Exploring the Flavonoid Biosynthesis Pathway of Two Ecotypes of Leymus chinensis Using Transcriptomic and Metabolomic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Transcriptome Analysis
2.2.1. Gene Expression and Functional Annotation
2.2.2. Differential Expression and Functional Enrichment Analysis
2.2.3. qRT-PCR
2.2.4. Metabolite Extraction from Fresh Foliage and Analysis via UPLC-MS
2.2.5. Identification and Statistical Analysis of Metabolites Using UHPLC-MS
2.2.6. Gradient Boosting Machine and Random Forest Regression
2.2.7. Association Evaluation between Transcriptomic and Metabolomic Data
3. Results
3.1. Metabolomic Analysis of Two Ecotypes of Leymus chinensis
3.2. Synopsis of RNA-Seq Analysis
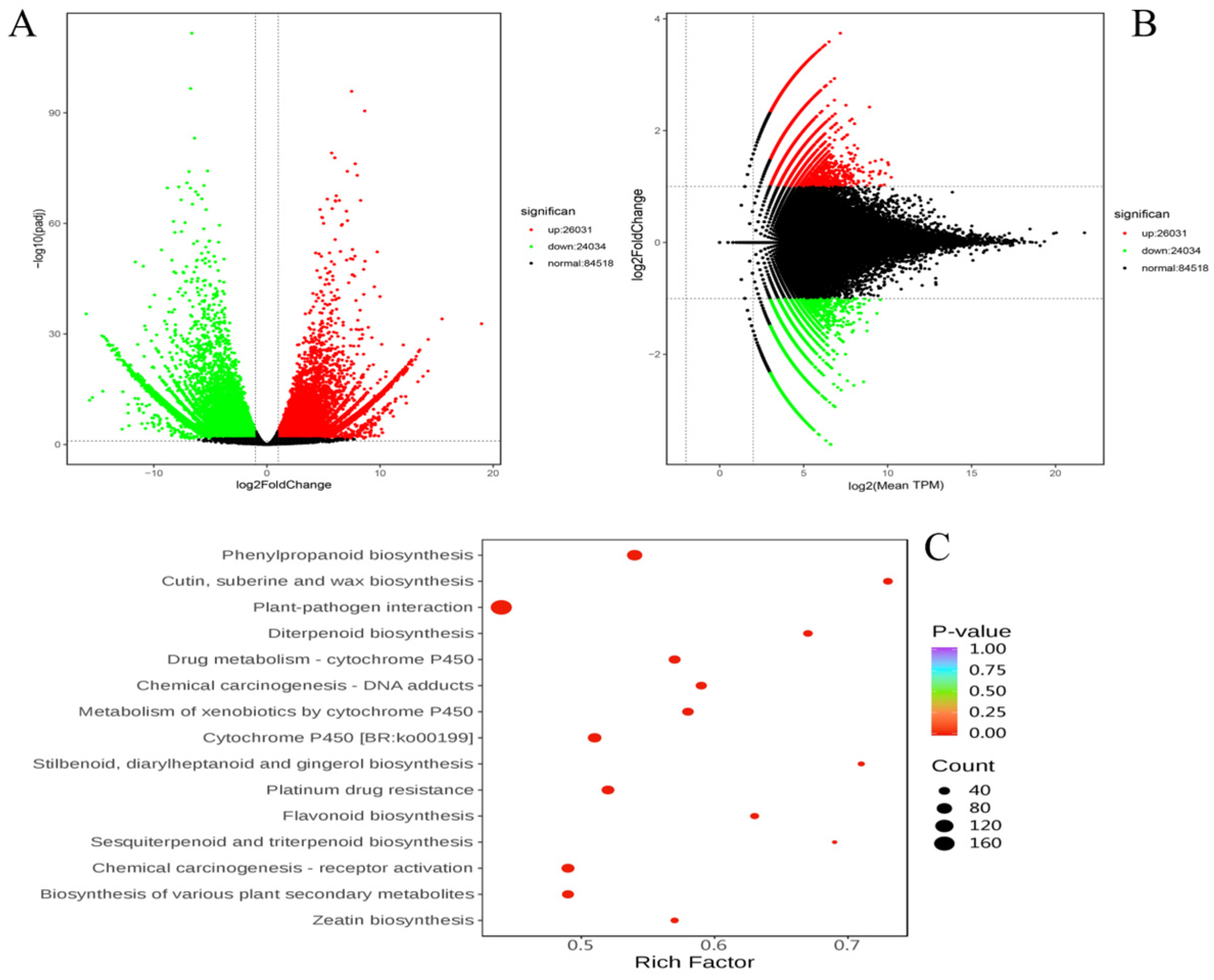
3.3. Differential Gene Expression Analysis
3.4. Integration of Machine Learning Models for Predictive Analysis
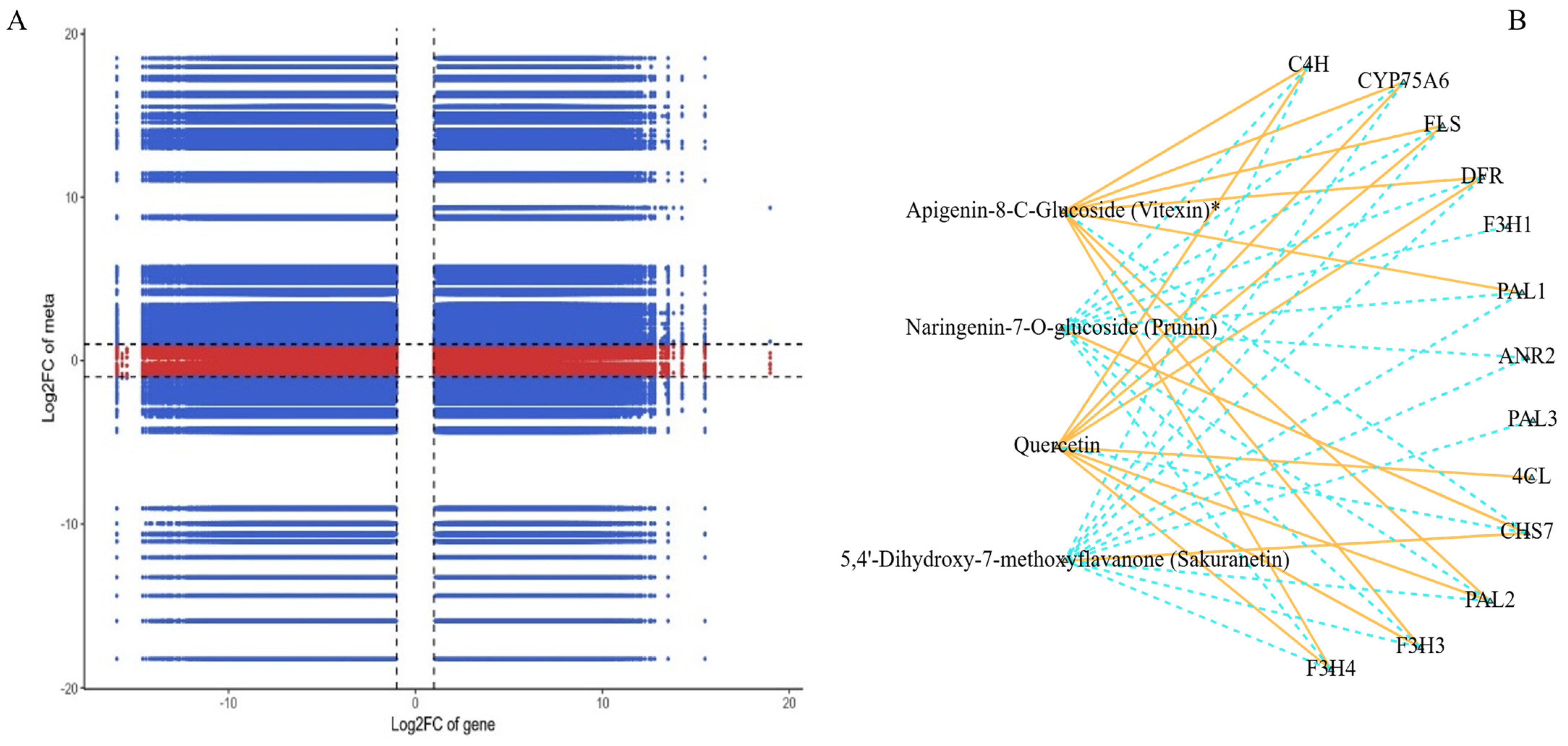
3.5. Association Study Comparing Transcriptomic and Metabolomic Data
3.6. qRT-PCR
4. Discussion
4.1. Differential Gene Expression Analysis
4.2. Metabolomic Profiling and Pathway Analysis
4.3. Integrating Gene Expression with Metabolism: Impacts on Plant Adaptation and Breeding
4.3.1. Gene Expression and Metabolite Accumulation
4.3.2. Detailed Insights from Supplementary Data
4.3.3. Pathway Enrichment and Biological Significance
4.3.4. Implications for Plant Adaptation and Breeding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Jia, J.; Zhao, P.; Guo, X.; Chen, S.; Qi, D.; Cheng, L.; Liu, G. LcMYB4, an unknown function transcription factor gene from sheepgrass, as a positive regulator of chilling and freezing tolerance in transgenic Arabidopsis. BMC Plant Biol. 2020, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Shi, B.; Zhong, S.; Chai, H.; Li, S.; Wang, Y.; Henry, H.A.L.; Ma, J.-Y.; Sun, W. Drought sensitivity of aboveground productivity in Leymus chinensis meadow steppe depends on drought timing. Oecologia 2019, 191, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Shi, Y.; Mu, C.; Wang, J. Differences in Organic Solute and Metabolites of Leymus chinensis in Response to Different Intensities of Salt and Alkali Stress. Plants 2023, 12, 1916. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, Y.; Liu, Q.; Deng, S.; Jin, X.; Yin, Y.; Guo, J.; Li, N.; Liu, Y.; Han, S.; et al. A Na2CO3-Responsive Chitinase Gene From Leymus chinensis Improve Pathogen Resistance and Saline-Alkali Stress Tolerance in Transgenic Tobacco and Maize. Front. Plant Sci. 2020, 11, 504. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Bhakta, S.; Negi, S.; Tak, H.; Singh, S.; Ganapathi, T.R. MusaATAF2-like protein regulates shoot development and multiplication by inducing cytokinin hypersensitivity and flavonoid accumulation in banana plants. Plant Cell Rep. 2022, 41, 1197–1208. [Google Scholar] [CrossRef]
- Chen, X.-L.; Sun, M.-C.; Chong, S.-L.; Si, J.-P.; Wu, L.-S. Transcriptomic and Metabolomic Approaches Deepen Our Knowledge of Plant–Endophyte Interactions. Front. Plant Sci. 2022, 12, 700200. [Google Scholar] [CrossRef] [PubMed]
- Bag, S.; Mondal, A.; Majumder, A.; Mondal, S.K.; Banik, A. Flavonoid mediated selective cross-talk between plants and beneficial soil microbiome. Phytochem. Rev. 2022, 21, 1739–1760. [Google Scholar] [CrossRef]
- George, V.C.; Dellaire, G.; Rupasinghe, H.V. Plant flavonoids in cancer chemoprevention: Role in genome stability. J. Nutr. Biochem. 2017, 45, 1–14. [Google Scholar] [CrossRef]
- Jiang, N.; Doseff, A.I.; Grotewold, E. Flavones: From Biosynthesis to Health Benefits. Plants 2016, 5, 27. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef] [PubMed]
- Tuenter, E.; Creylman, J.; Verheyen, G.; Pieters, L.; Van Miert, S. Development of a classification model for the antigenotoxic activity of flavonoids. Bioorganic Chem. 2020, 98, 103705. [Google Scholar] [CrossRef]
- Arikan, B.; Yildiztugay, E.; Ozfidan-Konakci, C. Protective role of quercetin and kaempferol against oxidative damage and photosynthesis inhibition in wheat chloroplasts under arsenic stress. Physiol. Plant. 2023, 175, e13964. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Li, J.; Chen, X.; Fu, X.; Sun, S.; Wu, Q. Chalcone Derivatives: Role in Anticancer Therapy. Biomolecules 2021, 11, 894. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, G.; Huang, T.; Zhang, T.; Lin, J.; Song, L.; Zhou, G.; Ma, X.; Ge, Y.; Xu, Y.; et al. Exogenous tannic acid relieves imidacloprid-induced oxidative stress in tea tree by activating antioxidant responses and the flavonoid biosynthetic pathway. Ecotoxicol. Environ. Saf. 2023, 266, 115557. [Google Scholar] [CrossRef]
- Ksila, M.; Ghzaiel, I.; Pires, V.; Ghrairi, T.; Masmoudi-Kouki, O.; Latruffe, N.; Vervandier-Fasseur, D.; Vejux, A.; Lizard, G. Characterization of Cell Death Induced by Imine Analogs of Trans-Resveratrol: Induction of Mitochondrial Dysfunction and Overproduction of Reactive Oxygen Species Leading to, or Not, Apoptosis without the Increase in the S-Phase of the Cell Cycle. Molecules 2023, 28, 3178. [Google Scholar] [CrossRef]
- Hao, Q.; Henning, S.M.; Magyar, C.E.; Said, J.; Zhong, J.; Rettig, M.B.; Vadgama, J.V.; Wang, P. Enhanced Chemoprevention of Prostate Cancer by Combining Arctigenin with Green Tea and Quercetin in Prostate-Specific Phosphatase and Tensin Homolog Knockout Mice. Biomolecules 2024, 14, 105. [Google Scholar] [CrossRef] [PubMed]
- Díaz, L.; Del Río, J.A.; Pérez-Gilabert, M.; Ortuño, A. Involvement of an extracellular fungus laccase in the flavonoid metabolism in Citrus fruits inoculated with Alternaria alternata. Plant Physiol. Biochem. 2015, 89, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Fontana, R.; Caproni, A.; Sicurella, M.; Manfredini, S.; Baldisserotto, A.; Marconi, P. Effects of Flavonoids and Phenols from Moringa oleifera Leaf Extracts on Biofilm Processes in Xanthomonas campestris pv. campestris. Plants 2023, 12, 1508. [Google Scholar] [CrossRef]
- Chen, Y.; Li, W.; Jia, K.; Liao, K.; Liu, L.; Fan, G.; Zhang, S.; Wang, Y. Metabolomic and transcriptomice analyses of flavonoid biosynthesis in apricot fruits. Front. Plant Sci. 2023, 14, 1210309. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Bautista, R.J.H.; Sandhu, M.A.; Hussein, O.E. Beneficial Effects of Citrus Flavonoids on Cardiovascular and Metabolic Health. Oxidative Med. Cell. Longev. 2019, 2019, 5484138. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Ran, L.; Chen, N.; Fan, X.; Ren, D.; Yi, L. Polarity-dependent extraction of flavonoids from citrus peel waste using a tailor-made deep eutectic solvent. Food Chem. 2019, 297, 124970. [Google Scholar] [CrossRef]
- Yi, L.; Ma, S.; Ren, D. Phytochemistry and bioactivity of Citrus flavonoids: A focus on antioxidant, anti-inflammatory, anticancer and cardiovascular protection activities. Phytochem. Rev. 2017, 16, 479–511. [Google Scholar] [CrossRef]
- Zhang, K.; Qin, Y.; Sun, W.; Shi, H.; Zhao, S.; He, L.; Li, C.; Zhao, J.; Pan, J.; Wang, G.; et al. Phylogenomic Analysis of Cytochrome P450 Gene Superfamily and Their Association with Flavonoids Biosynthesis in Peanut (Arachis hypogaea L.). Genes 2023, 14, 1944. [Google Scholar] [CrossRef]
- Martinez-Alonso, A.; Yepes-Molina, L.; Guarnizo, A.L.; Carvajal, M. Modification of Gene Expression of Tomato Plants through Foliar Flavonoid Application in Relation to Enhanced Growth. Genes 2023, 14, 2208. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhai, J.; Zhang, J.; Li, H.; Niu, X.; Liu, Y.; Ren, Y.; Du, H.; Zhu, J. Transcriptomic and Physiological Analyses of Pigment Accumulation in Eucommia ulmoides ‘Hongye’. Phyton 2022, 91, 1027–1044. [Google Scholar] [CrossRef]
- Datir, S.; Regan, S. Advances in Physiological, Transcriptomic, Proteomic, Metabolomic, and Molecular Genetic Approaches for Enhancing Mango Fruit Quality. J. Agric. Food Chem. 2023, 71, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zhang, K.; Wu, J.; Li, X.; Zhou, G.; Wan, Y. Integrated metabolomic and transcriptomic analysis revealed the flavonoid biosynthesis and regulation in Areca catechu. Phytochem. Anal. 2023, 34, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Liu, G.; Qin, J.; Wan, X.; Guo, A.; Wei, H.; Chen, Y.; Lian, B.; Zhong, F.; Zhang, J. Genomic and transcriptomic studies on flavonoid biosynthesis in Lagerstroemia indica. BMC Plant Biol. 2024, 24, 171. [Google Scholar] [CrossRef]
- Rajendran, N.; Subramaniam, S.; Christena, L.R.; Muthuraman, M.S.; Subramanian, N.S.; Pemiah, B.; Sivasubramanian, A. Antimicrobial flavonoids isolated from Indian medicinal plant Scutellaria oblonga inhibit biofilms formed by common food pathogens. Nat. Prod. Res. 2016, 30, 2002–2006. [Google Scholar] [CrossRef]
- Pei, T.; Yan, M.; Huang, Y.; Wei, Y.; Martin, C.; Zhao, Q. Specific Flavonoids and Their Biosynthetic Pathway in Scutellaria baicalensis. Front. Plant Sci. 2022, 13, 866282. [Google Scholar] [CrossRef]
- Patni, B.; Bhattacharyya, M.; Pokhriyal, A. The role of signaling compounds in enhancing rice allelochemicals for sustainable agriculture: An overview. Planta 2023, 258, 90. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, D.; Amaro, R.; Erban, A.; Mauri, N.; Soares, F.; Rego, C.; Martínez-Zapater, J.M.; Mithöfer, A.; Kopka, J.; Fortes, A.M. Transcriptional, hormonal, and metabolic changes in susceptible grape berries under powdery mildew infection. J. Exp. Bot. 2021, 72, 6544–6569. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Liu, X.; Li, C.; Chu, Q.; Cheng, S.; Su, L.; Shao, D.; Guo, X.; He, Z.; Zhou, X. Effect of light intensity on celery growth and flavonoid synthesis. Front. Plant Sci. 2024, 14, 1326218. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, D.K.; Heilmann, R.M.; Paital, B.; Patel, A.; Yadav, V.K.; Wong, D.; Jergens, A.E. Oxidative stress, hormones, and effects of natural antioxidants on intestinal inflammation in inflammatory bowel disease. Front. Endocrinol. 2023, 14, 1217165. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Wang, X.; Chai, X.; Gao, B.; Deng, C.; Günther, C.S.; Wu, T.; Zhang, X.; Xu, X.; Han, Z.; Wang, Y. Multi-omics analysis reveals the mechanism of bHLH130 responding to low-nitrogen stress of apple rootstock. Plant Physiol. 2023, 191, 1305–1323. [Google Scholar] [CrossRef]
- Zhao, Q.; Zeng, D.; Luo, Z.; Chen, A.; Xu, G.; Li, Y. Flavonoids Mediate the Modulation of Phosphate Uptake and Phosphate-Starvation Signaling in Tobacco. J. Plant Growth Regul. 2023, 42, 7229–7239. [Google Scholar] [CrossRef]
- Ma, R.; Sun, X.; Yang, C.; Fan, Y. Integrated transcriptome and metabolome provide insight into flavonoid variation in goji berries (Lycium barbarum L.) from different areas in China. Plant Physiol. Biochem. 2023, 199, 107722. [Google Scholar] [CrossRef]
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Rajpal, V.R.; Vyhnanek, T.; Topal, A.; Raina, S.N.; Gezgin, S. Insight into the Boron Toxicity Stress-Responsive Genes in Boron-Tolerant Triticum dicoccum Shoots Using RNA Sequencing. Agronomy 2023, 13, 631. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhong, L.; Xiao, W.; Du, Y.; Han, G.; Yan, Z.; He, D.; Zheng, C. Transcriptomics combined with physiological analysis reveals the mechanism of cadmium uptake and tolerance in Ligusticum chuanxiong Hort. under cadmium treatment. Front. Plant Sci. 2023, 14, 1263981. [Google Scholar] [CrossRef]
- Pandey, A.; Khan, M.K.; Hamurcu, M.; Brestic, M.; Topal, A.; Gezgin, S. Insight into the Root Transcriptome of a Boron-Tolerant Triticum zhukovskyi Genotype Grown under Boron Toxicity. Agronomy 2022, 12, 2421. [Google Scholar] [CrossRef]
- Kvastad, L.; Carlberg, K.; Larsson, L.; Villacampa, E.G.; Stuckey, A.; Stenbeck, L.; Mollbrink, A.; Zamboni, M.; Magnusson, J.P.; Basmaci, E.; et al. The spatial RNA integrity number assay for in situ evaluation of transcriptome quality. Commun. Biol. 2021, 4, 57. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, Y.; Tao, H.; Li, L.; He, Y.; Zhang, X.; Zhu, Y.; Hong, G. Analysis of the Flavonoidome Reveals the Different Health-Promoting Flavonoid Characteristics in Fruit. Antioxidants 2023, 12, 1665. [Google Scholar] [CrossRef]
- Huang, J.; Wang, L.; Tang, B.; Ren, R.; Shi, T.; Zhu, L.; Deng, J.; Liang, C.; Wang, Y.; Chen, Q. Integrated Transcriptomics and Widely Targeted Metabolomics Analyses Provide Insights Into Flavonoid Biosynthesis in the Rhizomes of Golden Buckwheat (Fagopyrum cymosum). Front. Plant Sci. 2022, 13, 803472. [Google Scholar] [CrossRef]
- Remali, J.; Sahidin, I.; Aizat, W.M. Xanthone Biosynthetic Pathway in Plants: A Review. Front. Plant Sci. 2022, 13, 809497. [Google Scholar] [CrossRef]
- Peniche-Pavía, H.A.; Guzmán, T.J.; Magaña-Cerino, J.M.; Gurrola-Díaz, C.M.; Tiessen, A. Maize Flavonoid Biosynthesis, Regulation, and Human Health Relevance: A Review. Molecules 2022, 27, 5166. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gu, X.; Guo, L.; Zhang, X.; Li, C. Integrated metabolomics and transcriptomics analysis reveals γ-aminobutyric acid enhances the ozone tolerance of wheat by accumulation of flavonoids. J. Hazard. Mater. 2024, 465, 133202. [Google Scholar] [CrossRef] [PubMed]
- Gouot, J.C.; Smith, J.P.; Holzapfel, B.P.; Walker, A.R.; Barril, C. Grape berry flavonoids: A review of their biochemical responses to high and extreme high temperatures. J. Exp. Bot. 2019, 70, 397–423. [Google Scholar] [CrossRef]
- Pandith, S.A.; Ramazan, S.; Khan, M.I.; Reshi, Z.A.; Shah, M.A. Chalcone synthases (CHSs): The symbolic type III polyketide synthases. Planta 2019, 251, 15. [Google Scholar] [CrossRef]
- Jia, Y.; Li, B.; Zhang, Y.; Zhang, X.; Xu, Y.; Li, C. Evolutionary dynamic analyses on monocot flavonoid 3′-hydroxylase gene family reveal evidence of plant-environment interaction. BMC Plant Biol. 2019, 19, 347. [Google Scholar] [CrossRef]
- Wu, J.; Lv, S.; Zhao, L.; Gao, T.; Yu, C.; Hu, J.; Ma, F. Advances in the study of the function and mechanism of the action of flavonoids in plants under environmental stresses. Planta 2023, 257, 108. [Google Scholar] [CrossRef]



| Model | MAE | RMSE | R2 | Top Feature Importance | Top 5 Features’ Cumulative Importance | Training Time (Seconds) | Prediction Time (Seconds/Sample) |
|---|---|---|---|---|---|---|---|
| GBM | 0.35 | 0.45 | 0.85 | 0.15 | 0.7 | 120 | 0.002 |
| Random Forest | 0.38 | 0.48 | 0.82 | 0.12 | 0.65 | 90 | 0.001 |
| Pathway | Upregulated DEGs | Downregulated DEGs | Total DEGs | Biological Significance |
|---|---|---|---|---|
| Flavonoid biosynthesis | 15 | 9 | 24 | Enhances UV protection, pathogen resistance, and antioxidant capacity |
| Plant–pathogen interaction | 73 | 79 | 152 | Mediates immune responses, crucial for pathogen defense |
| Cytochrome P450 | 20 | 18 | 38 | Involved in biosynthesis of secondary metabolites and detoxification processes |
| Phenylpropanoid biosynthesis | 12 | 14 | 26 | Key pathway for production of flavonoids, lignin, and other phenylpropanoids |
| Wax biosynthesis | 10 | 7 | 17 | Contributes to cuticle formation, protecting against desiccation and pathogens |
| Suberine and cutin biosynthesis | 8 | 5 | 13 | Important for barrier formation, providing resistance to environmental stress |
| Diterpenoid biosynthesis | 5 | 3 | 8 | Produces compounds involved in defense and growth regulation |
| Drug metabolism by cytochrome P450 | 6 | 7 | 13 | Metabolizes exogenous compounds, contributing to detoxification |
| Gene/Metabolite | GBM Importance Score | Random Forest Importance Score | Biological Role |
|---|---|---|---|
| Chalcone synthase (CHS) | 0.15 | 0.1 | Catalyzes the first step in flavonoid biosynthesis |
| Flavonoid 3′-hydroxylase (F3′H) | 0.12 | 0.14 | Hydroxylation of flavonoids, affecting bioactivity |
| Quercetin | 0.1 | 0.12 | Antioxidant, protects against oxidative stress |
| Kaempferol | 0.08 | 0.09 | Antioxidant, involved in UV protection |
| Phenylpropanoid pathway | Enriched | Enriched | Responds to biotic and abiotic stresses |
| Flavone/flavonol pathway | Enriched | Enriched | Drives variations in flavonoid profiles |
| Parameter | GG Ecotype | YG Ecotype | Significance (p-Value) |
|---|---|---|---|
| Total flavonoid content (mg/g) | 3.36 | 2.98 | <0.01 |
| Chlorophyll content (mg/g) | 0.26 | 0.21 | <0.05 |
| Quercetin (mg/g) | 0.12 | 0.08 | <0.05 |
| Kaempferol (mg/g) | 0.09 | 0.06 | <0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Naren, G.; Han, C.; Elsheery, N.I.; Zhang, L. Exploring the Flavonoid Biosynthesis Pathway of Two Ecotypes of Leymus chinensis Using Transcriptomic and Metabolomic Analysis. Agronomy 2024, 14, 1839. https://doi.org/10.3390/agronomy14081839
Wu H, Naren G, Han C, Elsheery NI, Zhang L. Exploring the Flavonoid Biosynthesis Pathway of Two Ecotypes of Leymus chinensis Using Transcriptomic and Metabolomic Analysis. Agronomy. 2024; 14(8):1839. https://doi.org/10.3390/agronomy14081839
Chicago/Turabian StyleWu, Haiyan, Gaowa Naren, Chenxu Han, Nabil I. Elsheery, and Lingang Zhang. 2024. "Exploring the Flavonoid Biosynthesis Pathway of Two Ecotypes of Leymus chinensis Using Transcriptomic and Metabolomic Analysis" Agronomy 14, no. 8: 1839. https://doi.org/10.3390/agronomy14081839
APA StyleWu, H., Naren, G., Han, C., Elsheery, N. I., & Zhang, L. (2024). Exploring the Flavonoid Biosynthesis Pathway of Two Ecotypes of Leymus chinensis Using Transcriptomic and Metabolomic Analysis. Agronomy, 14(8), 1839. https://doi.org/10.3390/agronomy14081839






