Genome-Wide Identification and Functional Characterization Under Abiotic Stress of Melatonin Biosynthesis Enzyme Family Genes in Poncirus trifoliata
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Melatonin Biosynthetic Pathway Genes and Proteins in Poncirus trifoliata (L.)
2.2. Physical and Chemical Properties and Evolutionary Relationship Analysis of Melatonin Biosynthesis Family Genes
2.3. Plant Materials and Stress Treatment
2.4. RNA Extraction and Quantitative Real-Time PCR Validation
2.5. Statistical Analysis
3. Results
3.1. Genome-Wide Mining Identified Genes Involved in Melatonin Biosynthesis in Trifoliate Orange
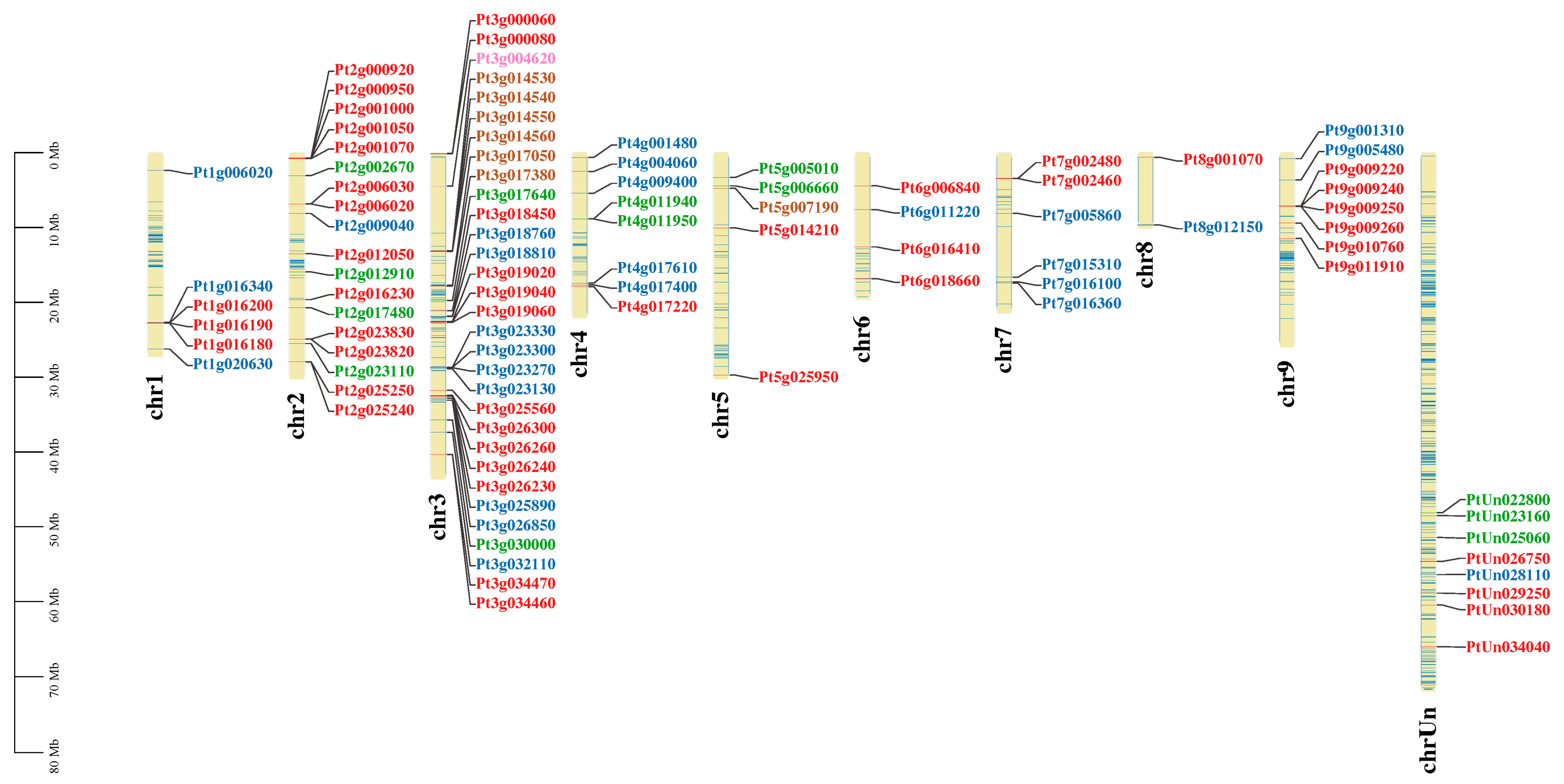
3.2. The Chromosomal Location of Melatonin Biosynthesis Gene Family Members in Trifoliate Orange
3.3. Phylogenetic Relationships Among Gene Family Members
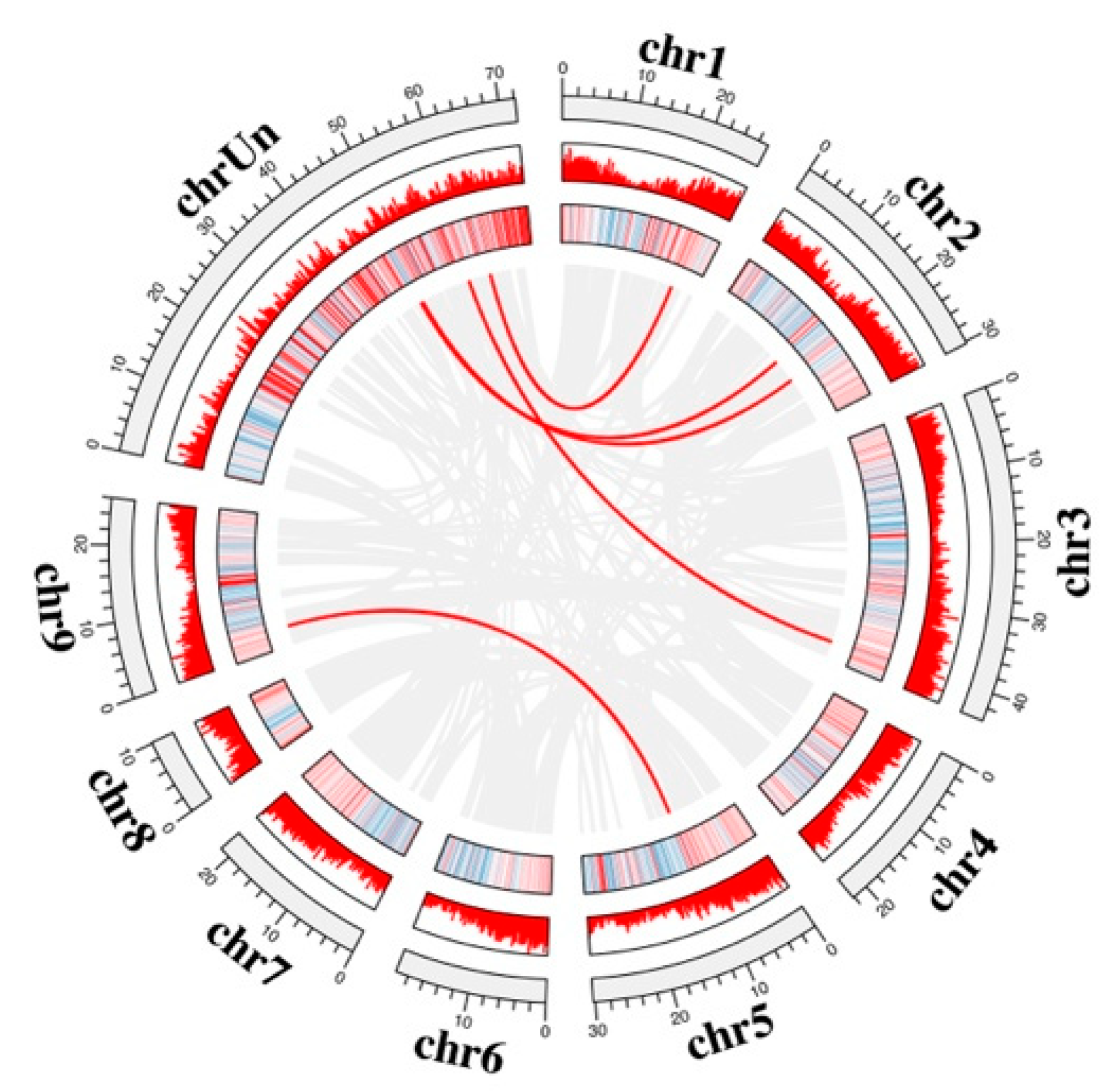
3.4. Gene Structure Collinearity Analysis of Gene Family Members
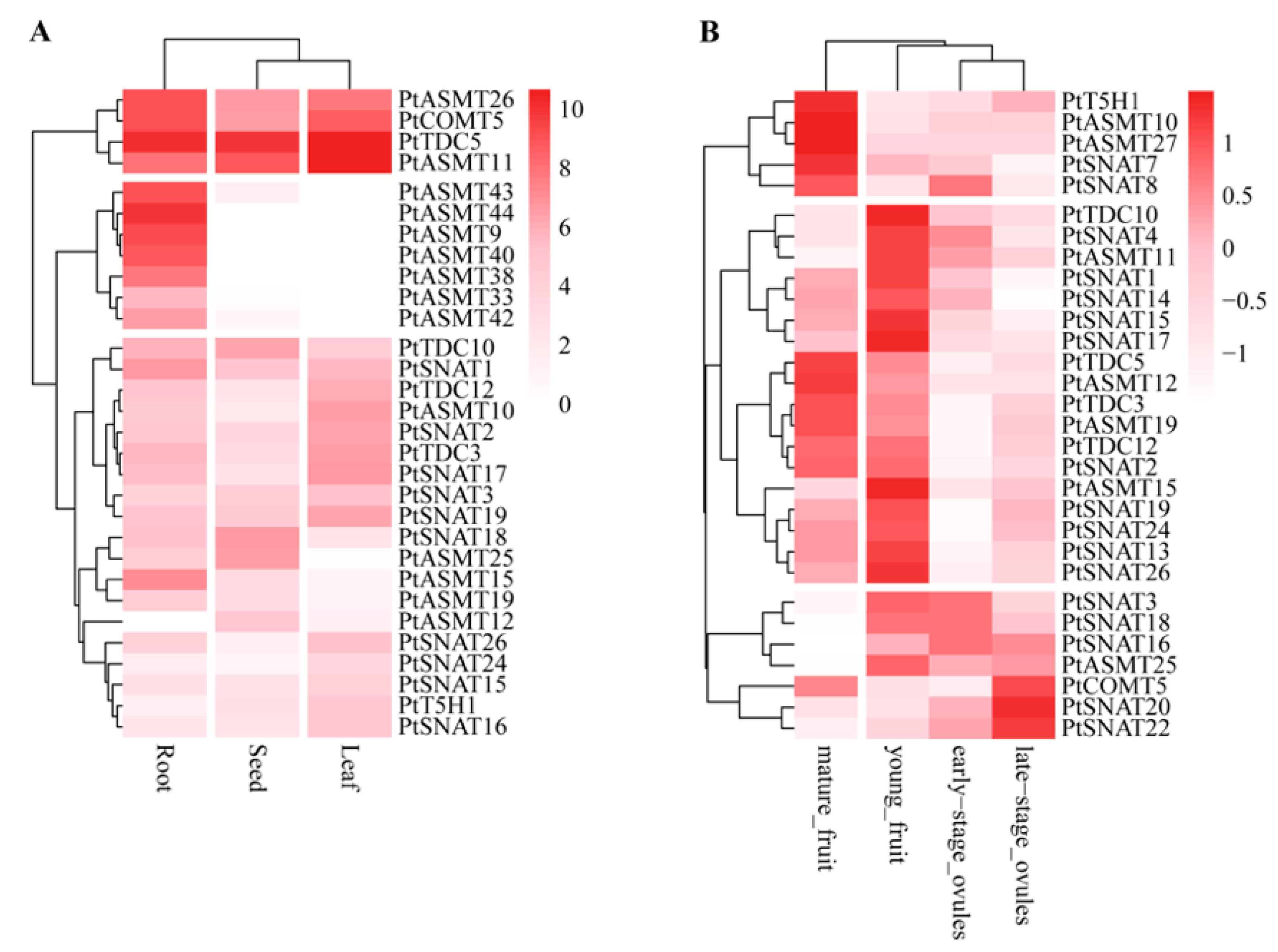
3.5. Expression of Melatonin Biosynthesis Genes in Tissues During Growth and Development in Trifoliate Orange
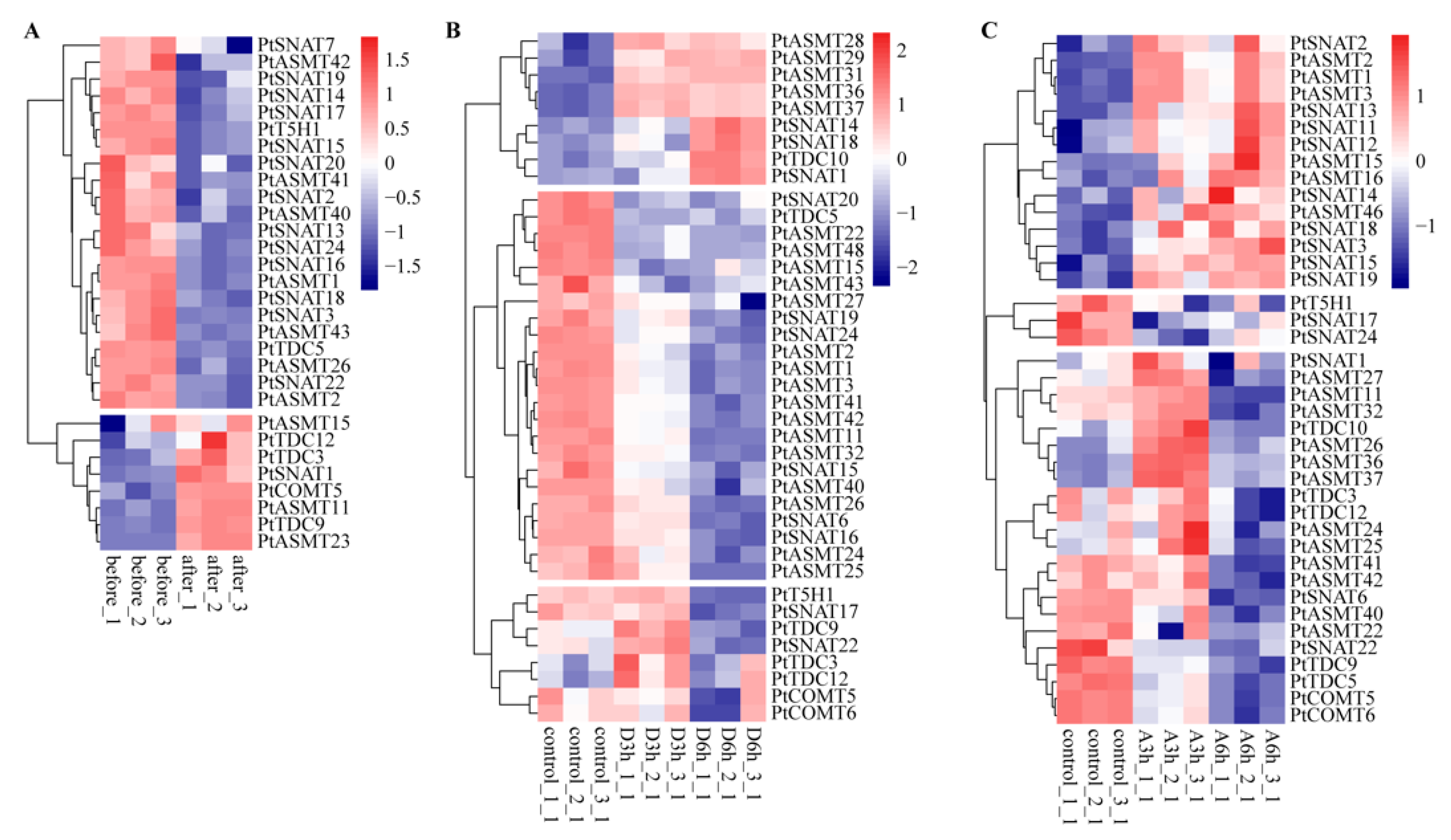
3.6. Gene Expression Analysis of Melatonin Biosynthesis-Related Genes Involved in the Abiotic Response
3.7. Gene Expression Analysis of Melatonin Biosynthesis-Related Genes in Response to ABA Treatment
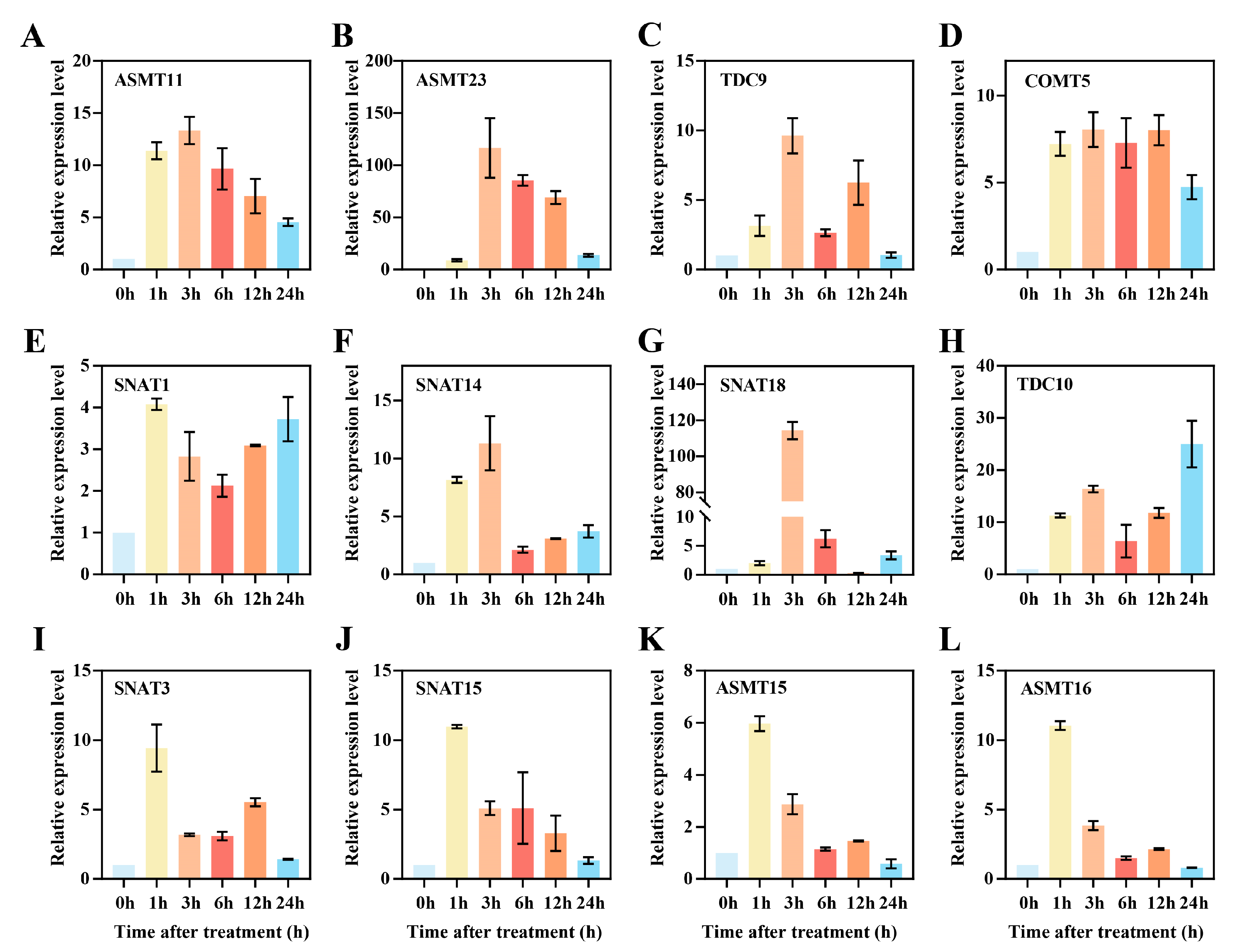
3.8. qRT-PCR Analysis of Candidate Genes Under Stress Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Dahro, B.; Li, C.; Liu, J.-H. Overlapping responses to multiple abiotic stresses in citrus: From mechanism understanding to genetic improvement. Hortic. Adv. 2023, 1, 4. [Google Scholar] [CrossRef]
- Ming, R.; Zhang, Y.; Wang, Y.; Khan, M.; Dahro, B.; Liu, J.H. The JA-responsive MYC2-BADH-like transcriptional regulatory module in Poncirus trifoliata contributes to cold tolerance by modulation of glycine betaine biosynthesis. New Phytol. 2021, 229, 2730–2750. [Google Scholar] [CrossRef]
- Dahro, B.; Wang, Y.; Khan, M.; Zhang, Y.; Fang, T.; Ming, R.; Li, C.; Liu, J.H. Two AT-Hook proteins regulate A/NINV7 expression to modulate sucrose catabolism for cold tolerance in Poncirus trifoliata. New Phytol. 2022, 235, 2331–2349. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Qu, J.; Wang, Y.; Fang, T.; Xiao, W.; Wang, Y.; Zhang, Y.; Khan, M.; Chen, Q.; Xu, X.; et al. Transcriptome and metabolome atlas reveals contributions of sphingosine and chlorogenic acid to cold tolerance in Citrus. Plant Physiol. 2024, 196, 634–650. [Google Scholar] [CrossRef]
- Hassan, M.K.; Ahammed, G.J.; Sun, S.C.; Li, M.Q.; Yin, H.Q.; Zhou, J. Melatonin inhibits cadmium translocation and enhances plant tolerance by regulating sulfur uptake and assimilation in Solanum lycopersicum L. J. Agric. Food Chem. 2019, 67, 10563–10576. [Google Scholar] [CrossRef]
- Sun, C.; Liu, L.; Wang, L.; Li, B.; Jin, C.; Lin, X. Melatonin: A master regulator of plant development and stress responses. J. Integr. Plant Biol. 2021, 63, 126–145. [Google Scholar] [CrossRef]
- Li, D.; Guo, Y.; Zhang, D.; He, S.; Gong, J.; Ma, H.; Gao, X.; Wang, Z.; Jiang, L.; Dun, X.; et al. Melatonin Represses Oil and Anthocyanin Accumulation in Seeds. Plant Physiol. 2020, 183, 898–914. [Google Scholar] [CrossRef]
- Tiwari, R.K.; Lal, M.K.; Kumar, R.; Chourasia, K.N.; Naga, K.C.; Kumar, D.; Das, S.K.; Zinta, G. Mechanistic insights on melatonin-mediated drought stress mitigation in plants. Physiol. Plant 2021, 172, 1212–1226. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.W.; Liu, M.; Zhao, D.; Du, P.; Yan, L.; Liu, D.; Shi, Q.; Yang, C.; Qin, G.; Gong, B. Melatonin confers saline-alkali tolerance in tomato by alleviating nitrosative damage and S-nitrosylation of H+-ATPase 2. Plant Cell 2025, 37, koaf035. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; You, J.; Li, J.; Wang, Y.; Chan, Z. Melatonin promotes Arabidopsis primary root growth in an IAA-dependent manner. J. Exp. Bot. 2021, 72, 5599–5611. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, L.X.; Chen, R.J.; Lu, Y.; Zhang, E.Y.; Miao, J.; Zuo, Z.H.; Zhao, Y.; Zhu, M.; Zhang, Z.; Li, P.; et al. OsCOMT, encoding a caffeic acid O-methyltransferase in melatonin biosynthesis, increases rice grain yield through dual regulation of leaf senescence and vascular development. Plant Biotechnol. J. 2022, 20, 1122–1139. [Google Scholar] [CrossRef]
- Zhao, D.; Luan, Y.; Shi, W.; Tang, Y.; Huang, X.; Tao, J. Melatonin enhances stem strength by increasing lignin content and secondary cell wall thickness in herbaceous peony. J. Exp. Bot. 2022, 73, 5974–5991. [Google Scholar] [CrossRef]
- Byeon, Y.; Choi, G.H.; Lee, H.Y.; Back, K. Melatonin biosynthesis requires N-acetylserotonin methyltransferase activity of caffeic acid O-methyltransferase in rice. J. Exp. Bot. 2015, 66, 6917–6925. [Google Scholar] [CrossRef]
- Wei, Y.X.; Liu, G.Y.; Bai, Y.J.; Xia, F.Y.; He, C.Z.; Shi, H.T. Two transcriptional activators of N-acetylserotonin O-methyltransferase 2 and melatonin biosynthesis in cassava. J. Exp. Bot. 2017, 68, 4997–5006. [Google Scholar] [CrossRef]
- Wei, Y.X.; Liu, G.Y.; Chang, Y.L.; Lin, D.Z.; Reiter, R.J.; He, C.Z.; Shi, H. Melatonin biosynthesis enzymes recruit WRKY transcription factors to regulate melatonin accumulation and transcriptional activity on W-box in cassava. J. Pineal Res. 2018, 65, e12487. [Google Scholar] [CrossRef]
- Bhowal, B.; Bhattacharjee, A.; Goswami, K.; Sanan-Mishra, N.; Singla-Pareek, S.L.; Kaur, C.; Sopory, S. Serotonin and Melatonin Biosynthesis in Plants: Genome-Wide Identification of the Genes and Their Expression Reveal a Conserved Role in Stress and Development. Int. J. Mol. Sci. 2021, 22, 11034. [Google Scholar] [CrossRef]
- Liu, G.F.; Hu, Q.; Zhang, X.; Jiang, J.F.; Zhang, Y.; Zhang, Z.X. Melatonin biosynthesis and signal transduction in plants in response to environmental conditions. J. Exp. Bot. 2022, 73, 5818–5827. [Google Scholar] [CrossRef]
- Xiao, X.C.; Liu, M.Y.; Jiang, M.Q.; Chen, Y.; Xue, X.D.; Zhou, C.Z.; Wu, X.-J.; Wu, J.-N.; Guo, Y.-S.; Yeh, K.-W.; et al. Whole-genome Identification and Expression Analysis of SNAT, ASMT and COMT Families of Melatonin Synthesis Pathway in Dimocarpus longan. Acta Hortic. Sin. 2023, 49, 1031–1046. [Google Scholar]
- Zheng, S.; Zhu, Y.; Liu, C.; Fan, W.; Xiang, Z.; Zhao, A. Genome-wide identification and characterization of genes involved in melatonin biosynthesis in Morus notabilis (wild mulberry). Phytochemistry 2021, 189, 112819. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tan, D.X.; Liang, D.; Chang, C.; Jia, D.; Ma, F. Melatonin mediates the regulation of ABA metabolism, free-radical scavenging, and stomatal behavior in two Malus species under drought stress. J. Exp. Bot. 2025, 66, 669–680. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, H.; Chen, D.; Hao, G. Genetic and evolutionary dissection of melatonin response signaling facilitates the regulation of plant growth and stress responses. J. Pineal. Res. 2023, 74, e12850. [Google Scholar] [CrossRef]
- Gao, T.; Zhang, D.; Shen, W.; Xu, S.; Jia, X.; Liu, X.; Tan, K.; Zhou, Y.; Zhang, Z.; Ma, F.; et al. MdASMT9-mediated melatonin biosynthesis enhances basal thermotolerance in apple plants. Plant Cell Environ. 2024, 47, 751–764. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, Y.; Wang, Y.; Xiao, W.; Khan, M.; Fang, T.; Ming, R.-H.; Dahro, B.; Liu, J.-H.; Jiang, L. The ABF4-bHLH28-COMT5 module regulates melatonin synthesis and root development for drought tolerance in citrus. Plant J. 2025, 121, e70078. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin and its relationship to plant hormones. Ann. Bot. 2018, 121, 195–207. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernandez-Ruiz, J. Melatonin: A new plant hormone and/or a plant master regulator? Trends. Plant Sci. 2019, 24, 38–48. [Google Scholar] [PubMed]
- Ma, W.; Xu, L.; Gao, S.; Lyu, X.; Cao, X.; Yao, Y. Melatonin alters the secondary metabolite profile of grape berry skin by promoting VvMYB14-mediated ethylene biosynthesis. Hortic. Res. 2021, 8, 43. [Google Scholar] [PubMed]
- Wu, H.; Fu, B.; Sun, P.P.; Xiao, C.; Liu, J.H. A NAC Transcription Factor Represses Putrescine Biosynthesis and Affects Drought Tolerance. Plant Physiol. 2016, 172, 1532–1547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, J.; Khan, M.; Wang, Y.; Xiao, W.; Fang, T.; Qu, J.; Xiao, P.; Li, C.; Liu, J.-H. Transcription factors ABF4 and ABR1 synergistically regulate amylase-mediated starch catabolism in drought tolerance. Plant Physiol. 2023, 191, 591–609. [Google Scholar] [CrossRef]
- Wei, T.; Wang, Y.; Liu, J.H. Comparative transcriptome analysis reveals synergistic and disparate defense pathways in the leaves and roots of trifoliate orange (Poncirus trifoliata) autotetraploids with enhanced salt tolerance. Hortic. Res. 2020, 7, 88. [Google Scholar] [CrossRef]
- Liang, S.; Xu, S.; Qu, D.; Yang, L.; Wang, J.; Liu, H.; Xin, W.; Zou, D.; Zheng, H. Identification and Functional Analysis of the Caffeic Acid O-Methyltransferase (COMT) Gene Family in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 8491. [Google Scholar] [CrossRef]
- Zhang, J.; Yao, Z.; Zhang, R.; Mou, Z.; Yin, H.; Xu, T.; Zhao, D.; Chen, S. Genome-Wide Identification and Expression Profile of the SNAT Gene Family in Tobacco (Nicotiana tabacum). Front. Genet. 2020, 11, 591984. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Han, Z.; Zhang, W.; He, L.; Shi, Z.; Ma, X.; Zhou, J.; Si, Z.; Hu, Y.; Zhang, T. Enhancing melatonin biosynthesis in crops through synthetic genetic circuits: A strategy for nutritional fortification in soybean and stress resistance in cotton. Plant Biotechnol. J. 2025. ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Powell, H.R.; Islam, S.A.; David, A.; Sternberg, M.J.E. Phyre2.2: A Community Resource for Template-based Protein Structure Prediction. J. Mol. Biol. 2025, 437, 168960. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rui, C.; Fan, Y.; Xu, N.; Zhang, H.; Wang, J.; Sun, L.; Dai, M.; Ni, K.; Chen, X.; et al. Identification of SNAT Family Genes Suggests GhSNAT3D Functional Reponse to Melatonin Synthesis Under Salinity Stress in Cotton. Front. Mol. Biosci. 2022, 9, 843814. [Google Scholar] [CrossRef]
- Wang, T.; Yang, J.; Cao, J.; Zhang, Q.; Liu, H.; Li, P.; Huang, Y.; Qian, W.; Bi, X.; Wang, H.; et al. MsbZIP55 regulates salinity tolerance by modulating melatonin biosynthesis in alfalfa. Plant Biotechnol. J. 2025, 23, 2125–2139. [Google Scholar] [CrossRef]
- Song, Z.; Yang, Q.; Dong, B.; Li, N.; Wang, M.; Du, T.; Liu, N.; Niu, L.; Jin, H.; Meng, D.; et al. Melatonin enhances stress tolerance in pigeon pea by promoting flavonoid enrichment, particularly luteolin in response to salt stress. J. Exp. Bot. 2022, 73, 5992–6008. [Google Scholar] [CrossRef]


| Name | Arabidopsis | Rice | Trifoliate Orange |
|---|---|---|---|
| TDC | 9 | 18 | 13 |
| T5H | 1 | 1 | 1 |
| SNAT | 24 | 23 | 27 |
| ASMT | 14 | 22 | 48 |
| COMT | 2 | 8 | 7 |
| Total | 50 | 72 | 96 |
| Gene Name | Gene ID | Molecular Weight (kDa) | PI | Protein Length (aa) | Predicted Subcellular Localization | Gene Name | Gene ID | Molecular Weight (kDa) | PI | Protein Length (aa) | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PtASMT1 | Pt1g016180 | 41.13 | 5.86 | 367 | cysk | PtTDC1 | Pt2g002670 | 58.42 | 6.44 | 520 | chlo |
| PtASMT2 | Pt1g016190 | 47.28 | 6.3 | 427 | cyto | PtTDC2 | Pt2g012910 | 55.4 | 6.51 | 500 | cyto |
| PtASMT3 | Pt1g016200 | 33.83 | 8.17 | 300 | cyto | PtTDC3 | Pt2g017480 | 56.29 | 5.31 | 495 | cyto |
| PtASMT4 | Pt2g000920 | 31.49 | 6.95 | 287 | cyto | PtTDC4 | Pt2g023110 | 55.88 | 6.44 | 507 | nucl |
| PtASMT5 | Pt2g000950 | 31.46 | 6.95 | 287 | cyto | PtTDC5 | Pt3g017640 | 56.28 | 6.27 | 499 | cyto |
| PtASMT6 | Pt2g001000 | 31.47 | 7.66 | 287 | cyto | PtTDC6 | Pt3g030000 | 68.56 | 6.47 | 611 | plas |
| PtASMT7 | Pt2g001050 | 31.47 | 7.66 | 287 | cyto | PtTDC7 | Pt4g011940 | 57.83 | 6.86 | 518 | chlo |
| PtASMT8 | Pt2g001070 | 31.46 | 6.72 | 287 | cyto | PtTDC8 | Pt4g011950 | 54.82 | 7.28 | 492 | chlo |
| PtASMT9 | Pt2g006020 | 40.71 | 4.95 | 363 | cysk | PtTDC9 | Pt5g005010 | 63.44 | 6.77 | 568 | chlo |
| PtASMT10 | Pt2g006030 | 41.99 | 5.81 | 377 | nucl | PtTDC10 | Pt5g006660 | 53.49 | 6.3 | 477 | cyto |
| PtASMT11 | Pt2g012050 | 39.4 | 6.13 | 357 | cyto | PtTDC11 | PtUn022800 | 55.4 | 6.51 | 500 | cyto |
| PtASMT12 | Pt2g016230 | 39.66 | 5.7 | 357 | cyto | PtTDC12 | PtUn023160 | 56.3 | 5.31 | 495 | cyto |
| PtASMT13 | Pt2g023820 | 40.22 | 5.25 | 360 | cyto | PtTDC13 | PtUn025060 | 38.89 | 7.23 | 355 | nucl |
| PtASMT14 | Pt2g023830 | 38.83 | 6.83 | 347 | cyto | PtT5H1 | Pt3g004620 | 58.34 | 6.58 | 514 | chlo |
| PtASMT15 | Pt2g025240 | 40.47 | 5.43 | 360 | cyto | PtSNAT1 | Pt1g006020 | 18.47 | 10.15 | 165 | cyto |
| PtASMT16 | Pt2g025250 | 49.14 | 6.4 | 437 | cysk | PtSNAT2 | Pt1g016340 | 32.82 | 10.29 | 282 | chlo |
| PtASMT17 | Pt3g000060 | 39.23 | 6.51 | 354 | cysk | PtSNAT3 | Pt1g020630 | 31.25 | 7.04 | 280 | chlo |
| PtASMT18 | Pt3g000080 | 39.23 | 6.51 | 354 | cysk | PtSNAT4 | Pt2g009040 | 23.71 | 6.51 | 206 | cyto |
| PtASMT19 | Pt3g018450 | 38.86 | 5.89 | 353 | cyto | PtSNAT5 | Pt3g018760 | 30.98 | 9.45 | 272 | chlo |
| PtASMT20 | Pt3g019020 | 25.91 | 6.72 | 237 | cyto | PtSNAT6 | Pt3g018810 | 30.96 | 9.34 | 272 | chlo |
| PtASMT21 | Pt3g019040 | 39.44 | 6.24 | 358 | cyto | PtSNAT7 | Pt3g023130 | 40.93 | 8.3 | 359 | cyto |
| PtASMT22 | Pt3g019060 | 39.05 | 5.55 | 353 | cyto | PtSNAT8 | Pt3g023270 | 10.22 | 6.51 | 92 | chlo |
| PtASMT23 | Pt3g025560 | 26.98 | 9.28 | 250 | cyto | PtSNAT9 | Pt3g023300 | 16.46 | 10.15 | 146 | cyto |
| PtASMT24 | Pt3g026230 | 29.7 | 5.59 | 271 | chlo | PtSNAT10 | Pt3g023330 | 49.14 | 9.56 | 431 | chlo |
| PtASMT25 | Pt3g026240 | 56.01 | 5.19 | 511 | plas | PtSNAT11 | Pt3g025890 | 28.76 | 9.28 | 254 | chlo |
| PtASMT26 | Pt3g026260 | 39.32 | 4.99 | 357 | cyto | PtSNAT12 | Pt3g026850 | 28.76 | 9.28 | 254 | chlo |
| PtASMT27 | Pt3g026300 | 45.66 | 7.71 | 405 | chlo | PtSNAT13 | Pt3g032110 | 25.19 | 9.64 | 217 | chlo |
| PtASMT28 | Pt3g034460 | 39.8 | 6.51 | 359 | cyto | PtSNAT14 | Pt4g001480 | 23.5 | 7.95 | 209 | chlo |
| PtASMT29 | Pt3g034470 | 23.46 | 5.55 | 212 | cyto | PtSNAT15 | Pt4g004060 | 34.71 | 8.92 | 309 | nucl |
| PtASMT30 | Pt4g017220 | 30.71 | 6.25 | 279 | cyto | PtSNAT16 | Pt4g009400 | 26.28 | 10.75 | 237 | chlo |
| PtASMT31 | Pt5g014210 | 40.3 | 6.37 | 357 | cyto | PtSNAT17 | Pt4g017400 | 31.44 | 10.39 | 286 | chlo |
| PtASMT32 | Pt5g025950 | 45.58 | 6.24 | 411 | chlo | PtSNAT18 | Pt4g017610 | 22.85 | 10.38 | 207 | nucl |
| PtASMT33 | Pt6g006840 | 39.33 | 5.83 | 354 | cyto | PtSNAT19 | Pt6g011220 | 27.57 | 5.8 | 248 | chlo |
| PtASMT34 | Pt6g016410 | 27.25 | 5.87 | 249 | cyto | PtSNAT20 | Pt7g005860 | 46.24 | 9.45 | 408 | cyto |
| PtASMT35 | Pt6g018660 | 37.36 | 5.7 | 334 | chlo | PtSNAT21 | Pt7g015310 | 27.64 | 9.7 | 244 | nucl |
| PtASMT36 | Pt7g002460 | 36.37 | 6.4 | 325 | cyto | PtSNAT22 | Pt7g016100 | 45.72 | 8.31 | 417 | cyto |
| PtASMT37 | Pt7g002480 | 36.37 | 6.4 | 325 | cyto | PtSNAT23 | Pt7g016360 | 47.15 | 8.76 | 420 | cyto |
| PtASMT38 | Pt8g001070 | 38.56 | 5.8 | 347 | cysk | PtSNAT24 | Pt8g012150 | 44.19 | 9.23 | 387 | cyto |
| PtASMT39 | Pt9g009220 | 33.66 | 6.96 | 308 | nucl | PtSNAT25 | Pt9g001310 | 45.54 | 8.88 | 400 | chlo |
| PtASMT40 | Pt9g009240 | 28.24 | 6.83 | 255 | cysk | PtSNAT26 | Pt9g005480 | 19.66 | 5.11 | 175 | nucl |
| PtASMT41 | Pt9g009250 | 32.48 | 5.95 | 296 | cyto | PtSNAT27 | PtUn028110 | 49.07 | 9.46 | 431 | cyto |
| PtASMT42 | Pt9g009260 | 32.46 | 6.12 | 296 | cyto | PtCOMT1 | Pt3g014530 | 25.55 | 7.13 | 233 | cyto |
| PtASMT43 | Pt9g010760 | 39.49 | 5.63 | 357 | cyto | PtCOMT2 | Pt3g014540 | 31.71 | 6.95 | 290 | cyto |
| PtASMT44 | Pt9g011910 | 39.68 | 5.63 | 357 | cysk | PtCOMT3 | Pt3g014550 | 23.26 | 6.67 | 214 | extr |
| PtASMT45 | PtUn026750 | 41.24 | 6.06 | 373 | cyto | PtCOMT4 | Pt3g014560 | 30.46 | 6.51 | 280 | cyto |
| PtASMT46 | PtUn029250 | 39.8 | 6.51 | 359 | cyto | PtCOMT5 | Pt3g017050 | 40.02 | 5.83 | 367 | cyto |
| PtASMT47 | PtUn030180 | 30.07 | 6.3 | 275 | cyto | PtCOMT6 | Pt3g017380 | 34.06 | 8.21 | 310 | cyto |
| PtASMT48 | PtUn034040 | 39.03 | 5.55 | 353 | cyto | PtCOMT7 | Pt5g007190 | 33.83 | 6.79 | 312 | nucl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; He, L.; Song, F.; Wang, Z.; Ma, X.; Xiao, C.; Song, X.; Fan, Y.; Wang, C.; Xie, Y.; et al. Genome-Wide Identification and Functional Characterization Under Abiotic Stress of Melatonin Biosynthesis Enzyme Family Genes in Poncirus trifoliata. Agronomy 2025, 15, 2246. https://doi.org/10.3390/agronomy15102246
Zhu J, He L, Song F, Wang Z, Ma X, Xiao C, Song X, Fan Y, Wang C, Xie Y, et al. Genome-Wide Identification and Functional Characterization Under Abiotic Stress of Melatonin Biosynthesis Enzyme Family Genes in Poncirus trifoliata. Agronomy. 2025; 15(10):2246. https://doi.org/10.3390/agronomy15102246
Chicago/Turabian StyleZhu, Jian, Ligang He, Fang Song, Zhijing Wang, Xiaofang Ma, Cui Xiao, Xin Song, Yanjie Fan, Ce Wang, Yun Xie, and et al. 2025. "Genome-Wide Identification and Functional Characterization Under Abiotic Stress of Melatonin Biosynthesis Enzyme Family Genes in Poncirus trifoliata" Agronomy 15, no. 10: 2246. https://doi.org/10.3390/agronomy15102246
APA StyleZhu, J., He, L., Song, F., Wang, Z., Ma, X., Xiao, C., Song, X., Fan, Y., Wang, C., Xie, Y., Jiang, Y., Wu, L., & Zhang, Y. (2025). Genome-Wide Identification and Functional Characterization Under Abiotic Stress of Melatonin Biosynthesis Enzyme Family Genes in Poncirus trifoliata. Agronomy, 15(10), 2246. https://doi.org/10.3390/agronomy15102246






