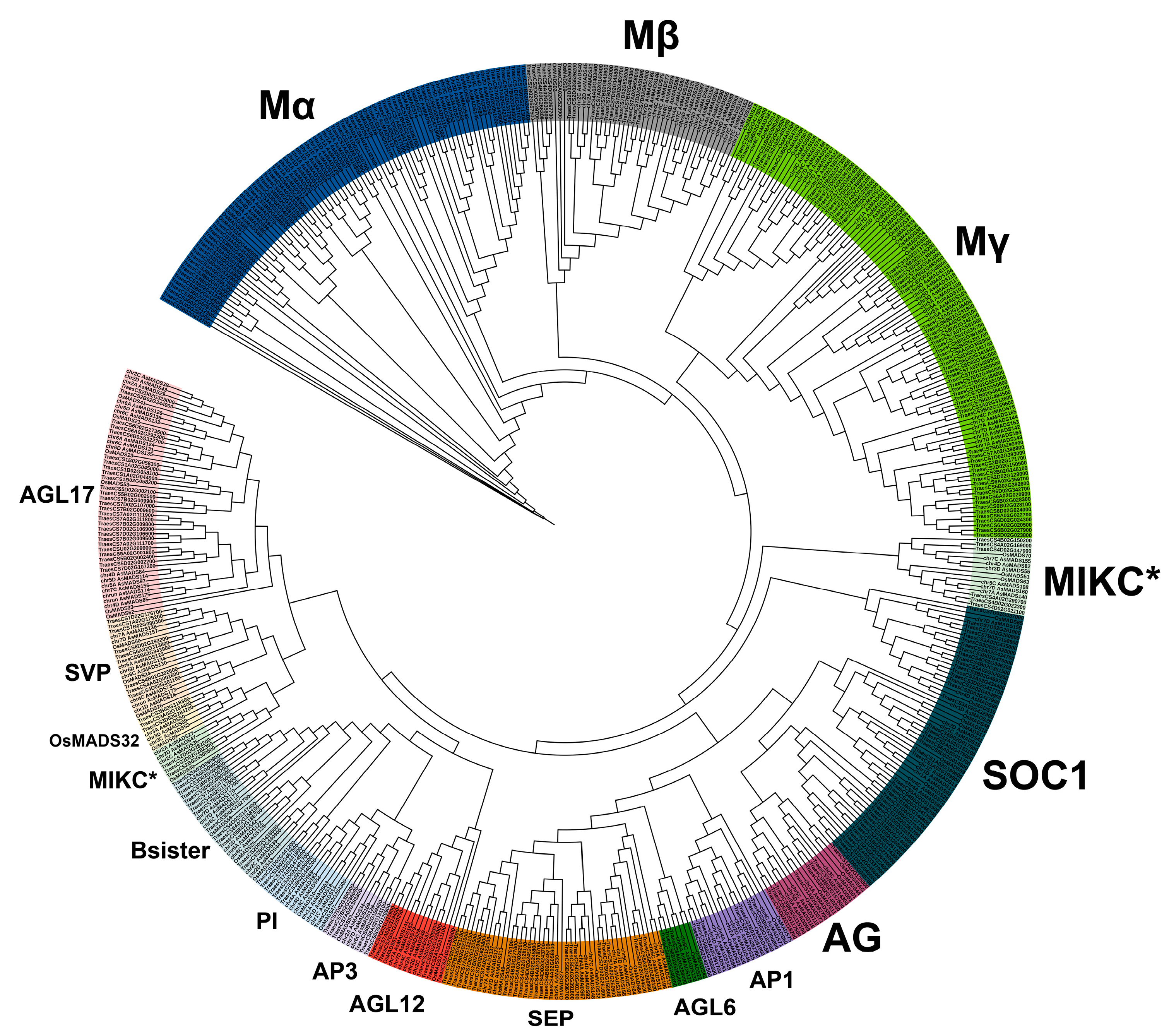
3.2. Phylogenetic Analysis of AsMADS-box Genes
3.3. Gene Structure Analysis, Conserved Motif and Domain Identification
3.4. Cis-Regulatory Element Distribution of AsMADS-box Genes
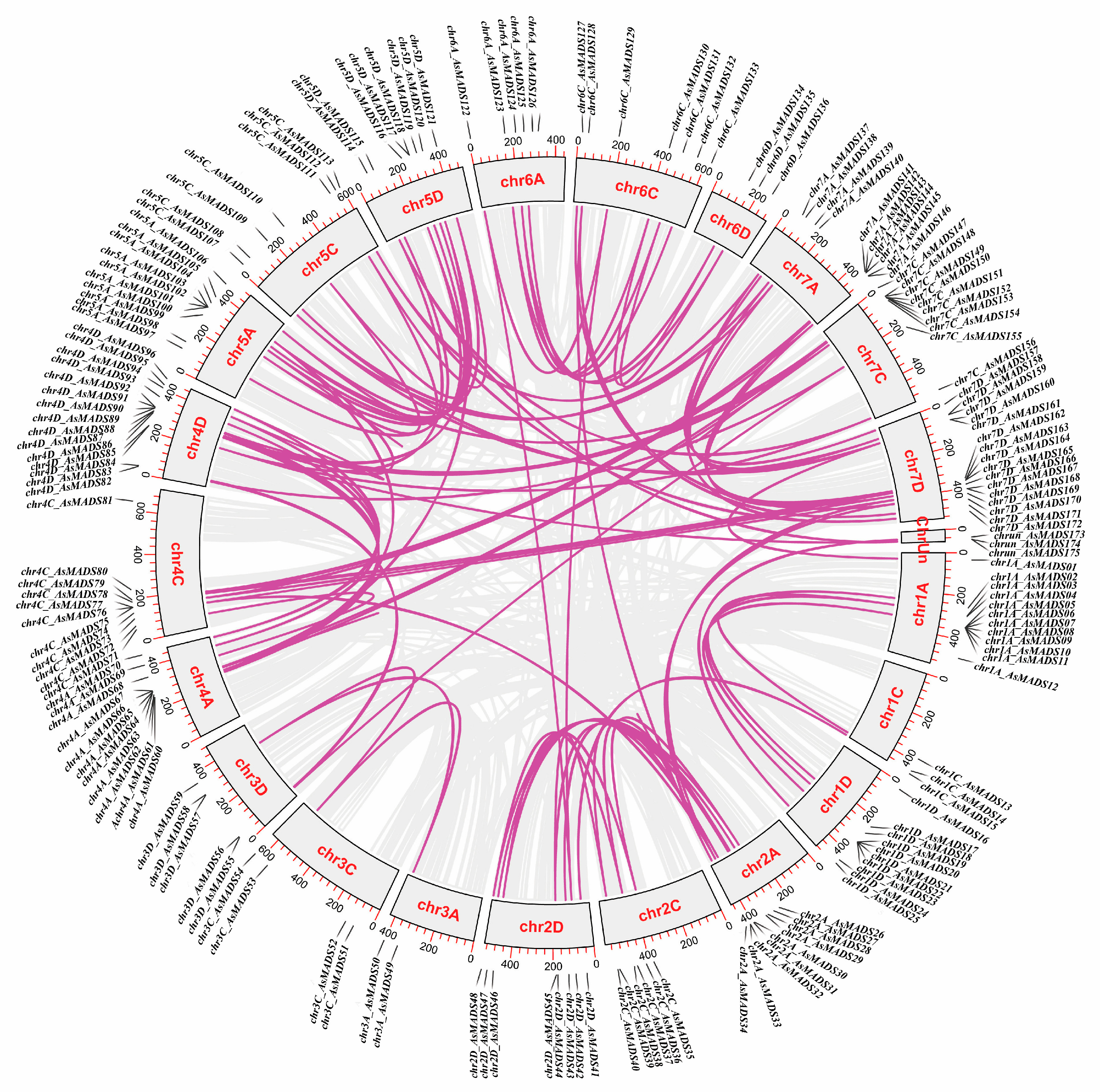
3.5. Gene Duplication Analysis and Orthologous Identification Between the Common Oat and the Other Four Species
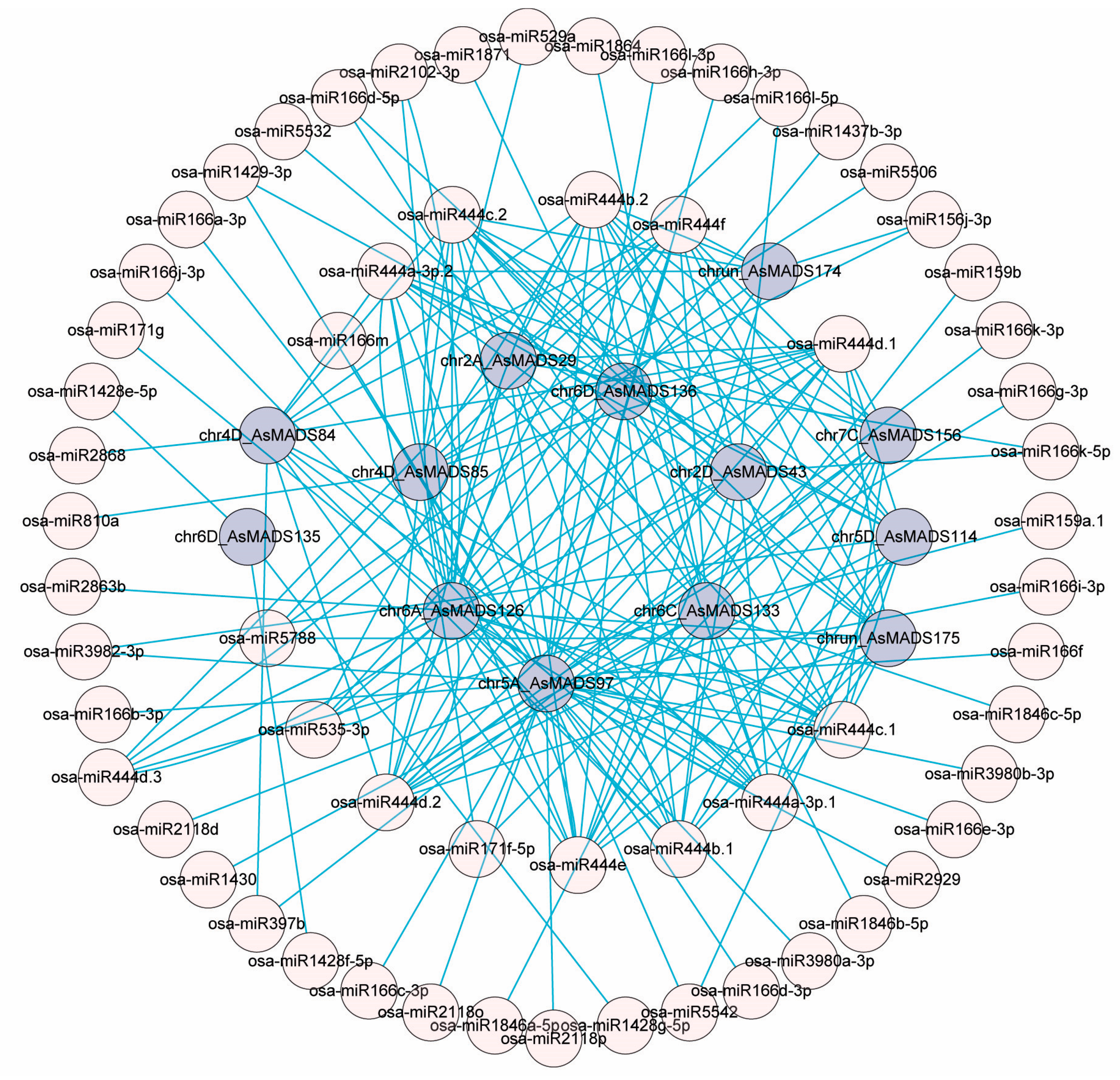
3.6. MiRNA Prediction of AsMADS-box Genes from the AGL17 Subclade
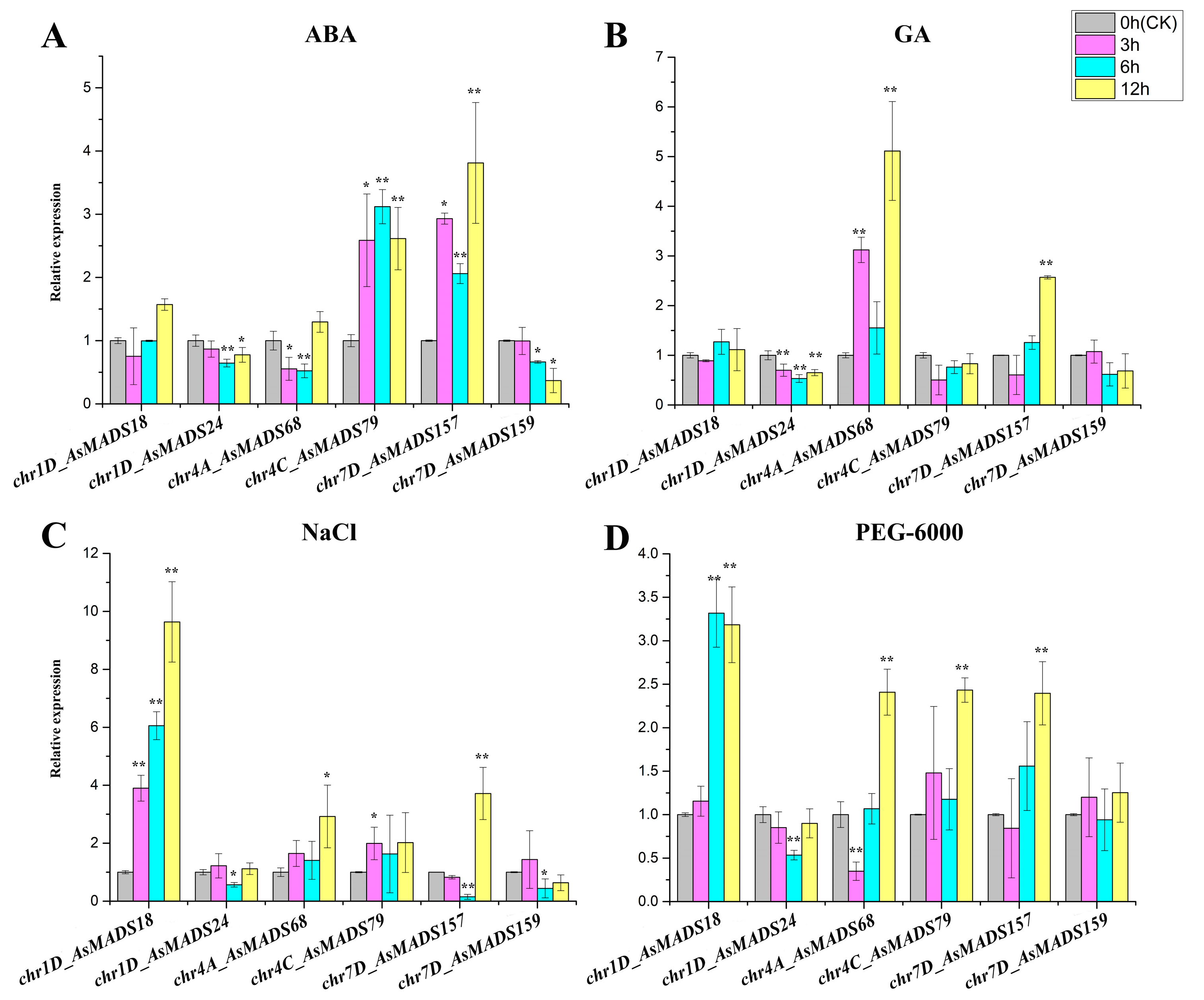
3.7. Analysis of AsMADS-box Gene Expression Patterns Under Hormone and Abiotic Stress Conditions
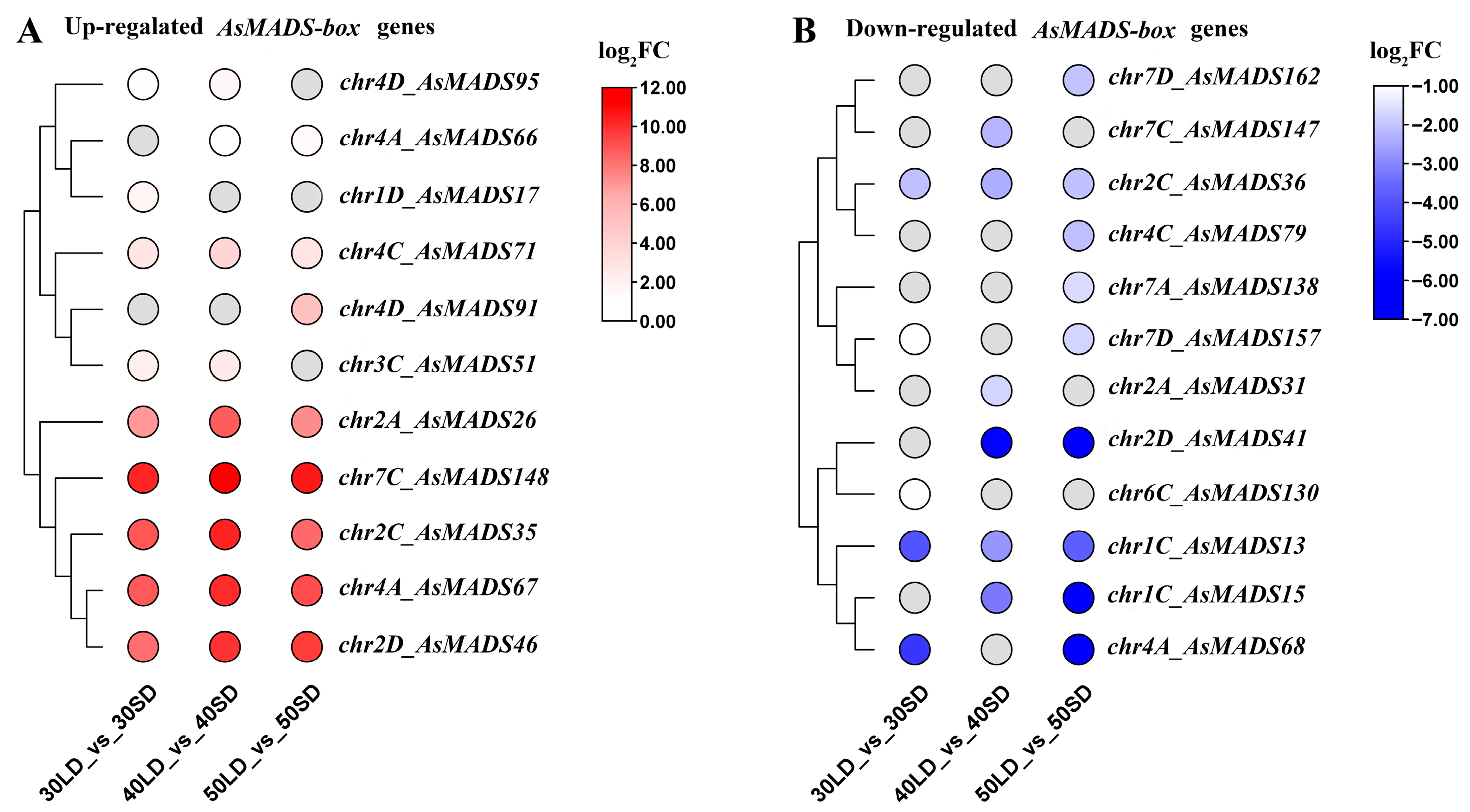
3.8. Expression Analysis of AsMADS-box Genes Under in Long-Day and Short-Day Conditions
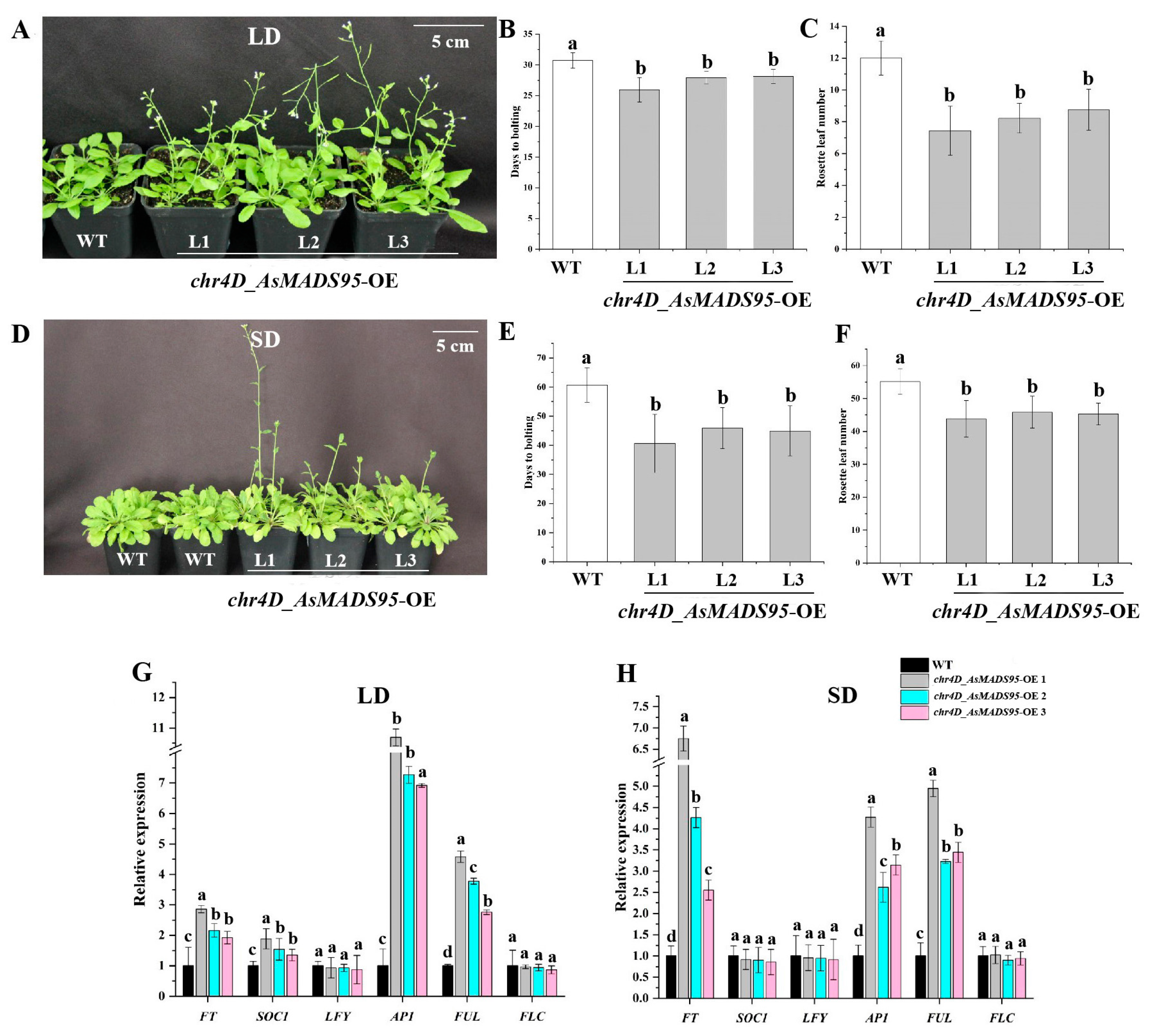
3.9. chr4D_AsMADS95 Promotes Flowering Under LD and SD Photoperiod Conditions
4. Discussion
4.1. The AsMADS-box Gene Family Has Undergone Significant Expansion in the Common Oat
4.2. AsMADS-box Genes May Be Involved in Hormone Signalling and Abiotic Stress Responses
4.3. AsMADS-box Genes Can Participate in Responses Associated with Flowering in the Common Oat Under Long-Day and Short-Day Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yu, H.; Xia, L.; Zhu, J.; Xie, X.; Wei, Y.; Li, X.; He, X.; Luo, C. Genome-wide analysis of the MADS-box gene family in mango and ectopic expression of MiMADS77 in Arabidopsis results in early flowering. Gene 2025, 935, 149054. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Niels, A.; Suzanne, D.B.; Hilda, V.M.; Angenent, G.C.; Van, D.A.D.J. Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 131. [Google Scholar] [CrossRef] [PubMed]
- Käppel, S.; Eggeling, R.; Rümpler, F.; Groth, M.; Melzer, R.; Theißen, G. DNA-binding properties of the MADS-domain transcription factor SEPALLATA3 and mutant variants characterized by SELEX-seq. Plant Mol. Biol. 2021, 105, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani Marghashi, M.; Bagheri, H.; Gholami, M. Genome-wide study of flowering-related MADS-box genes family in Cardamine hirsuta. 3 Biotech 2020, 10, 518. [Google Scholar] [CrossRef]
- Duan, K.; Fu, H.; Fang, D.; Wang, K.; Zhang, W.; Liu, H.; Sahu, S.K.; Chen, X. Genome-wide analysis of the MADS-box gene family in Holoparasitic Plants (Balanophora subcupularis and Balanophora fungosa var. globosa). Front. Plant Sci. 2022, 13, 846697. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Paul, P.; Joshi, S.; Tian, R.; Diogo Junior, R.; Chakrabarti, M.; Perry, S.E. The MADS-domain factor AGAMOUS-Like 18 promotes somatic embryogenesis. Plant Physiol. 2022, 188, 1617–1631. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.; Zhang, R.; Yang, C.; Zhang, X.; Chang, S.; Chen, Q.; Rossi, V.; Zhao, L.; Xiao, J.; et al. Type I MADS-box transcription factor TaMADS-GS regulates grain size by stabilizing cytokinin signalling during endosperm cellularization in wheat. Plant Biotechnol. J. 2024, 22, 200–215. [Google Scholar] [CrossRef]
- Zhang, A.; He, H.; Li, Y.; Wang, L.; Liu, Y.; Luan, X.; Wang, J.; Liu, H.; Liu, S.; Zhang, J.; et al. MADS-box subfamily gene GmAP3 from glycine max regulates early flowering and flower development. Int. J. Mol. Sci. 2023, 24, 2751. [Google Scholar] [CrossRef]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, L.L.; Xu, Z.S.; Fu, L.; Pang, H.X.; Ma, Y.Z.; Min, D.H. Genome-wide analysis of MADS-Box genes in foxtail millet (Setaria italica L.) and functional assessment of the role of SiMADS51 in the drought stress response. Front. Plant Sci. 2021, 12, 659474. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhu, G.P.; Li, F.D.; Wang, L.; Chen, C.; Zhao, H. MIKC-type MADS-box gene family discovery and evolutionary investigation in Rosaceae Plants. Agronomy 2023, 13, 1695. [Google Scholar] [CrossRef]
- Duan, W.; Song, X.; Liu, T.; Huang, Z.; Ren, J.; Hou, X.; Li, Y. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet. Genom. 2015, 290, 239–255. [Google Scholar] [CrossRef]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis new openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Gao, L. Evolutionary Dynamics and expression divergence of the MADS-Box gene family during recent speciation of AA-genome Oryza Species. Plants 2025, 14, 379. [Google Scholar] [CrossRef]
- Tahmasebi, S.; Jonoubi, P.; Majdi, M.; Majd, A.; Heidari, P. Genome-wide characterization and expression profiling of MADS-box family genes during organ development and drought stress in Camelina sativa L. Sci. Rep. 2025, 15, 9327. [Google Scholar] [CrossRef]
- Bouché, F.; Lobet, G.; Tocquin, P.; Périlleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2015, 44, D1167–D1171. [Google Scholar] [CrossRef]
- Meng, Q.; Gao, Y.N.; Cheng, H.; Liu, Y.; Yuan, L.N.; Song, M.R.; Li, Y.R.; Zhao, Z.X.; Hou, X.F.; Tan, X.M.; et al. Molecular mechanism of interaction between SHORT VEGETATIVE PHASE and APETALA1 in Arabidopsis thaliana. Plant Physiol. Biochem. 2025, 220, 109512. [Google Scholar] [CrossRef]
- Wang, D.; Chai, Y.; Chen, S. OsMADS22 interacts with OsMADS50 to regulate floral transition in rice. Biochem. Biophys. Res. Commun. 2025, 757, 151607. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, H.; Wang, D.; Wang, Y.; Jiang, H.; Chen, J. Integrative transcriptome and metabolome analyses reveal the molecular mechanism of re-flowering induction in Hydrangea macrophylla. J. Plant Physiol. 2025, 308, 154492. [Google Scholar] [CrossRef]
- Feng, T.; Wang, L.; Li, L.; Liu, Y.; Chong, K.; Theißen, G.; Meng, Z. OsMADS14 and NF-YB1 cooperate in the direct activation of OsAGPL2 and Waxy during starch synthesis in rice endosperm. New Phytol. 2022, 234, 7–92. [Google Scholar] [CrossRef]
- Tan, X.M.; Li, Y.R.; Song, M.R.; Yuan, L.N.; Zhao, Z.X.; Liu, Y.; Meng, Q.; Huang, X.; Ma, Y.Y.; Xu, Z.Q. The molecular mechanism of interaction between SEPALLATA3 and APETALA1 in Arabidopsis thaliana. Plant Direct 2025, 9, e70052. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Kim, S.Y.; Susila, H.; Nasim, Z.; Youn, G.; Ahn, J.H. FLOWERING LOCUS M isoforms differentially affect the subcellular localization and stability of SHORT VEGETATIVE PHASE to regulate temperature-responsive flowering in Arabidopsis. Mol. Plant 2022, 15, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Yoon, H.; Seo, P.J. The AGL6–ELF3–FT circuit controls flowering time in Arabidopsis. Plant Signal Behav. 2024, 19, 2358684. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, B.; Wu, Q.; Min, Q.; Zeng, R.; Xie, Z.; Huang, J. OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PLoS Genet. 2021, 17, e1009699. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, F.; Hong, Y.; Yao, J.; Ren, Z.; Shi, H.; Zhu, J. The flowering repressor SVP confers drought resistance in Arabidopsis by regulating abscisic acid catabolism. Mol. Plant 2018, 11, 1184–1197. [Google Scholar] [CrossRef]
- Raingeval, M.; Leduque, B.; Baduel, P.; Edera, A.; Roux, F.; Colot, V.; Quadrana, L. Retrotransposon-driven environmental regulation of FLC leads to adaptive response to herbicide. Nat. Plants 2024, 10, 1672–1681. [Google Scholar] [CrossRef]
- Voogd, C.; Brian, L.A.; Wu, R.; Wang, T.; Allan, A.C.; Varkonyi-Gasic, E. A MADS-box gene with similarity to FLC is induced by cold and correlated with epigenetic changes to control budbreak in kiwifruit. New Phytol. 2022, 233, 2111–2126. [Google Scholar] [CrossRef]
- FAO. Available online: http://faostat.fao.org (accessed on 19 August 2025).
- Jiang, P.; Kang, Z.; Zhao, S.; Meng, N.; Liu, M.; Tan, B. Effect of dynamic high-pressure microfluidizer on physicochemical and microstructural properties of whole-grain oat pulp. Foods 2023, 12, 2747. [Google Scholar] [CrossRef]
- Zhang, M.; Jiang, Y.; Dong, H.; Shan, X.; Tian, J.; Sun, M.; Ma, F.; Ren, C.; Yuan, Y. Transcriptomic response for revealing the molecular mechanism of oat flowering under different photoperiods. Front. Plant Sci. 2023, 14, 1279107. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, Q.; Li, Y.; Hao, P.B.; Wei, H.L.; Fu, X.K.; Lu, J.H.; Ma, L.; Yu, S.X.; Wang, H.T. GhGAI interacts with GhAP1 and regulates flowering pathway in upland cotton. Ind. Crops Prod. 2023, 192, 116110. [Google Scholar] [CrossRef]
- Luo, X.; Liu, B.; Xie, L.; Wang, K.; Xu, D.; Tian, X.; Xie, L.; Li, L.; Ye, X.; He, Z.; et al. The TaSOC1-TaVRN1 module integrates photoperiod and vernalization signals to regulate wheat flowering. Plant Biotechnol. J. 2024, 22, 635–649. [Google Scholar] [CrossRef]
- Wang, F.; Zhou, Z.; Zhu, L.; Gu, Y.; Guo, B.; Lv, C.; Zhu, J.; Xu, R. Genome-wide analysis of the MADS-box gene family involved in salt and waterlogging tolerance in barley (Hordeum vulgare L.). Front. Plant Sci. 2023, 14, 1178065. [Google Scholar] [CrossRef]
- Zhao, D.; Chen, Z.; Xu, L.; Zhang, L.; Zou, Q. Genome-wide analysis of the MADS-Box gene family in maize: Gene structure, evolution, and relationships. Genes 2021, 12, 1956. [Google Scholar] [CrossRef]
- Nan, J.; An, J.; Yang, Y.; Zhao, G.; Yang, X.; Liu, H.; Han, B. Genome-wide identification of the MADS-box gene family in Avena sativa and its role in photoperiod-insensitive oat. PeerJ 2024, 12, e16759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jiang, Y.; Dong, H.; Shan, X.; Chen, H.; Li, X.; Wang, C.; Ba, S.; Li, J.; Guo, L.; et al. Comparative transcriptome analysis of oat varieties with different flowering performances under a short-day photoperiod. BMC Plant Biol. 2025, 25, 622. [Google Scholar] [CrossRef]
- Cui, J.; Li, Y.; Liu, H.; Jiang, X.; Zhang, L.; Dai, H.; Wang, X.; He, F.; Li, M.; Kang, J. Genome-wide identification and expression analysis of CBF/DREB1 gene family in Medicago sativa L. and functional verification of MsCBF9 affecting flowering time. BMC Plant Biol. 2025, 25, 87. [Google Scholar] [CrossRef]
- Kamal, N.; Tsardakas Renhuldt, N.; Bentzer, J.; Gundlach, H.; Haberer, G.; Juhász, A.; Lux, T.; Bose, U.; Tye-Din, J.A.; Lang, D.; et al. The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 2022, 606, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Raza, Q.; Riaz, A.; Atif, R.M.; Hussain, B.; Rana, I.A.; Ali, Z.; Budak, H.; Alaraidh, I.A. Genome-wide diversity of MADS-Box genes in bread wheat is associated with its rapid global adaptability. Front. Genet. 2022, 12, 818880. [Google Scholar] [CrossRef]
- Lu, S.N.; Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2019, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2021, 40, e49. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kumari, A.; Kumar, M.; Tyagi, A.; Maheshwari, C.; Meena, N.L. Nutritional Composition of Oats and Its Comparison with Other Major Cereal Crops. Oat (Avena sativa), 1st ed.; CRS Press: Boca Raton, FL, USA, 2024; pp. 69–103. [Google Scholar]
- Abdullah-Zawawi, M.R.; Ahmad-Nizammuddin, N.F.; Govender, N.; Harun, S.; Mohd-Assaad, N.; Mohamed-Hussein, Z.A. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice. Sci. Rep. 2021, 11, 19678. [Google Scholar] [CrossRef]
- Fatima, M.; Ma, X.; Zhang, J.; Ming, R. Genome-wide analysis of MADS-box genes and their expression patterns in unisexual flower development in dioecious spinach. Sci. Rep. 2024, 14, 18635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, W.; Wang, X.; Duan, Y.; Li, Y.; Wang, Y.; Jiang, Q.; Liao, B.; Zhou, S.; Li, Y. Genome-wide analyses of MADS-Box Genes reveal their involvement in seed development and oil accumulation of tea-oil tree (Camellia oleifera). Int. J. Genom. 2024, 2024, 3375173. [Google Scholar] [CrossRef]
- Cao, J.; Gong, Y.; Zou, M.; Li, H.; Chen, S.; Ma, C. Genome-wide identification and salt stress response analysis of the MADS-box transcription factors in sugar beet. Physiol. Plant. 2024, 176, e70001. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Guo, Q.; Liu, S.; Wang, D.; Zuo, D.; Niu, T.; Wei, D.; Guo, L.; Hou, X. Genome-wide characterization of the MADS-box gene family in Paeonia ostii and expression analysis of genes related to floral organ development. BMC Genom. 2025, 26, 49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, Y.; Zhang, Y.; Yang, F. Genome-wide investigation of MADS-box genes in flower development and environmental acclimation of Lumnitzera littorea (Jack) Voigt. Int. J. Mol. Sci. 2025, 26, 1680. [Google Scholar] [CrossRef]
- Wang, Z.; Chang, J.; Han, J.; Yin, M.; Wang, X.; Ren, Z.; Wang, L. Genome-wide reidentification and expression analysis of MADS-Box gene family in Cucumber. Int. J. Mol. Sci. 2025, 26, 3800. [Google Scholar] [CrossRef]
- Abdelsattar, M.; Nassar, A.E.; Mousa, K.H.; Hussein, A.; El-Baghdady, M.M.S.; Radwan, K.H.; Ibrahim, M.S.; El Allali, A.; Hamwieh, A.; Alsamman, A.M.; et al. Genome-wide identification, characterization, and expression analysis of the MADS-box gene family in grass pea (Lathyrus sativus) under salt stress conditions. BMC Genom. 2025, 26, 519. [Google Scholar]
- Ma, J.; Yang, Y.; Luo, W.; Yang, C.; Ding, P.; Liu, Y.; Qiao, L.; Chang, Z.; Geng, H.; Wang, P.; et al. Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0181443. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, X.; Liu, Q.; Lei, Z.; Feng, C. Genome-wide identification and expression analyses of the MADS-box gene during flowering in Primulina huaijiensis. Plants 2025, 14, 1843. [Google Scholar] [CrossRef]
- Sun, J.; Zhou, Z.; Meng, F.; Wen, M.; Liu, A.; Yu, A. Characterization analyses of MADS-box genes highlighting their functions with seed development in Ricinus communis. Front. Plant Sci. 2025, 16, 1589915. [Google Scholar] [CrossRef]
- Chen, R.; Ma, J.; Luo, D.; Hou, X.; Ma, F.; Zhang, Y.; Meng, Y.; Zhang, H.; Guo, W. CaMADS, a MADS-box transcription factor from pepper, plays an important role in the response to cold, salt, and osmotic stress. Plant Sci. 2019, 280, 164–174. [Google Scholar] [PubMed]
- Wu, J.; Yu, C.; Huang, L.; Gan, Y. A rice transcription factor, OsMADS57, positively regulates high salinity tolerance in transgenic Arabidopsis thaliana and Oryza sativa plants. Physiol. Plant. 2021, 173, 1120–1135. [Google Scholar] [CrossRef]
- Mostafa, K.; Yerlikaya, B.A.; Abdulla, M.F.; Aydin, A.; Yerlikaya, S.; Kavas, M. Genome-wide analysis of PvMADS in common bean and functional characterization of PvMADS31 in Arabidopsis thaliana as a player in abiotic stress responses. Plant Genome 2024, 17, e20432. [Google Scholar]
- Chojnacka, A.; Smoczynska, A.; Bielewicz, D.; Pacak, A.; Hensel, G.; Kumlehn, J.; Maciej Karlowski, W.; Grabsztunowicz, M.; Sobieszczuk-Nowicka, E.; Jarmolowski, A.; et al. PEP444c encoded within the MIR444c gene regulates microRNA444c accumulation in barley. Physiol. Plant. 2023, 175, e14018. [Google Scholar] [CrossRef]
- Pachamuthu, K.; Hari Sundar, V.; Narjala, A.; Singh, R.R.; Das, S.; Avik Pal, H.C.Y.; Shivaprasad, P.V. Nitrate-dependent regulation of miR444-OsMADS27 signalling cascade controls root development in rice. J. Exp. Bot. 2022, 73, 3511–3530. [Google Scholar]
- Yang, H.; Zhang, P.; Guo, D.; Wang, N.; Lin, H.; Wang, X.; Niu, L. Transcriptional repressor AGL79 positively regulates flowering time in Arabidopsis. J. Plant Physiol. 2023, 285, 153985. [Google Scholar] [CrossRef]
- Lim, C.J.; Park, K.S.; Ali, A.; Park, J.; Ryou, S.M.; Shen, M.; Khan, H.A.; Bader, Z.E.; Zareen, S.; Bae, M.J.; et al. Negative regulation of floral transition in Arabidopsis by HOS15-PWR-HDA9 complex. Front. Plant Sci. 2023, 13, 1105988. [Google Scholar] [CrossRef]
- Shin, W.J.; Nam, A.H.; Kim, J.Y.; Kwak, J.S.; Song, J.T.; Seo, H.S. Intronic long noncoding RNA, RICE FLOWERING ASSOCIATED (RIFLA), regulates OsMADS56-mediated flowering in rice. Plant Sci. 2022, 320, 111278. [Google Scholar] [CrossRef]
- Min, Y.; He, S.; Wang, X.; Hu, H.; Wei, S.; Ge, A.; Jiang, L.; Yang, S.; Guo, Y.; Liu, Z.; et al. Transcription factors BnaC09.FUL and BnaC06.WIP2 antagonistically regulate flowering time under long-day conditions in Brassica napus. J. Genet. Genom. 2025, 52, 650–665. [Google Scholar] [CrossRef]
- Deng, Q.; Lu, H.; Liu, D.; Huang, Y.; Feng, J.; Wei, D.; Wang, Z.; Tang, Q. Modulation of flowering by an alternatively spliced AGL18-1 transcript in Brassica juncea. Crop J. 2025, 13, 456–467. [Google Scholar] [CrossRef]
- Yao, X.; Li, K.; Wang, S.; Ji, X.; Zhang, Y.; Zhu, H. GmMADS66 regulates flowering time under photoperiod dependent pathway in Arabidopsis. Plant Cell Tissue Organ Cult. 2025, 160, 14. [Google Scholar] [CrossRef]
- Ma, Y.; Dong, J.; Yang, W.; Chen, L.; Wu, W.; Li, W.; Zhou, L.; Wang, J.; Chen, J.; Yang, T.; et al. OsFLZ2 interacts with OsMADS51 to fine-tune rice flowering time. Development 2022, 149, dev200862. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Wang, C.-L.; Jiang, Y.; Feng, B.; Dong, H.-X.; Chen, H.; Li, X.-Y.; Shan, X.-H.; Tian, J.; Xu, W.-W.; et al. Genome-Wide Identification of MADS-box Family Genes and Analysis of Their Expression Patterns in the Common Oat (Avena sativa L.). Agronomy 2025, 15, 2286. https://doi.org/10.3390/agronomy15102286
Zhang M, Wang C-L, Jiang Y, Feng B, Dong H-X, Chen H, Li X-Y, Shan X-H, Tian J, Xu W-W, et al. Genome-Wide Identification of MADS-box Family Genes and Analysis of Their Expression Patterns in the Common Oat (Avena sativa L.). Agronomy. 2025; 15(10):2286. https://doi.org/10.3390/agronomy15102286
Chicago/Turabian StyleZhang, Man, Chun-Long Wang, Yuan Jiang, Bo Feng, Hai-Xiao Dong, Hao Chen, Xue-Ying Li, Xiao-Hui Shan, Juan Tian, Wei-Wei Xu, and et al. 2025. "Genome-Wide Identification of MADS-box Family Genes and Analysis of Their Expression Patterns in the Common Oat (Avena sativa L.)" Agronomy 15, no. 10: 2286. https://doi.org/10.3390/agronomy15102286
APA StyleZhang, M., Wang, C.-L., Jiang, Y., Feng, B., Dong, H.-X., Chen, H., Li, X.-Y., Shan, X.-H., Tian, J., Xu, W.-W., Yuan, Y.-P., Ren, C.-Z., & Guo, L.-C. (2025). Genome-Wide Identification of MADS-box Family Genes and Analysis of Their Expression Patterns in the Common Oat (Avena sativa L.). Agronomy, 15(10), 2286. https://doi.org/10.3390/agronomy15102286




