Improvement of Citrus Yield Prediction Using UAV Multispectral Images and the CPSO Algorithm
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Collection
2.2.1. Yield Data Acquisition
2.2.2. Multispectral Image Acquisition and Processing
2.3. Selections of Vegetable Indices and Texture Features
2.4. Models and Analysis Methods
2.4.1. Machine Learning Models
2.4.2. Compound Coded Particle Swarm Optimization (CPSO)
2.4.3. Shapley Additive Explanations (SHAP)
2.5. Statistical Indicators
3. Results
3.1. Screening of Vegetation Indices and Texture Features
3.2. Using Machine Learning Models to Predict Citrus Fruit Yield
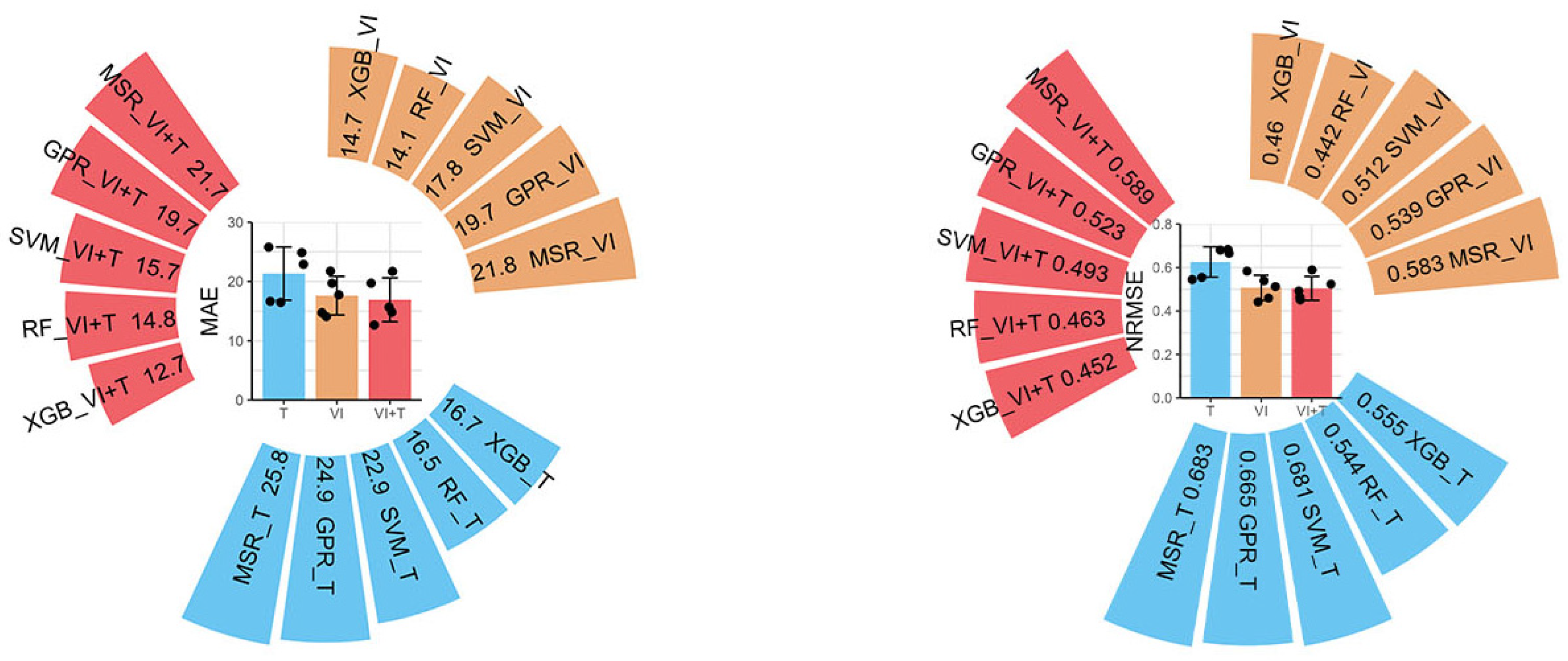
3.2.1. Prediction Models of Citrus Fruit Number in Three Combinations
3.2.2. Prediction Models of Citrus Fruit Quality in Three Combinations
3.3. Using CPSO-Coupled XGB and SVM Models to Predict Citrus Fruit Yield
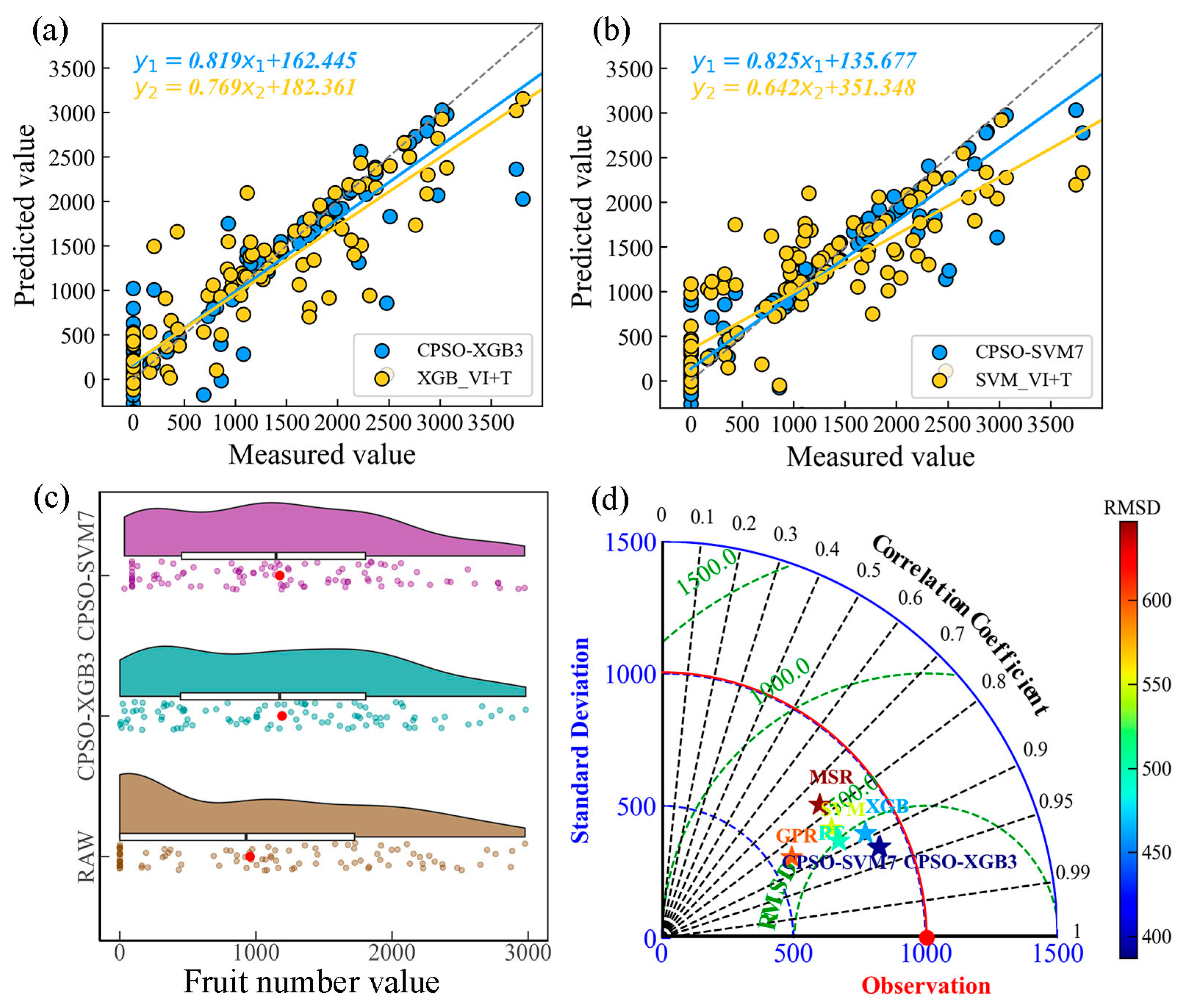
3.3.1. Comparison of CPSO-Optimized Models for Citrus Fruit Number Prediction
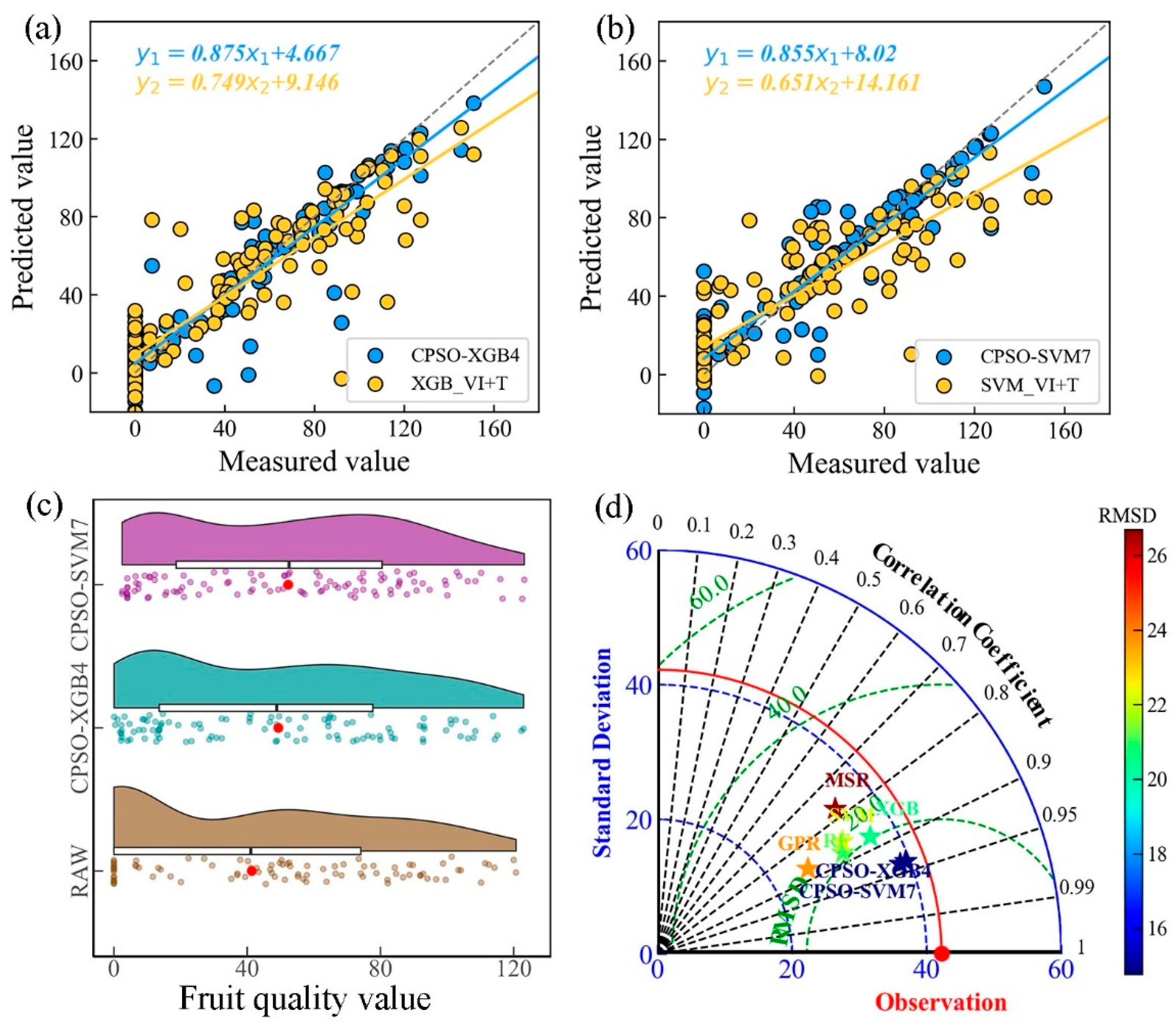
3.3.2. Comparison of CPSO-Optimized Models for Citrus Fruit Quality Prediction
3.4. Analysis of Input Features
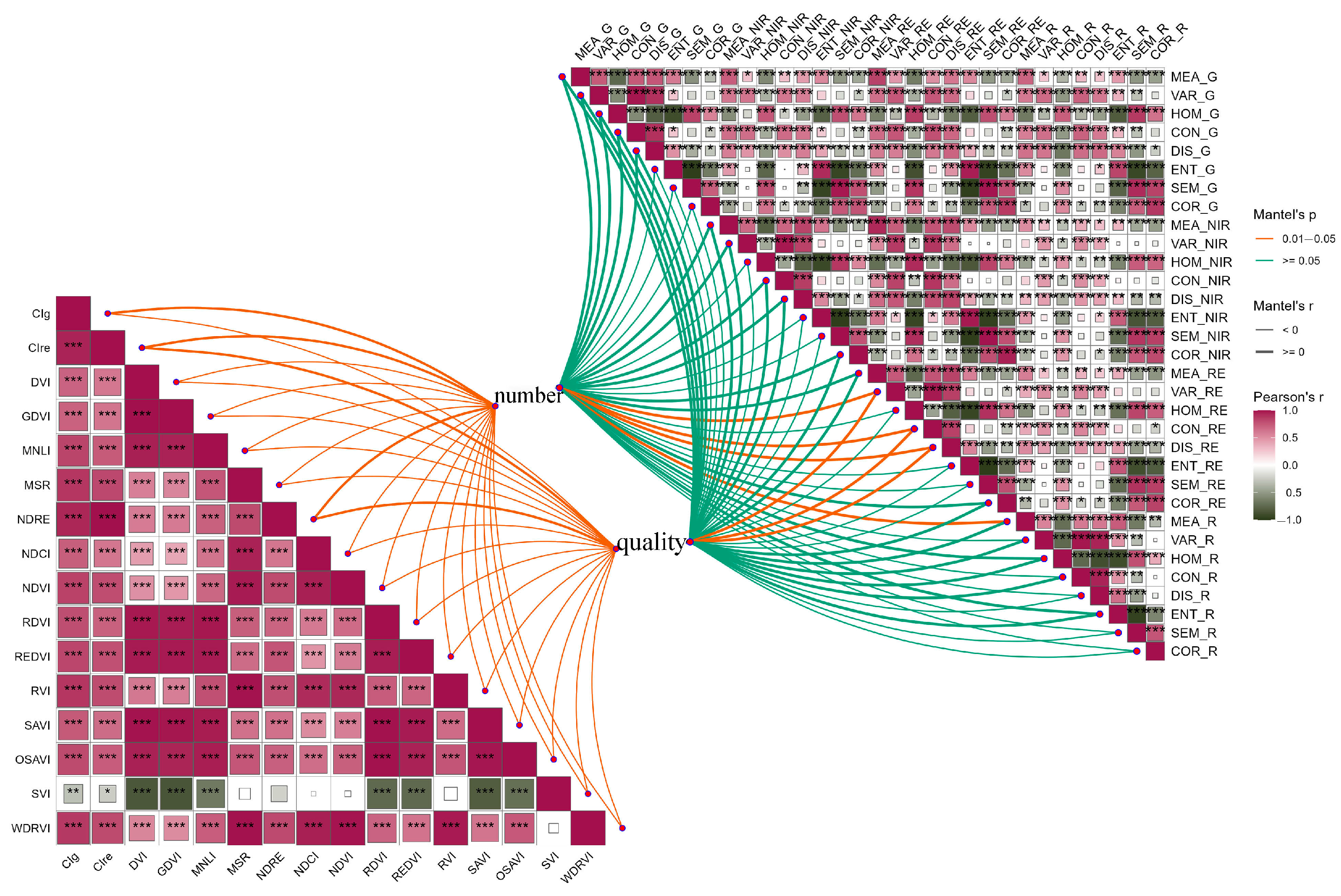
3.4.1. Correlation Analysis
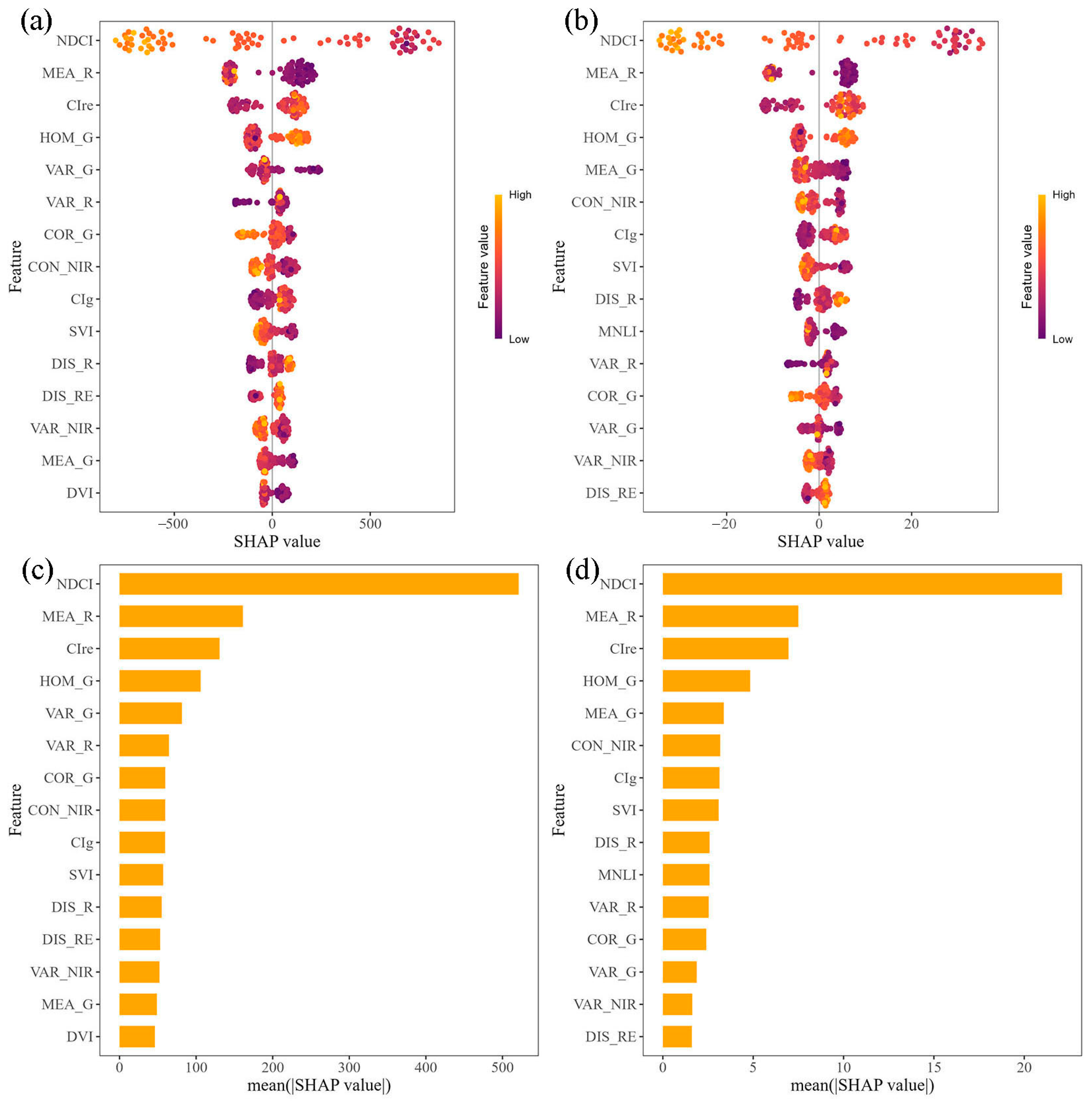
3.4.2. SHAP Analysis
4. Discussion
4.1. Comparison of Different Machine Learning Models
4.2. The Prediction Advantage of Vegetation Indices Combined with Texture Features
4.3. The Prediction Advantage of CPSO-Coupled Models
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Heying, E.; Tanumihardjo, S.A. History, global distribution, and nutritional importance of citrus fruits. Compr. Rev. Food Sci. Saf. 2012, 11, 530–545. [Google Scholar] [CrossRef]
- Saini, R.K.; Ranjit, A.; Sharma, K.; Prasad, P.; Shang, X.; Gowda, K.G.M.; Keum, Y.S. Bioactive compounds of citrus fruits: A review of composition and health benefits of carotenoids, flavonoids, limonoids, and terpenes. Antioxidants 2022, 11, 239. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, X.; Yang, H.; Xu, C.; Wang, S.; Hu, Z. Comparison of fruit mastication trait among different citrus reticulata blanco cv. kinokuni varieties (lines). J. South. Agric. 2023, 54, 3657–3664. [Google Scholar]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Guebsi, R.; Mami, S.; Chokmani, K. Drones in Precision Agriculture: A Comprehensive Review of Applications, Technologies, and Challenges. Drones 2024, 8, 686. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, Z.; Zhao, F.; Liu, J.; Li, Z.; Wang, X.; Gao, Y. An estimation of the leaf nitrogen content of apple tree canopies based on multispectral unmanned aerial vehicle imagery and machine learning methods. Agronomy 2024, 14, 552. [Google Scholar] [CrossRef]
- Zhang, Y.; Ta, N.; Guo, S.; Chen, Q.; Zhao, L.; Li, F.; Chang, Q. Combining spectral and textural information from UAV RGB images for leaf area index monitoring in kiwifruit orchard. Remote Sens. 2022, 14, 1063. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sens. Environ. 2020, 237, 111599. [Google Scholar] [CrossRef]
- Sanches, G.M.; Duft, D.G.; Kölln, O.T.; Luciano, A.C.D.S.; De Castro, S.G.Q.; Okuno, F.M.; Franco, H.C.J. The potential for RGB images obtained using unmanned aerial vehicle to assess and predict yield in sugarcane fields. Int. J. Remote Sens. 2018, 39, 5402–5414. [Google Scholar] [CrossRef]
- Taşan, S.; Cemek, B.; Taşan, M.; Cantürk, A. Estimation of eggplant yield with machine learning methods using spectral vegetation indices. Comput. Electron. Agric. 2022, 202, 107367. [Google Scholar] [CrossRef]
- Lukas, V.; Huňady, I.; Kintl, A.; Mezera, J.; Hammerschmiedt, T.; Sobotková, J.; Brtnický, M.; Elbl, J. Using UAV to identify the optimal vegetation index for yield prediction of oil seed rape (Brassica napus L.) at the flowering stage. Remote Sens. 2022, 14, 4953. [Google Scholar] [CrossRef]
- Van Beek, J.; Tits, L.; Somers, B.; Deckers, T.; Verjans, W.; Bylemans, D.; Janssens, P.; Coppin, P. Temporal dependency of yield and quality estimation through spectral vegetation indices in pear orchards. Remote Sens. 2015, 7, 9886–9903. [Google Scholar] [CrossRef]
- Rotili, D.H.; de Voil, P.; Eyre, J.; Serafin, L.; Aisthorpe, D.; Maddonni, G.Á.; Rodríguez, D. Untangling genotype x management interactions in multi-environment on-farm experimentation. Field Crop Res. 2020, 255, 107900. [Google Scholar] [CrossRef]
- Khaki, S.; Wang, L.; Archontoulis, S.V. A CNN-RNN framework for crop yield prediction. Front. Plant Sci. 2019, 10, 1750. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.Q.; Zhang, C.J.; Xu, B.; Zhu, Y.; Zhao, F.; Han, S.; Yang, G.; Yang, H. Predicting individual apple tree yield using UAV multi-source remote sensing data and ensemble learning. Comput. Electron. Agric. 2022, 201, 107275. [Google Scholar] [CrossRef]
- Rahman, M.M.; Robson, A.; Bristow, M. Exploring the potential of high resolution WorldView-3 imagery for estimating yield of mango. Remote Sens. 2018, 10, 1866. [Google Scholar] [CrossRef]
- Kang, Y.; Wang, Y.; Fan, Y.; Wu, H.; Zhang, Y.; Yuan, B.; Li, H.; Wang, S.; Li, Z. Wheat yield estimation based on unmanned aerial vehicle multispectral images and texture feature indices. Agriculture 2024, 14, 167. [Google Scholar] [CrossRef]
- Zhang, J.; Cheng, J.; Liu, C.; Wu, Q.; Xiong, S.; Yang, H.; Chang, S.; Fu, Y.; Yang, M.; Zhang, S.; et al. Enhanced crop leaf area index estimation via random forest regression: Bayesian optimization and feature selection approach. Remote Sens. 2024, 16, 3917. [Google Scholar] [CrossRef]
- Wei, L.; Huang, C.; Zhong, Y.; Wang, Z.; Hu, X.; Lin, L. Inland waters suspended solids concentration retrieval based on PSO-LSSVM for UAV-borne hyperspectral remote sensing imagery. Remote Sens. 2019, 11, 1455. [Google Scholar] [CrossRef]
- Zeng, L.; Peng, G.; Meng, R.; Man, J.; Li, W.; Xu, B.; Lv, Z.; Sun, R. Wheat yield prediction based on unmanned aerial vehicles-collected red-green-blue imagery. Remote Sens. 2021, 13, 2937. [Google Scholar] [CrossRef]
- Olson, D.; Chatterjee, A.; Franzen, D.W.; Day, S.S. Relationship of Drone-Based Vegetation Indices with Corn and Sugarbeet Yields. Agron. J. 2019, 111, 2545–2557. [Google Scholar] [CrossRef]
- Bellis, E.S.; Hashem, A.A.; Causey, J.L.; Runkle, B.R.K.; Moreno-García, B.; Burns, B.W.; Green, V.S.; Burcham, T.N.; Reba, M.L.; Huang, X. Detecting Intra-Field Variation in Rice Yield With Unmanned Aerial Vehicle Imagery and Deep Learning. Front. Plant Sci. 2022, 13, 716506. [Google Scholar] [CrossRef] [PubMed]
- Murray, H.; Lucieer, A.; Williams, R. Texture-based classification of sub-Antarctic vegetation communities on Heard Island. Int. J. Appl. Earth Obs. 2010, 12, 138–149. [Google Scholar] [CrossRef]
- Kelcey, J.; Lucieer, A. Sensor correction and radiometric calibration of a 6-band multispectral imaging sensor for UAV remote sensing. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, 39, 393–398. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Viña, A.; Ciganda, V.; Rundquist, D.C.; Arkebauer, T.J. Remote estimation of canopy chlorophyll content in crops. Geophys. Res. Lett. 2005, 32, L08403. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Gong, P.; Pu, R.L.; Biging, G.S.; Larrieu, M.R. Estimation of forest leaf area index using vegetation indices derived from hyperion hyperspectral data. Ieee T. Geosci. Remote 2003, 41, 1355–1362. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of vegetation indices and a modified simple ratio for boreal applications. Can. J. Remote Sens. 2014, 22, 229–242. [Google Scholar] [CrossRef]
- Peng, Y.; Gitelson, A.A. Remote estimation of gross primary productivity in soybean and maize based on total crop chlorophyll content. Remote Sens. Environ. 2012, 117, 440–448. [Google Scholar] [CrossRef]
- Mishra, S.; Mishra, D.R. Normalized difference chlorophyll index:a novel model for remote estimation of chlorophyll-a concentration in turbid productive waters. Remote Sens. Environ. 2012, 117, 394–406. [Google Scholar] [CrossRef]
- Liu, H.Q.; Huete, A. A feedback based modification of the NDVI to minimize canopy background and atmospheric noise. IEEE T. Geosci. Remote 1995, 33, 457–465. [Google Scholar] [CrossRef]
- Roujean, J.L.; Breon, F.M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Cao, Q.; Miao, Y.X.; Wang, H.Y.; Huang, S.; Cheng, S.; Khosla, R.; Jiang, R. Non-destructive estimation of rice plant nitrogen status with crop circle multispectral active canopy sensor. Field Crop Res. 2013, 154, 133–144. [Google Scholar] [CrossRef]
- Birth, G.S.; McVey, G.R. Measuring the color of growing turf with a reflectance spectrophotometer. Agron. J. 1968, 60, 640–643. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Perry, J.C.R.; Lautenschlager, L.F. Functional equivalence of spectral vegetation indices. Remote Sens. Environ. 1984, 14, 169–182. [Google Scholar] [CrossRef]
- Gitelson, A.A. Wide dynamic range vegetation index for remote quantification of biophysical characteristics of vegetation. J. Plant Physiol. 2004, 161, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE T. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Vapnik, V.; Izmailov, R. Knowledge transfer in SVM and neural networks. Ann. Math. Artif. Intel. 2017, 81, 3–19. [Google Scholar] [CrossRef]
- Wang, J. An intuitive tutorial to gaussian process regression. Comput. Sci. Eng. 2023, 25, 4–11. [Google Scholar] [CrossRef]
- Liu, Y.; Heuvelink, G.B.M.; Bai, Z.; He, P.; Xu, X.; Ding, W.; Huang, S. Analysis of spatio-temporal variation of crop yield in China using stepwise multiple linear regression. Field Crop Res. 2021, 264, 108098. [Google Scholar] [CrossRef]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95-International Conference on Neural Networks, Perth, Australia, 27 November—1 December 1995; pp. 1942–1948. [Google Scholar]
- Mojtaba Ahmadieh, K.; Mohammad, T.; Mahdi Aliyari, S. A novel binary particle swarm optimization. In Proceedings of the 2007 Mediterranean Conference on Control & Automation, Athens, Greece, 27–29 June 2007; pp. 1–6. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Huang, J.-C.; Ko, K.-M.; Shu, M.-H.; Hsu, B.-M. Application and comparison of several machine learning algorithms and their integration models in regression problems. Neural Comput. Appl. 2019, 32, 5461–5469. [Google Scholar] [CrossRef]
- Pei, S.; Dai, Y.; Bai, Z.; Li, Z.; Zhang, F.; Yin, F.; Fan, J. Improved estimation of canopy water status in cotton using vegetation indices along with textural information from UAV-based multispectral images. Comput. Electron. Agric. 2024, 224, 109176. [Google Scholar] [CrossRef]
- Guimarães, N.; Sousa, J.J.; Couto, P.; Bento, A.; Pádua, L. Combining UAV-Based Multispectral and Thermal Infrared Data with Regression Modeling and SHAP Analysis for Predicting Stomatal Conductance in Almond Orchards. Remote Sens. 2024, 16, 1–19. [Google Scholar] [CrossRef]
- Mo, X.; Chen, C.; Riaz, M.; Moussa, M.G.; Chen, X.; Wu, S.; Tan, Q.; Sun, X.; Zhao, X.; Shi, L.; et al. Fruit characteristics of citrus trees grown under different soil cu levels. Plants 2022, 11, 2943. [Google Scholar] [CrossRef] [PubMed]
- Qiao, D.; Yang, J.; Bai, B.; Li, G.; Wang, J.; Li, Z.; Liu, J.; Liu, J. Non-destructive monitoring of peanut leaf area index by combing UAV spectral and textural characteristics. Remote Sens. 2024, 16, 2182. [Google Scholar] [CrossRef]
- Kwak, G.-H.; Park, N.-W. Impact of Texture Information on Crop Classification with Machine Learning and UAV Images. Appl. Sci. 2019, 9, 643. [Google Scholar] [CrossRef]
- Dhakal, R.; Maimaitijiang, M.; Chang, J.; Caffe, M. Utilizing Spectral, Structural and Textural Features for Estimating Oat Above-Ground Biomass Using UAV-Based Multispectral Data and Machine Learning. Sensors 2023, 23, 9708. [Google Scholar] [CrossRef] [PubMed]
- Honeycutt, C.E.; Plotnick, R. Image analysis techniques and gray-level co-occurrence matrices (GLCM) for calculating bioturbation indices and characterizing biogenic sedimentary structures. Comput. Geosci. 2008, 34, 1461–1472. [Google Scholar] [CrossRef]
- Yang, N.; Zhang, Z.; Zhang, J.; Guo, Y.; Yang, X.; Yu, G.; Bai, X.; Chen, J.; Chen, Y.; Shi, L.; et al. Improving estimation of maize leaf area index by combining of UAV-based multispectral and thermal infrared data: The potential of new texture index. Comput. Electron. Agric. 2023, 214, 108294. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, T.; Zhou, M.; Li, D.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Improved estimation of rice aboveground biomass combining textural and spectral analysis of UAV imagery. Precis. Agric. 2018, 20, 611–629. [Google Scholar] [CrossRef]
- Morales-Hernández, A.; Van Nieuwenhuyse, I.; Rojas Gonzalez, S. A survey on multi-objective hyperparameter optimization algorithms for machine learning. Artif. Intell. Rev. 2022, 56, 8043–8093. [Google Scholar] [CrossRef]
- Barbieri, M.C.; Grisci, B.I.; Dorn, M. Analysis and comparison of feature selection methods towards performance and stability. Expert Syst. Appl. 2024, 249, 123667. [Google Scholar] [CrossRef]


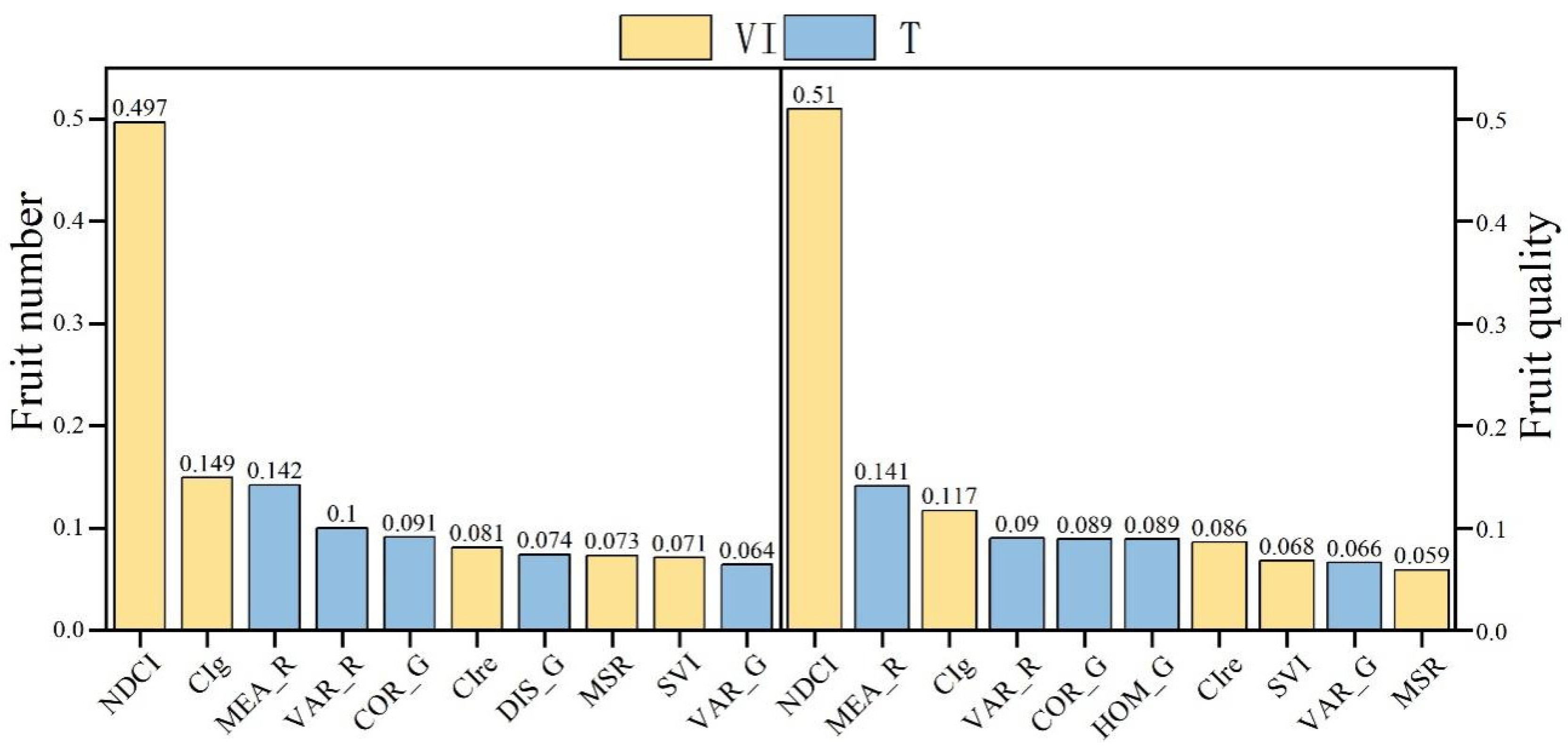
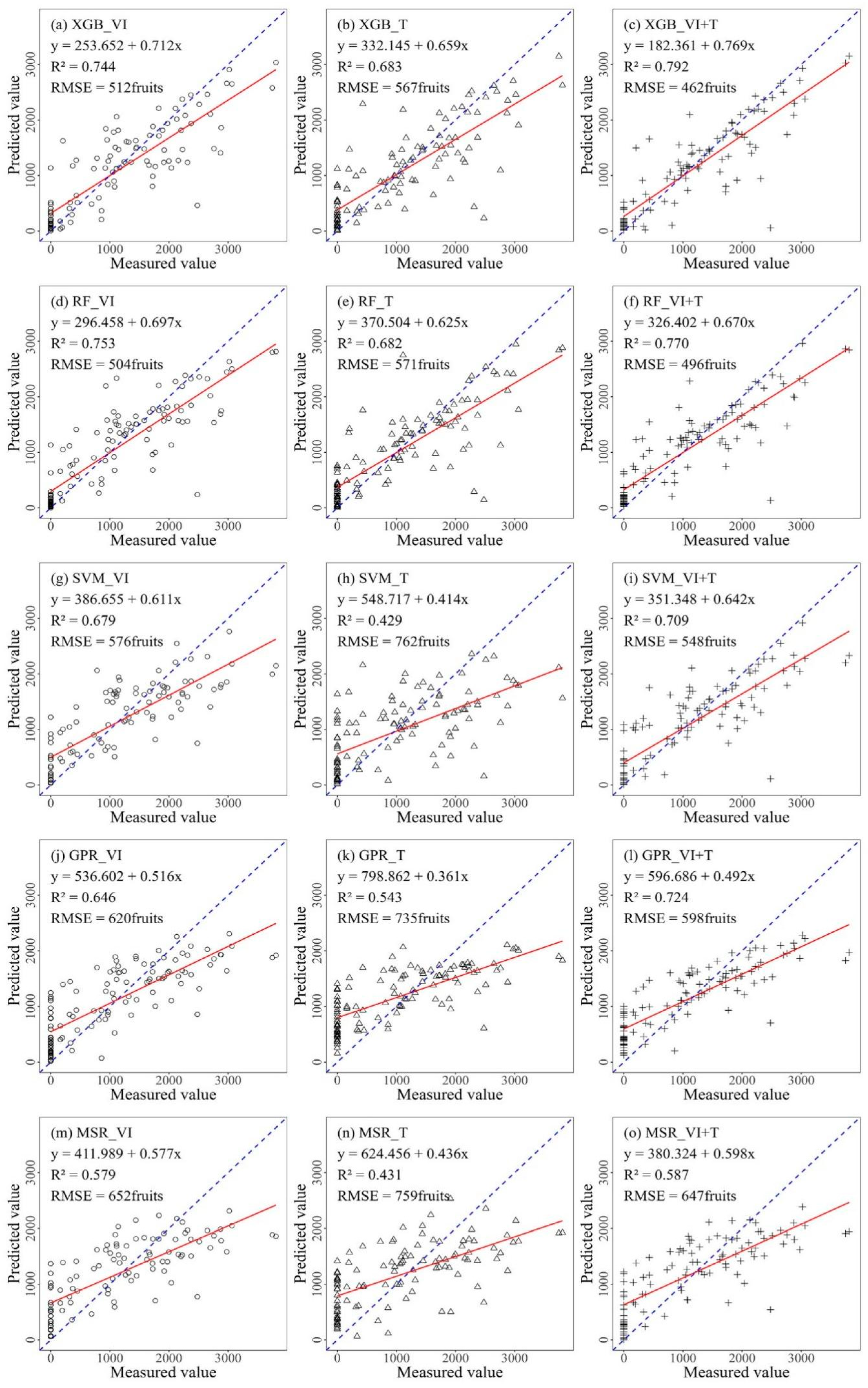
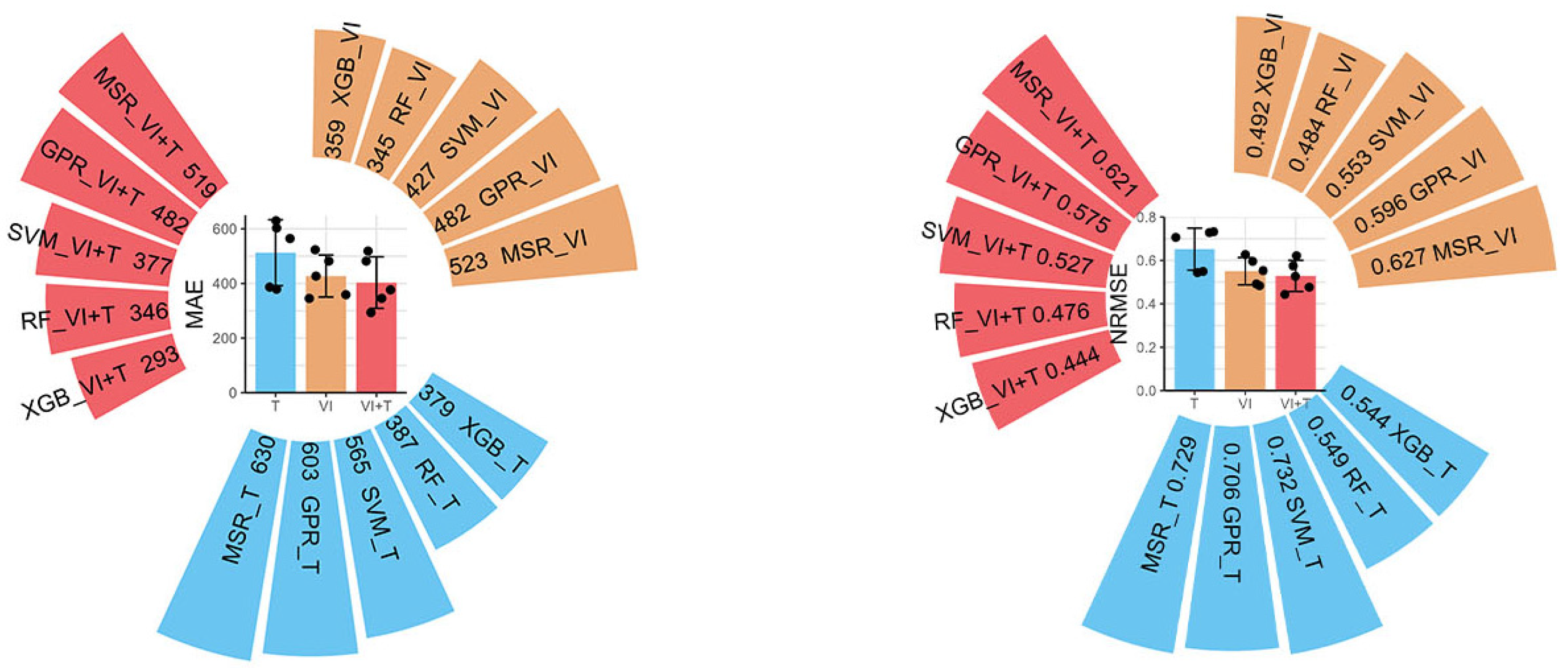
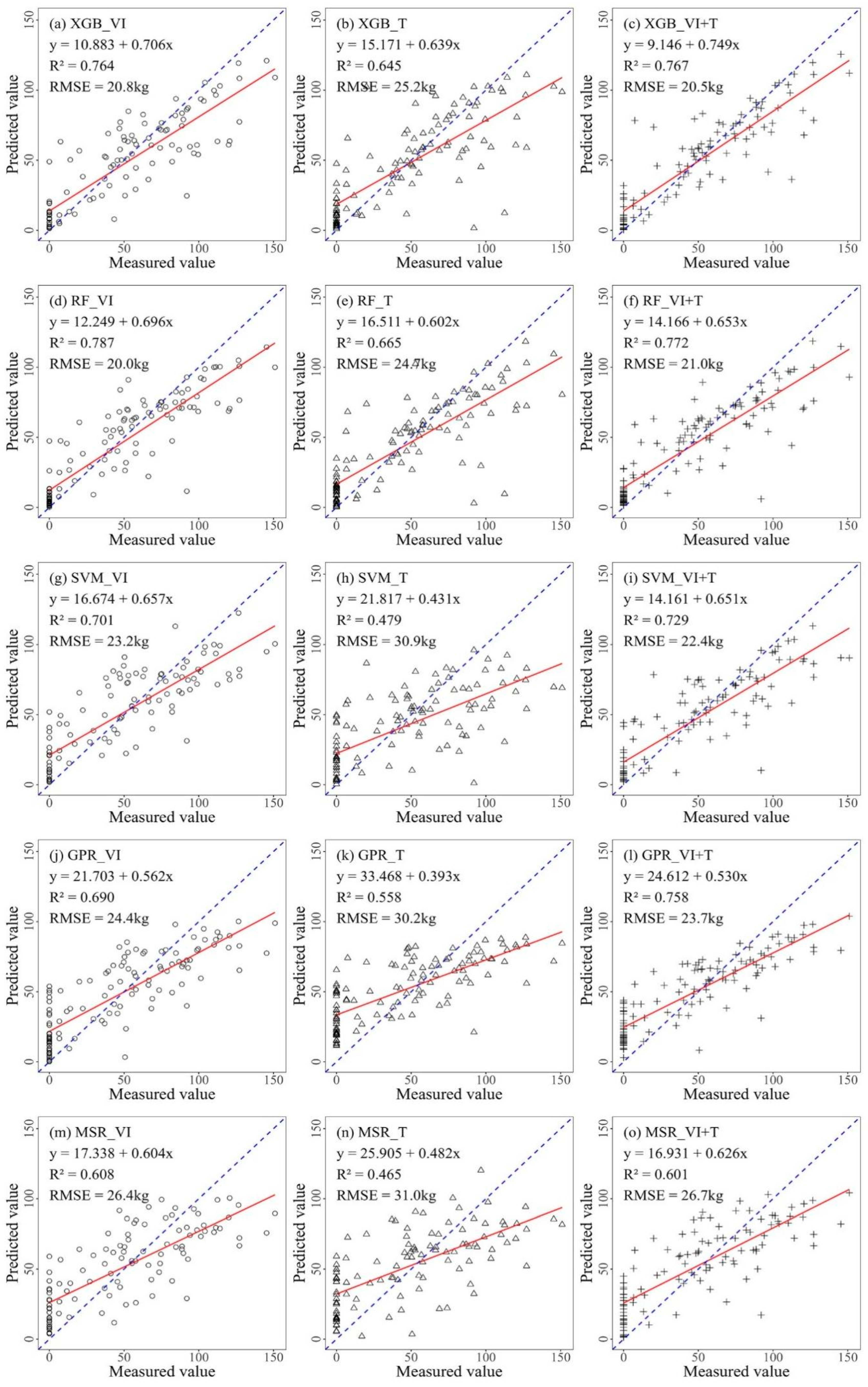
| Max | Min | Median | Mean | Standard Deviation | Coefficient of Variation | Kurtosis | Skewness | |
|---|---|---|---|---|---|---|---|---|
| Number/fruits | 3890 | 0 | 960 | 1041 | 1005 | 0.965 | −0.563 | 0.630 |
| Quality/kg | 151.0 | 0.0 | 44.5 | 45.4 | 42.2 | 0.931 | −0.902 | 0.472 |
| UAV | Description | Sensor | Description |
|---|---|---|---|
| Name | DJI M3M | Bands | Green (560 nm ± 16 nm) |
| Flight altitude | 50 m | Red (650 nm ± 16 nm) | |
| Flight speed | 4.4 m/s | Red Edge (730 nm ± 16 nm) | |
| Satellite systems | GPS + Galileo + BeiDou | NIR (860 nm ± 26 nm) | |
| Forward overlap | 80% | Pixel | 5 million |
| Side overlap | 80% | Image dimension | 2592 × 1944 |
| Field of view | 90° | Resolution | 2.31 cm/pixel |
| Shooting interval | 2 s | Image format | TIFF |
| No. | Vegetable Index | Equations | Reference |
|---|---|---|---|
| 1 | Green chlorophyll index | CIg = NIR/G − 1 | [25] |
| 2 | Red edge chlorophyll index | CIre = NIR/RE − 1 | [25] |
| 3 | Difference vegetation index | DVI = NIR − R | [26] |
| 4 | Green difference vegetation index | GDVI = NIR − G | [26] |
| 5 | Modified nonlinear index (MNLI) | MNLI = 1.5 × (NIR2 − R)/(NIR2 + R + 0.5) | [27] |
| 6 | Modified simple ratio (MSR) | MSR = (NIR/R − 1)/sqrt (NIR/R + 1) | [28] |
| 7 | Normalized difference red edge | NDRE = (NIR − RE)/(NIR + RE) | [29] |
| 8 | Normalized difference chlorophyll index | NDCI = (RE − R)/(RE + R) | [30] |
| 9 | Normalized difference vegetation index | NDVI = (NIR − R)/(NIR + R) | [31] |
| 10 | Renormalized difference vegetation index | RDVI = (NIR − R)/sqrt (NIR + R) | [32] |
| 11 | Red edge difference vegetation index | REDVI = NIR − RE | [33] |
| 12 | Ratio vegetation index | RVI = NIR/R | [34] |
| 13 | Soil-adjusted vegetation index | SAVI = 1.5 × (NIR − R)/(NIR + R + 0.5) | [35] |
| 14 | Optimized soil-adjusted vegetation index | OSAVI = (NIR − R)/(NIR + R + 0.16) | [36] |
| 15 | Spectrum vegetation index | SVI = (NIR − R)/(NIR + R)/NIR | [37] |
| 16 | Wide dynamic range vegetation index | WDRVI = (0.12 × NIR − R)/(0.12 × NIR + R) | [38] |
| No. | Texture Feature | Equations | Reference |
|---|---|---|---|
| 1 | Mean | [39] | |
| 2 | Variance | ||
| 3 | Homogeneity | ||
| 4 | Contrast | ||
| 5 | Dissimilarity | ||
| 6 | Entropy | ||
| 7 | Second moment | ||
| 8 | Correlation |
| Model | Number of Input | Input Factors | R2 | RMSE (Fruits) | MAE (Fruits) | NRMSE |
|---|---|---|---|---|---|---|
| CPSO-XGB1 | 3 | CIg, NDCI, COR_RE | 0.819 | 432 | 230 | 0.415 |
| CPSO-XGB2 | 4 | CIg, NDCI, MEA_G, COR_RE | 0.803 | 451 | 300 | 0.434 |
| CPSO-XGB3 | 5 | CIg, NDCI, COR_G, SEM_NIR, MEA_RE | 0.853 | 387 | 197 | 0.372 |
| CPSO-XGB4 | 6 | CIg, NDCI, SEM_G, MEA_RE, HOM_RE, COR_R | 0.811 | 441 | 244 | 0.424 |
| CPSO-XGB5 | 7 | CIg, NDCI, SEM_G, ENT_NIR, MEA_RE, VAR_RE, SEM_RE | 0.850 | 395 | 213 | 0.380 |
| CPSO-XGB6 | 8 | CIg, NDCI, COR_G, MEA_NIR, CON_NIR, DIS_NIR, MEA_RE, ENT_RE | 0.835 | 417 | 259 | 0.401 |
| CPSO-XGB7 | 9 | CIg, DVI, NDCI, MEA_G, VAR_G, COR_G, VAR_NIR, MEA_RE, DIS_R | 0.816 | 435 | 225 | 0.418 |
| CPSO-SVM1 | 3 | CIg, WDRVI, COR_G | 0.780 | 474 | 321 | 0.456 |
| CPSO-SVM2 | 4 | NDCI, RVI, MEA_G, VAR_R | 0.734 | 520 | 372 | 0.499 |
| CPSO-SVM3 | 5 | NDCI, NDVI, WDRVI, MEA_G, VAR_R | 0.733 | 520 | 367 | 0.500 |
| CPSO-SVM4 | 6 | CIg, NDCI, RVI, MEA_G, VAR_R, CON_R | 0.775 | 482 | 334 | 0.463 |
| CPSO-SVM5 | 7 | CIg, NDVI, MEA_G, COR_G, COR_NIR, MEA_R, VAR_R | 0.825 | 424 | 266 | 0.408 |
| CPSO-SVM6 | 8 | CIg, DVI, NDCI, RDVI, MEA_G, ENT_NIR, HOM_RE, VAR_R | 0.828 | 425 | 277 | 0.408 |
| CPSO-SVM7 | 9 | NDCI, NDVI, SVI, VAR_G, MEA_NIR, HOM_NIR, MEA_RE, VAR_RE, MEA_R | 0.852 | 391 | 234 | 0.375 |
| Model | Number of Input | Input Factors | R2 | RMSE (kg) | MAE (kg) | NRMSE |
|---|---|---|---|---|---|---|
| CPSO-XGB1 | 3 | CIg, NDCI, SEM_G | 0.749 | 21.3 | 16.5 | 0.47 |
| CPSO-XGB2 | 4 | CIre, NDCI, ENT_G, SEM_G | 0.83 | 17.8 | 12.7 | 0.392 |
| CPSO-XGB3 | 5 | CIg, DVI, NDCI, SEM_G, ENT_RE | 0.844 | 17 | 12 | 0.374 |
| CPSO-XGB4 | 6 | CIg, DVI, NDCI, MEA_G,DIS_NIR, SEM_RE | 0.878 | 14.8 | 9.3 | 0.326 |
| CPSO-XGB5 | 7 | CIg, CIre, NDCI, REDVI, ENT_RE, MEA_R, VAR_R | 0.746 | 21.6 | 17.6 | 0.477 |
| CPSO-XGB6 | 8 | CIg, NDCI, REDVI, MEA_G, HOM_G, COR_G, MEA_NIR, ENT_NIR | 0.867 | 15.6 | 10.2 | 0.344 |
| CPSO-XGB7 | 9 | CIg, MSR, NDCI, REDVI, HOM_G, ENT_G, DIS_RE, SEM_RE, CON_R | 0.874 | 15.4 | 9.7 | 0.34 |
| CPSO-SVM1 | 3 | CIg, NDCI, COR_G | 0.829 | 17.5 | 11.9 | 0.387 |
| CPSO-SVM2 | 4 | NDCI, WDRVI, MEA_G, VAR_R | 0.772 | 20.2 | 14.6 | 0.446 |
| CPSO-SVM3 | 5 | MSR, NDCI, RVI, MEA_G, MEA_R | 0.776 | 20.1 | 14.8 | 0.443 |
| CPSO-SVM4 | 6 | CIg, MSR, NDVI, MEA_G, MEA_R, VAR_R | 0.78 | 19.8 | 14.4 | 0.437 |
| CPSO-SVM5 | 7 | MSR, NDRE, NDCI, WDRVI, MEA_G, MEA_R, CON_R | 0.783 | 19.8 | 13.3 | 0.436 |
| CPSO-SVM6 | 8 | CIg, MNLI, RVI, SVI, ENT_G, SEM_NIR, MEA_R, SEM_R | 0.841 | 17 | 11.2 | 0.375 |
| CPSO-SVM7 | 9 | CIg, MNLI, SVI, WDRVI, SEM_NIR, MEA_RE, HOM_RE, MEA_R, VAR_R | 0.88 | 14.8 | 9.7 | 0.326 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Liu, X.; Dong, J.; Tan, J.; Wang, X.; Wang, X.; Wu, L. Improvement of Citrus Yield Prediction Using UAV Multispectral Images and the CPSO Algorithm. Agronomy 2025, 15, 171. https://doi.org/10.3390/agronomy15010171
Xu W, Liu X, Dong J, Tan J, Wang X, Wang X, Wu L. Improvement of Citrus Yield Prediction Using UAV Multispectral Images and the CPSO Algorithm. Agronomy. 2025; 15(1):171. https://doi.org/10.3390/agronomy15010171
Chicago/Turabian StyleXu, Wenhao, Xiaogang Liu, Jianhua Dong, Jiaqiao Tan, Xulei Wang, Xinle Wang, and Lifeng Wu. 2025. "Improvement of Citrus Yield Prediction Using UAV Multispectral Images and the CPSO Algorithm" Agronomy 15, no. 1: 171. https://doi.org/10.3390/agronomy15010171
APA StyleXu, W., Liu, X., Dong, J., Tan, J., Wang, X., Wang, X., & Wu, L. (2025). Improvement of Citrus Yield Prediction Using UAV Multispectral Images and the CPSO Algorithm. Agronomy, 15(1), 171. https://doi.org/10.3390/agronomy15010171






