Improved UNet Recognition Model for Multiple Strawberry Pests Based on Small Samples
Abstract
1. Introduction
2. Building the Strawberry Multi-Pest Dataset
2.1. Pest Image Collection
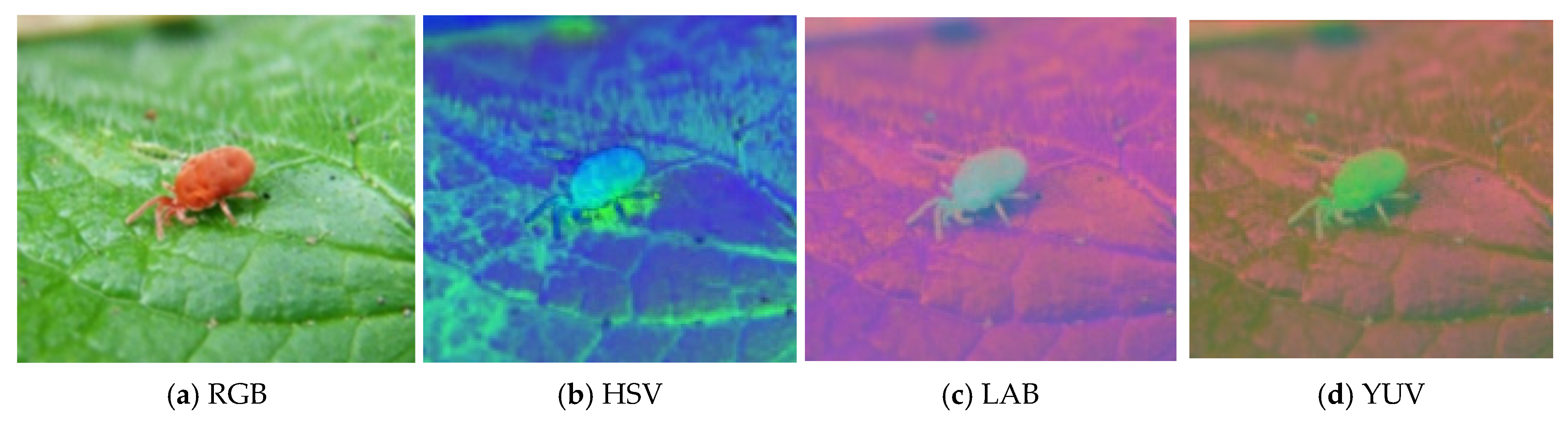
2.2. Image Color Space Conversion
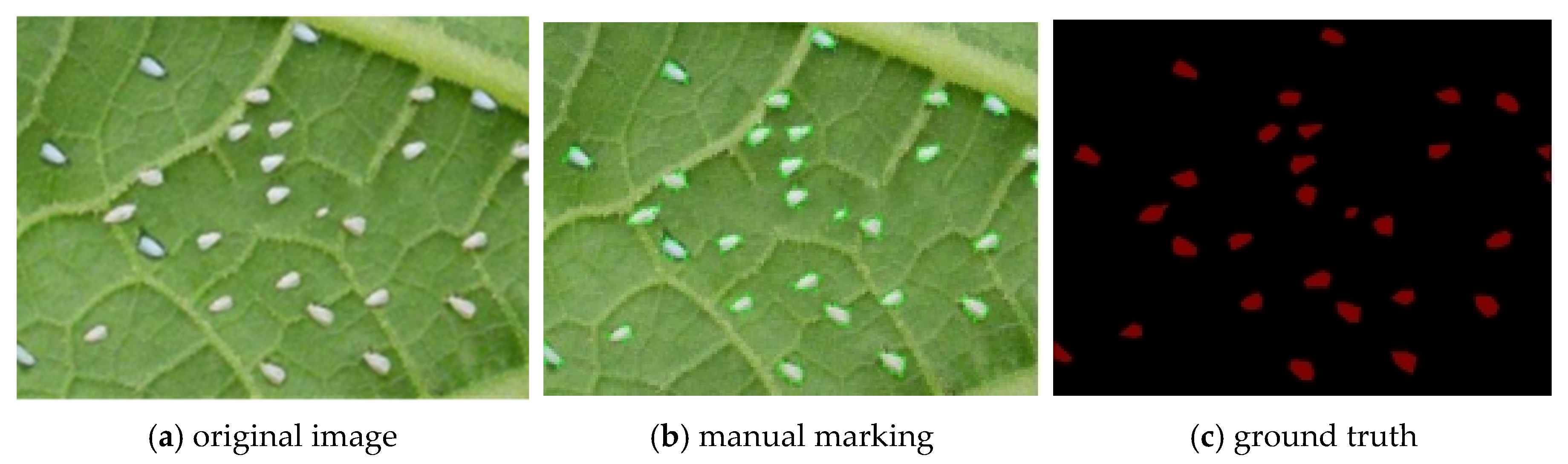
2.3. Pest Pixel Marker
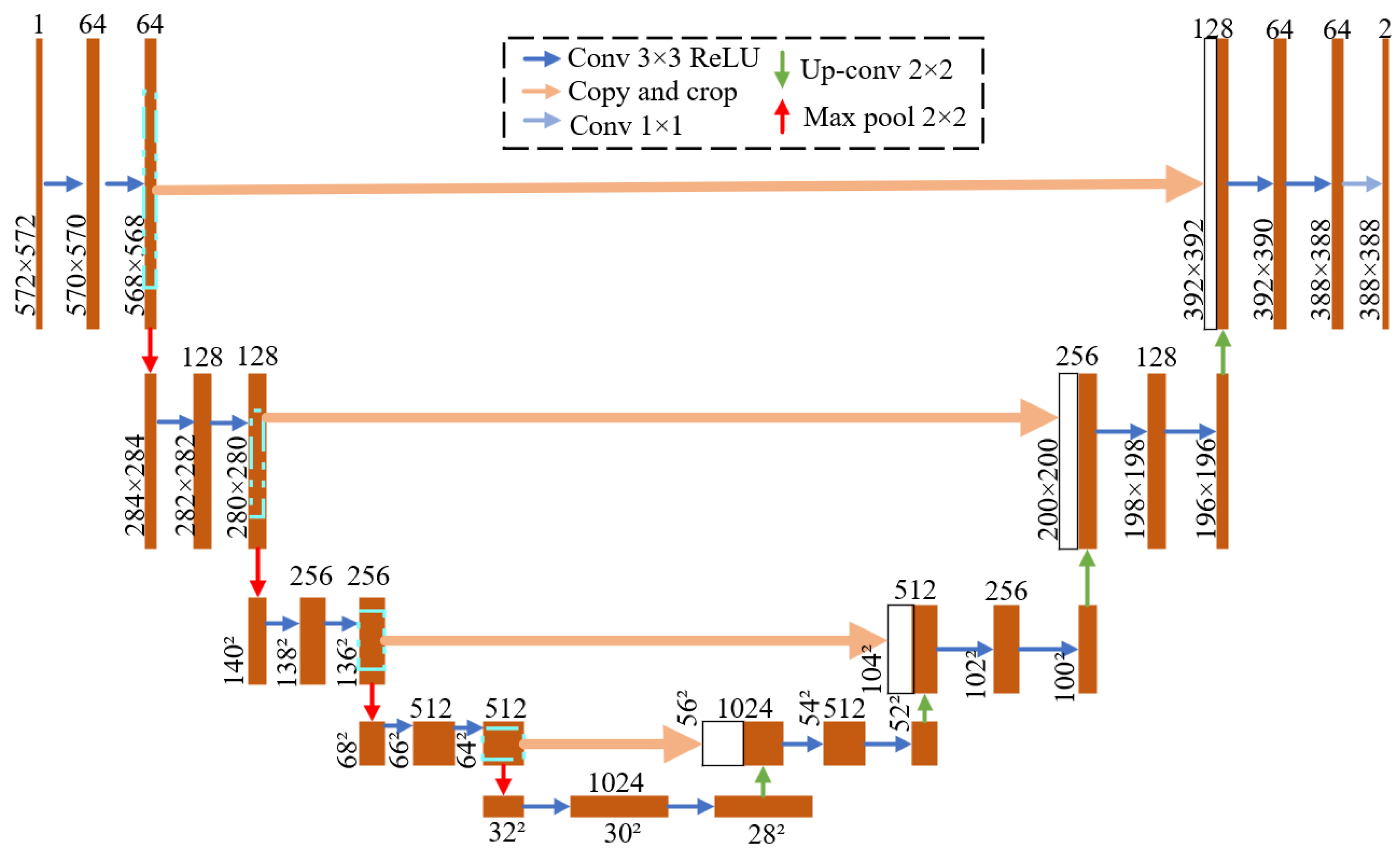
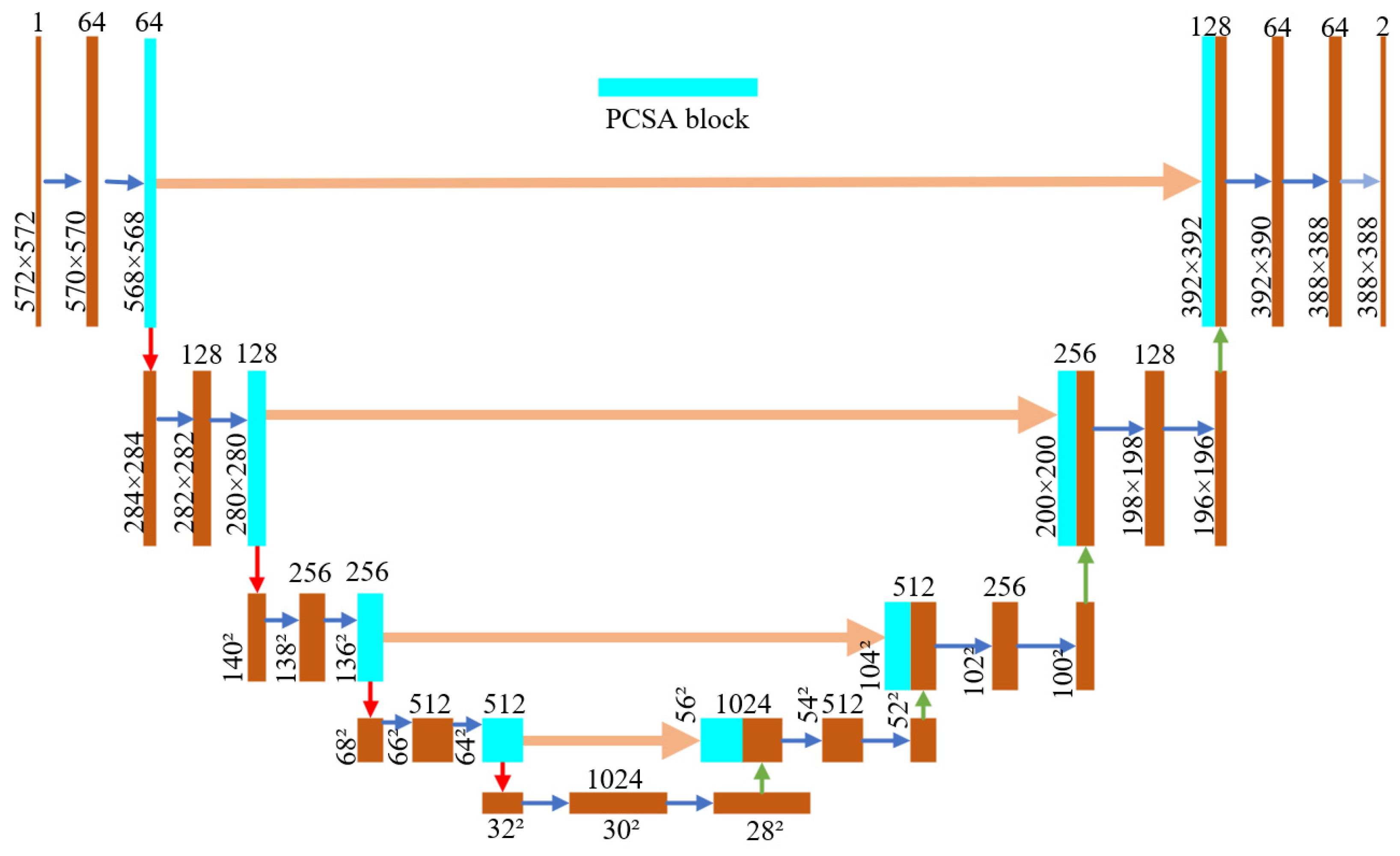
3. Pixel-Level Pest Segmentation Model Architecture
3.1. Backbone
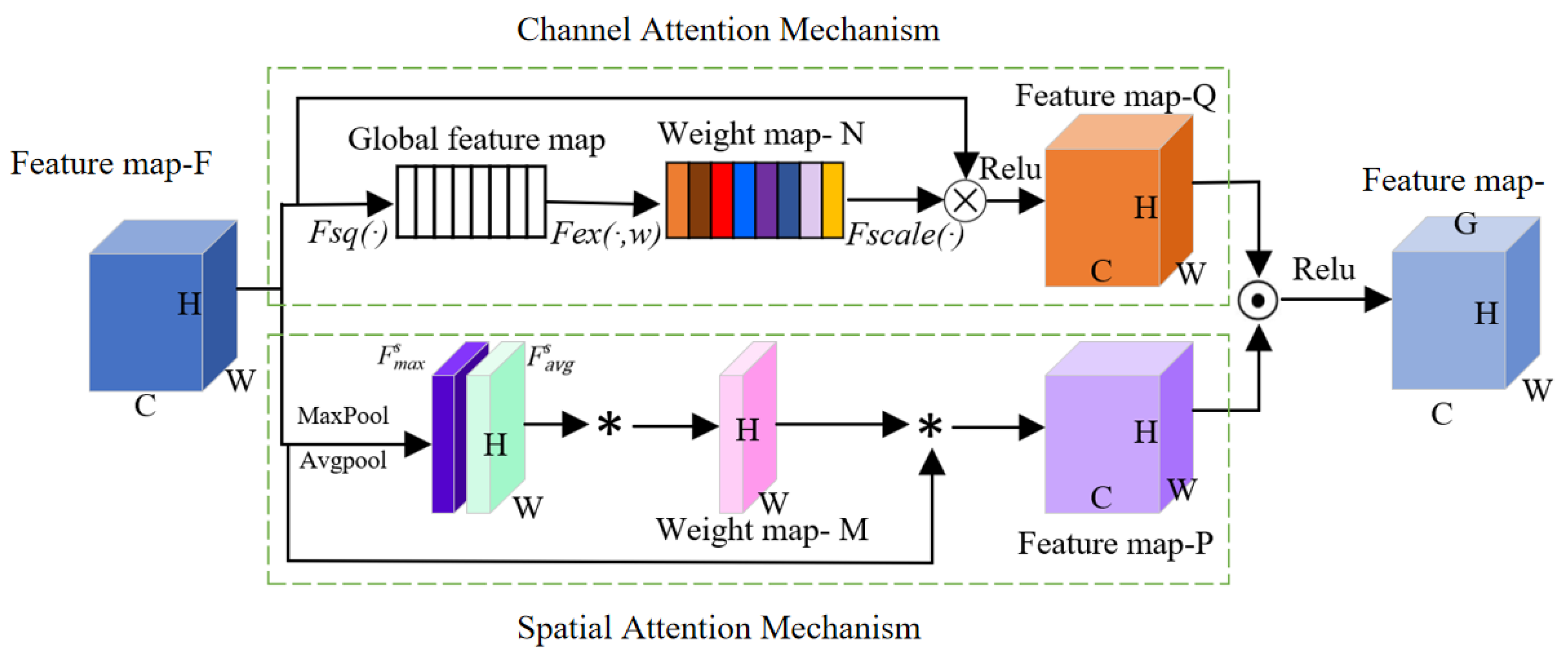
3.2. Channel–Space Parallel Attention (PCSA)
- (1)
- Feature map extraction based on channel attention
- (2)
- Spatial attention performs average pooling and max pooling operations on the original feature map F, generating two single-channel feature maps and . Then, these two feature maps are merged to generate the weight map M. The feature map F is weighted using the weight map M to produce the feature map P.
- (3)
- A dot product is performed between feature maps Q and P, and then the ReLU activation function is applied to obtain feature map G. The feature map G combines weight distributions across both channel and spatial dimensions, enabling it to highlight pest-specific regions while suppressing various types of interference. This allows the model to identify pests with greater accuracy. Meanwhile, when training on small datasets, we employ transfer learning and utilize regularization along with smaller-scale networks to minimize overfitting.
3.3. UNet Adds Attention Mechanism
3.4. Experimental Setup and Evaluation Index
4. Experimental Results and Discussion
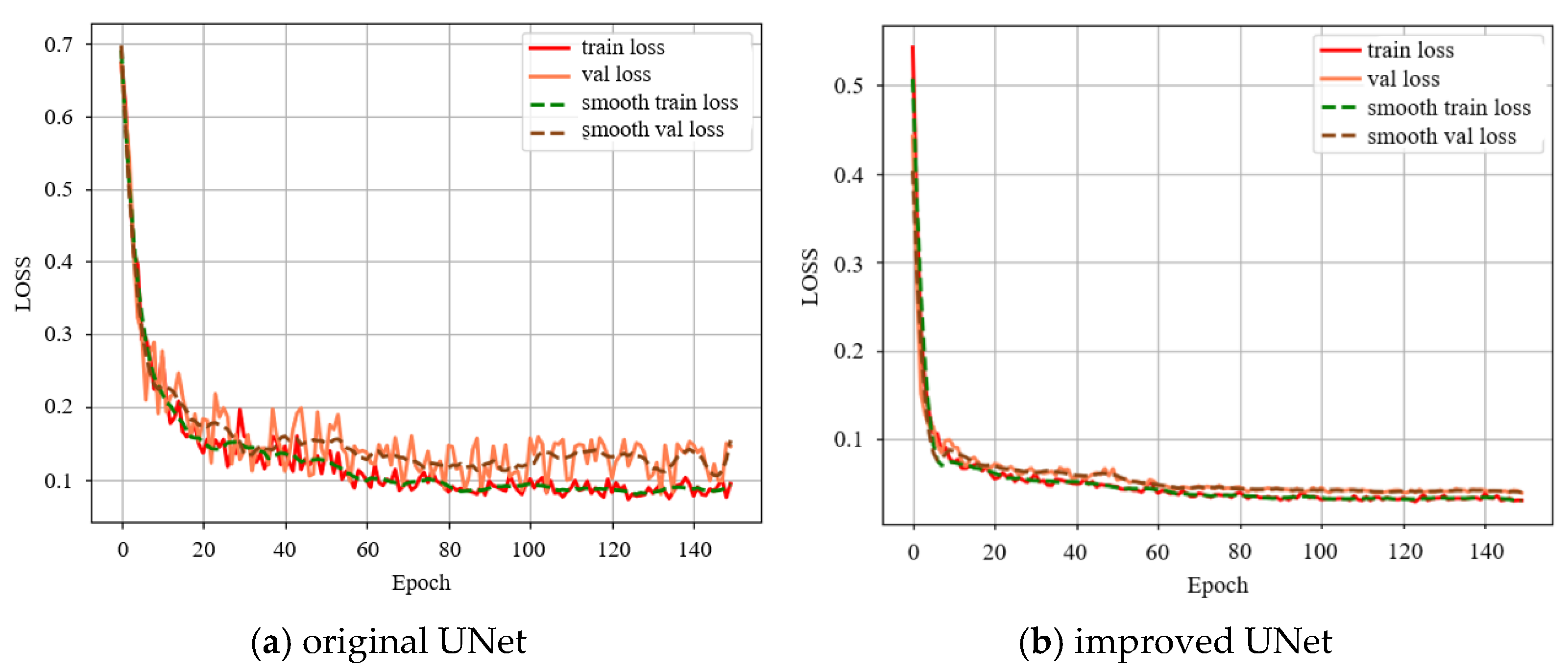
4.1. PCSA Enhancements to UNet Performance
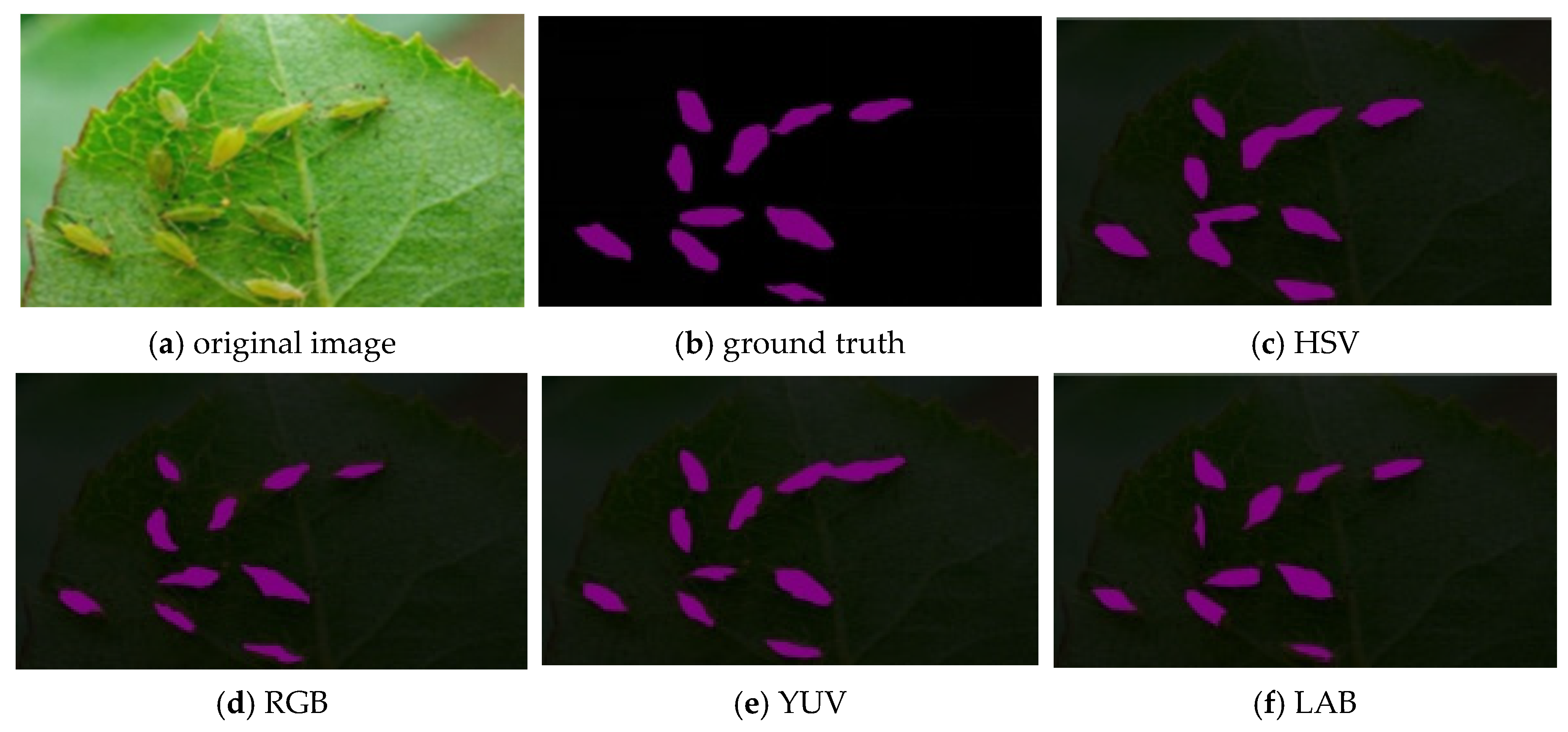
4.2. Comparison of Different Color Spaces for Pest Identification
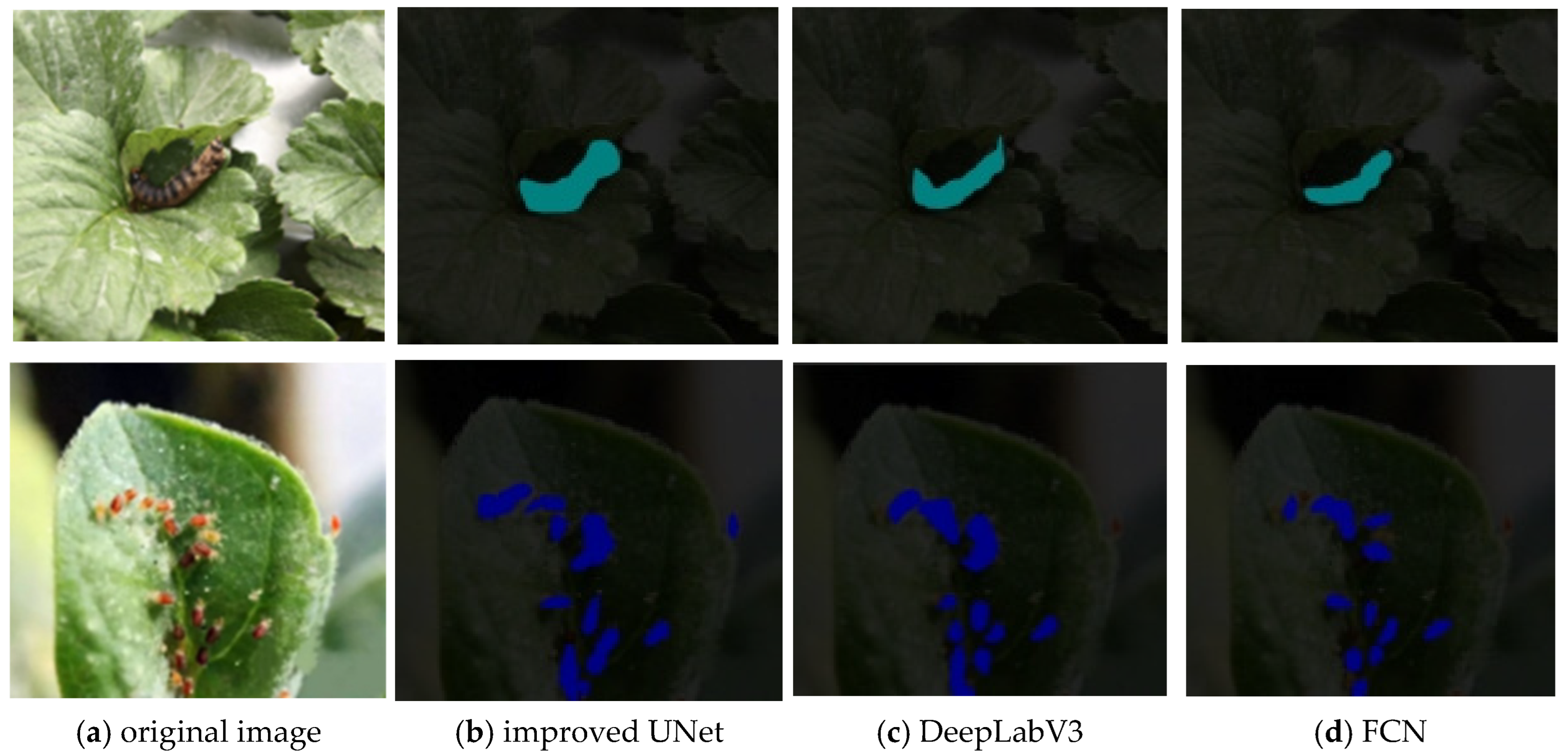
4.3. Performance Comparison of Various Semantic Segmentation Models
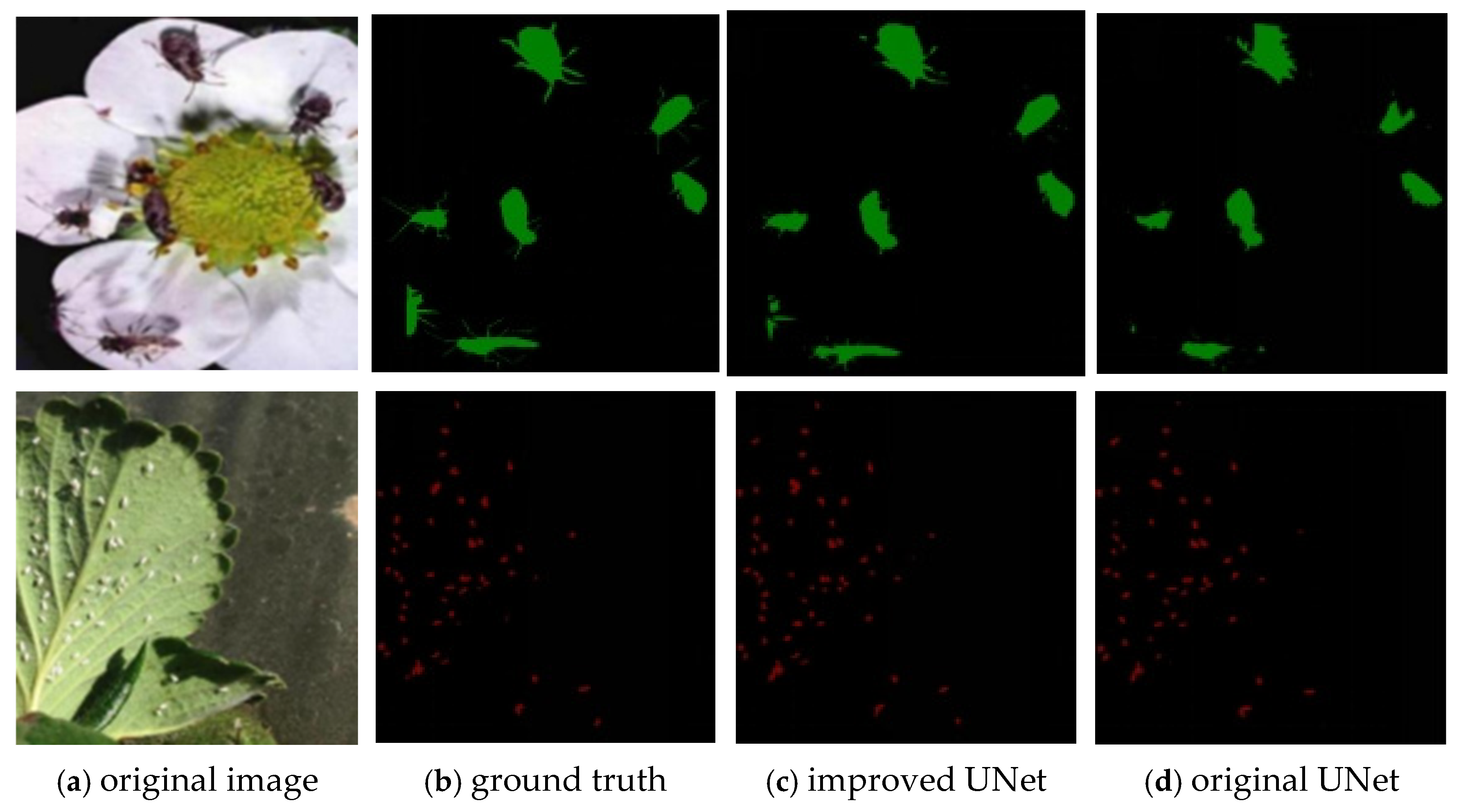
4.4. Validation of PCSA Effectiveness
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, Z.; Wang, W.; Chen, X.; Gehman, K.; Yang, H.; Yang, Y. Prediction of the global occurrence of maize diseases and estimation of yield loss under climate change. Pest Manag. Sci. 2024, 80, 5759–5770. [Google Scholar] [CrossRef]
- Zhao, S.; Peng, Y.; Liu, J.; Wu, S. Tomato leaf disease diagnosis based on improved convolution neural network by attention module. Agriculture 2021, 11, 651. [Google Scholar] [CrossRef]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited ‘FAO estimate’ of 25%. Crit. Rev. Food Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar] [CrossRef]
- Upadhyay, A.; Chandel, N.S.; Singh, K.P.; Chakraborty, S.K.; Nandede, B.M.; Kumar, M.; Subeesh, A.; Upendar, K.; Salem, A.; Elbeltagi, A. Deep learning and computer vision in plant disease detection: A comprehensive review of techniques, models, and trends in precision agriculture. Artif. Intell. Rev. 2025, 58, 92. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, N.; Ma, G.; Taha, M.F.; Mao, H.; Zhang, X.; Shi, Q. Detection of spores using polarization image features and BP neural network. Int. J. Agric. Biol. Eng. 2024, 17, 213–221. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Taha, M.F.; Chen, T.; Yang, N.; Zhang, J.; Mao, H. Detection method of fungal spores based on fingerprint characteristics of diffraction–polarization images. J. Fungi 2023, 9, 1131. [Google Scholar] [CrossRef]
- Liu, J.; Wu, S. Research Progress and Prospect of Strawberry Whole-process Farming Mechanization Technology and Equipment. Trans. Chin. Soc. Agric. Mach. 2021, 52, 1–16. [Google Scholar]
- Castro, P.; Bushakra, J.; Stewart, P.; Weebadde, C.; Wang, D.; Hancock, J.; Finn, C.; Luby, J.; Lewers, K. Genetic mapping of day-neutrality in cultivated strawberry. Mol. Breed. 2015, 35, 79. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, J.; Wu, S. Multiple disease detection method for greenhouse-cultivated strawberry based on multiscale feature fusion Faster R_CNN. Comput. Electron. Agric. 2022, 199, 107176. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, C.; Zhan, X.; Wang, Z. Fine-grained identification of crop pests using an enhanced ConvNeXt model. Trans. Chin. Soc. Agric. Eng. 2025, 41, 185–192. [Google Scholar]
- Batz, P.; Will, T.; Thiel, S.; Ziesche, T.M.; Joachim, C. From identification to forecasting: The potential of image recognition and artificial intelligence for aphid pest monitoring. Front. Plant Sci. 2023, 14, 1150748. [Google Scholar] [CrossRef] [PubMed]
- Pattnaik, G.; Parvathi, K. Automatic detection and classification of tomato pests using support vector machine based on HOG and LBP feature extraction technique. In Progress in Advanced Computing and Intelligent Engineering, Proceedings of the ICACIE 2019, Odisha, India, 20–22 December 2019; Springer: Singapore, 2020; Volume 2, pp. 49–55. [Google Scholar]
- Deng, L.; Wang, Y.; Han, Z.; Yu, R. Research on insect pest image detection and recognition based on bio-inspired methods. Biosyst. Eng. 2018, 169, 139–148. [Google Scholar] [CrossRef]
- Peng, H.; Xu, H.; Liu, H. Lightweight agricultural crops pest identification model using improved ShuffleNet V2. Trans. Chin. Soc. Agric. Eng. (Trans. CSAE) 2022, 38, 161–170. [Google Scholar]
- Dong, S.; Du, J.; Jiao, L.; Wang, F.; Liu, K.; Teng, Y.; Wang, R. Automatic crop pest detection oriented multiscale feature fusion approach. Insects 2022, 13, 554. [Google Scholar] [CrossRef]
- Ullah, N.; Khan, J.A.; Alharbi, L.A.; Raza, A.; Khan, W.; Ahmad, I. An efficient approach for crops pests recognition and classification based on novel DeepPestNet deep learning model. IEEE Access 2022, 10, 73019–73032. [Google Scholar] [CrossRef]
- Zhang, Z.; Rong, J.; Qi, Z.; Yang, Y.; Zheng, X.; Gao, J.; Li, W.; Yuan, T. A multi-species pest recognition and counting method based on a density map in the greenhouse. Comput. Electron. Agric. 2024, 217, 108554. [Google Scholar] [CrossRef]
- He, Y.; Zhou, Z.; Tian, L.; Liu, Y.; Luo, X. Brown rice planthopper (Nilaparvata lugens Stal) detection based on deep learning. Precis. Agric. 2020, 21, 1385–1402. [Google Scholar] [CrossRef]
- Tang, Y.; Luo, F.; Wu, P.; Tan, J.; Wang, L.; Niu, Q.; Li, H.; Wang, P. An improved YOLO network for small target insects detection in tomato fields. Comput. Electron. Agric. 2025, 239, 110915. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, L.; Yuan, Y. Few-shot agricultural pest recognition based on multimodal masked autoencoder. Crop Prot. 2025, 187, 106993. [Google Scholar] [CrossRef]
- Xing, S.; Lee, H.J. Crop pests and diseases recognition using DANet with TLDP. Comput. Electron. Agric. 2022, 199, 107144. [Google Scholar] [CrossRef]
- Dong, C.; Zhang, Z.; Yue, J.; Zhou, L. Automatic recognition of strawberry diseases and pests using convolutional neural network. Smart Agric. Technol. 2021, 1, 100009. [Google Scholar] [CrossRef]
- Gan, G.; Xiao, X.; Jiang, C.; Ye, Y.; He, Y.; Xu, Y.; Luo, C. Strawberry disease and pest identification and control based on Se-Resnext50 model. In Proceedings of the 2022 3rd International Conference on Computer Vision, Image and Deep Learning & International Conference on Computer Engineering and Applications (CVIDL & ICCEA), Changchun, China, 20–22 May 2022; pp. 237–243. [Google Scholar]
- Choi, Y.-W.; Kim, N.-e.; Paudel, B.; Kim, H.-t. Strawberry pests and diseases detection technique optimized for symptoms using deep learning algorithm. J. Bio-Environ. Control 2022, 31, 255–260. [Google Scholar] [CrossRef]
- Wang, D.; Deng, L.; Ni, J.; Gao, J.; Zhu, H.; Han, Z. Recognition pest by image-based transfer learning. J. Sci. Food Agric. 2019, 99, 4524–4531. [Google Scholar]
- Lottes, P.; Behley, J.; Milioto, A.; Stachniss, C. Fully convolutional networks with sequential information for robust crop and weed detection in precision farming. IEEE Robot. Autom. Lett. 2018, 3, 2870–2877. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Semantic image segmentation with deep convolutional nets and fully connected crfs. arXiv 2014, arXiv:1412.7062. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Tassis, L.M.; de Souza, J.E.T.; Krohling, R.A. A deep learning approach combining instance and semantic segmentation to identify diseases and pests of coffee leaves from in-field images. Comput. Electron. Agric. 2021, 186, 106191. [Google Scholar] [CrossRef]
- Bose, K.; Shubham, K.; Tiwari, V.; Patel, K.S. Insect image semantic segmentation and identification using unet and deeplab v3+. In ICT Infrastructure and Computing, Proceedings of the ICT4SD 2022, Goa, India, 29–30 July 2022; Springer: Singapore, 2022; pp. 703–711. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Janarthan, S.; Thuseethan, S.; Joseph, C.; Palanisamy, V.; Rajasegarar, S.; Yearwood, J. Efficient Attention-Lightweight Deep Learning Architecture Integration for Plant Pest Recognition. IEEE Trans. AgriFood Electron. 2025, 1–13. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Ye, W.; Lao, J.; Liu, Y.; Chang, C.-C.; Zhang, Z.; Li, H.; Zhou, H. Pine pest detection using remote sensing satellite images combined with a multi-scale attention-UNet model. Ecol. Inform. 2022, 72, 101906. [Google Scholar] [CrossRef]
- Zhang, J.; Cong, S.; Zhang, G.; Ma, Y.; Zhang, Y.; Huang, J. Detecting pest-infested forest damage through multispectral satellite imagery and improved UNet++. Sensors 2022, 22, 7440. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Wang, R.; Liu, Z.; Jiao, L.; Dong, S.; Zhang, L.; Du, J.; Hu, H. Mcunet: Multidimensional cognition unet for multi-class maize pest image segmentation. In Proceedings of the 2023 2nd International Conference on Robotics, Artificial Intelligence and Intelligent Control (RAIIC), Mianyang, China, 11–13 August 2023; pp. 340–346. [Google Scholar]

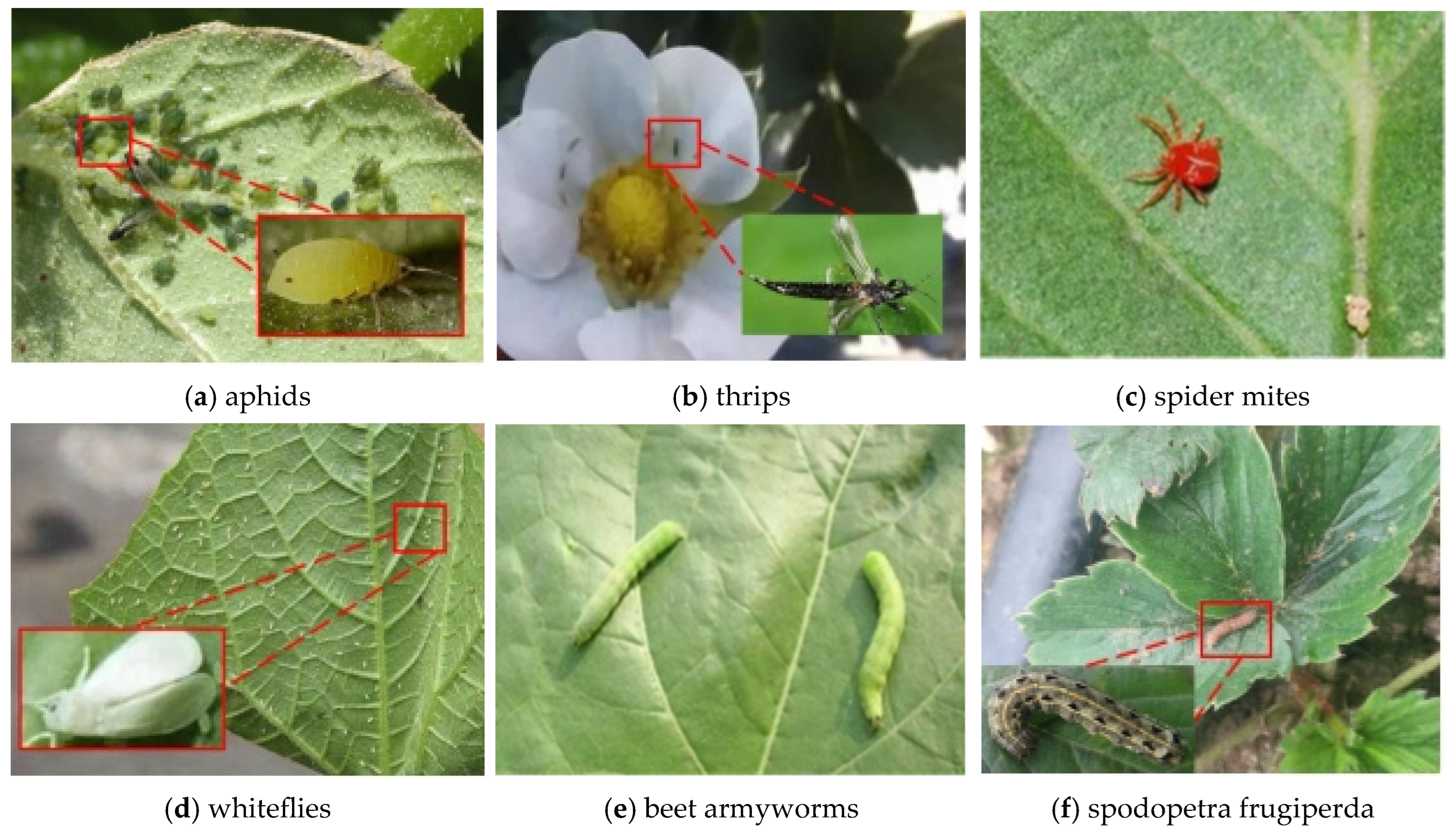
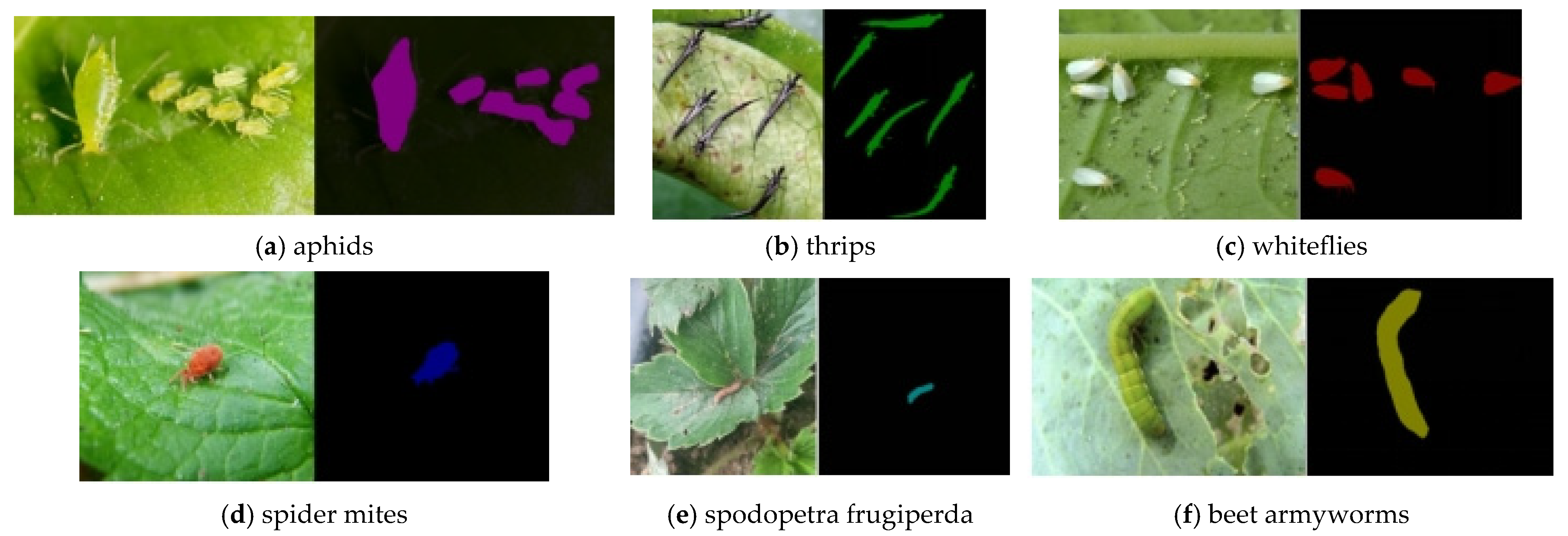
| Type | Color Features | Stage | Training | Testing |
|---|---|---|---|---|
| Aphids | Green (close to the leaves) | Adult | 96 | 24 |
| Thrips | Black | Adult | 64 | 16 |
| Whiteflies | White | Adult | 80 | 20 |
| Beet armyworms | Green (close to the leaves) | Larva | 80 | 20 |
| spodopetra frugiperda | Brown | Larva | 72 | 18 |
| Spider mites | Red | Adult | 88 | 22 |
| Total | 480 | 120 | ||
| Model | AP (%) | mAP (%) | IoU (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Aphids | Thrips | Whiteflies | Beet Armyworms | Spodopetra Frugiperda | Spider Mites | |||
| Original UNet | 83.15 | 84.09 | 82.75 | 88.03 | 88.67 | 87.36 | 85.68 | 75.3 |
| Improved UNet | 80.04 | 79.39 | 78.12 | 83.34 | 83.65 | 84.07 | 81.44 | 79.6 |
| No. | Color Space | IoU (%) | Recall (%) | Precision (%) |
|---|---|---|---|---|
| 1 | RGB | 79.6 | 87.1 | 87.5 |
| 2 | HSV | 84.8 | 89.9 | 91.8 |
| 3 | LAB | 79.7 | 86.6 | 87.0 |
| 4 | YUV | 80.1 | 86.2 | 87.9 |
| Model | Color Space | IoU (%) | Recall (%) | Precision (%) |
|---|---|---|---|---|
| Improved UNet | HSV | 84.8 | 89.9 | 91.8 |
| FCN | 79.8 | 84.5 | 86.4 | |
| DeepLabv3 | 81.1 | 86.6 | 88.4 |
| No. | Attention Mechanism | IoU (%) | Recall (%) | Precision (%) |
|---|---|---|---|---|
| 1 | PCSA | 84.8 | 89.9 | 91.8 |
| 2 | CBAM | 83.1 | 88.6 | 90.5 |
| 3 | SENet | 80.3 | 85.2 | 87.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Liu, J.; Hua, T.; Jiang, Y. Improved UNet Recognition Model for Multiple Strawberry Pests Based on Small Samples. Agronomy 2025, 15, 2252. https://doi.org/10.3390/agronomy15102252
Zhao S, Liu J, Hua T, Jiang Y. Improved UNet Recognition Model for Multiple Strawberry Pests Based on Small Samples. Agronomy. 2025; 15(10):2252. https://doi.org/10.3390/agronomy15102252
Chicago/Turabian StyleZhao, Shengyi, Jizhan Liu, Tianzheng Hua, and Yong Jiang. 2025. "Improved UNet Recognition Model for Multiple Strawberry Pests Based on Small Samples" Agronomy 15, no. 10: 2252. https://doi.org/10.3390/agronomy15102252
APA StyleZhao, S., Liu, J., Hua, T., & Jiang, Y. (2025). Improved UNet Recognition Model for Multiple Strawberry Pests Based on Small Samples. Agronomy, 15(10), 2252. https://doi.org/10.3390/agronomy15102252






