Abstract
This study constructs a model for the rapid identification of the origins of edible sunflower (Helianthus) using Kernel Extreme Learning Machine (KELM) with multi-source information fusion technology. Near-infrared spectroscopy (NIRS) and nuclear magnetic resonance spectroscopy (NMRS) were utilized to analyze 180 sunflower samples from the Xinjiang, Heilongjiang, and Inner Mongolia regions. Initially, the identification models for the origin of sunflowers using NIR and NMR data were compared between two algorithms: the Extreme Learning Machine (ELM) and KELM, combined with various spectral preprocessing methods. The experiment found that the NIR spectral model preprocessed with standard normal variate (SNV) using the KELM algorithm was the most accurate, achieving accuracies of 98.7% in the training set and 97.2% in the test set. The spin-echo NMR spectral model preprocessed with non-local means (NLMs) using the KELM algorithm was the second best, with accuracies of 98.4% in the training set and 96.4% in the test set. To further improve the accuracy of the identification models, innovative sunflower origin identification models were developed based on data layer fusion and feature layer fusion using NIRS and NMRS. In the data layer fusion model, the KELM algorithm model was optimal, achieving a test set accuracy and F1 score of 98.2% and 98.18%, respectively, an improvement of 1.0% over the best single data source model. In the feature layer fusion model, four types of feature-layer information-fusion identification models were established using two feature extraction algorithms, Competitive Adaptive Reweighted Sampling (CARS) and Variable Importance Projection (VIP), combined with joint feature and simple merging feature strategies. The CARS-KELM algorithm combined with the joint feature method was found to be the best, achieving 100% accuracy in both the training and test sets, an improvement of 2.8% over the best single data source model. Identifying the origin of edible sunflower using NIRS and NMRS is demonstrated as feasible by the results. The best single-spectrum sunflower origin identification model was achieved using the KELM algorithm with SNV preprocessing. The feature layer fusion method combining NIRS and NMRS data is suitable for handling the task of sunflower origin identification. This method significantly improves the recognition accuracy of the model compared to a single model, achieving fast and accurate origin identification of edible sunflowers. The research results provide a new method for rapid identification of sunflower origin.
1. Introduction
In recent years, the global cultivation area of edible sunflowers has continued to expand, with China now ranked sixth worldwide. The quality of sunflower cultivation is crucial for China’s food safety. Northern Chinese provinces like Heilongjiang, Xinjiang, and Inner Mongolia have cultivated the high-yielding and resistant “LongshikuiNo.1” sunflower variety, yet their nutritional and flavor profiles vary significantly due to regional environmental and climate disparities. Traditional identification methods, such as morphological classification techniques using microscopes, scanning electron microscopes, and paraffin sectioning, are complex when observing microscopic features and anatomical characteristics [1,2,3,4,5,6,7,8,9,10]. Modern molecular biology identification techniques, although accurate, involve high costs and require being operated by scientifically trained personnel [11,12,13,14,15], which do not meet the needs for rapid identification of sunflower origins.
Compared to traditional methods, studies have shown that near-infrared reflectance spectroscopy (NIRS) and nuclear magnetic resonance spectroscopy (NMRS) can measure specific chemical components such as fats, sugars, and amino acids and are used for identifying the origins of agricultural products, including applications in tea, grains, meats, and wines [16,17,18,19,20,21,22]. NIRS and NMRS can also be used for non-crop testing, demonstrating their versatility in analyzing various samples beyond agricultural products [23,24]. However, the chemical composition of sunflower samples is complex, and models based on single spectroscopic data cannot fully characterize the chemical information of the samples or meet the identification needs for sunflowers. Recently, origin identification models using machine learning combined with multi-source information have emerged. For instance, in the origin identification research involving multi-source data fusion, Luan, X. [25] and others innovatively established a model integrating near-infrared, mid-infrared, and Raman spectroscopy for rice origin identification. They found that Competitive Adaptive Reweighted Sampling (CARS) significantly improved the accuracy of the information fusion model compared to single-spectrum models. Dai, Y. [26] investigated the application of near-infrared (NIR) and Raman spectroscopy as well as low-level and intermediate-level data fusion in the classification of four rice species of similar origin. Low-level data fusion splices the two types of spectra together and applies the classification techniques. Mid-level data fusion involves selecting bands, extracting features from the spectra of each technique, and building a classification model. Moreover, Zhou, Y [27] and others used the Variable Importance Projection (VIP) strategy combined with the Random Forest algorithm (RF) to identify the origins of notoginseng, discovering that Fourier-transform near-infrared and near-infrared spectroscopy could reflect subtle differences between notoginseng from different origins and that all three levels of fusion strategies enhanced the accuracy of origin identification. Li, Y. [28] applied three data fusion strategies and showed that the advanced data fusion strategy of Fourier-transform mid-infrared spectroscopy and near-infrared spectroscopy can be used as a reliable tool for the correct geographic identification of notoginseng. These studies show that integrating multi-source data can effectively improve the classification performance of machine learning models. Through [29], we learned that in the current field of artificial intelligence, the frontiers focus on inverse learning, non-learning, adaptive machine learning, and deep personalization. However, when constructing models for identifying the origin of sunflowers using gradient learning techniques such as the backpropagation (BP) algorithm, manual configuration of step sizes and numerous iterations are necessitated. The BP algorithm operates by iteratively adjusting network weights and biases via gradient descent, calculating the error in the output layer, and subsequently propagating this error backward through the hidden layers. Traditional algorithms often involve complex architectures and numerous iterations, consuming a lot of time to tune model parameters and using substantial computational resources. Maragathavalli, P. [30] provided the idea of “Ensemble Methodologies” to improve accuracy by aggregating the predictions of weak learners so that strong learners can make accurate predictions. The Extreme Learning Machine (ELM) and Kernel Extreme Learning Machine (KELM) proposed by Huang and others effectively overcome these issues [31,32,33,34,35,36,37,38,39].
To date, studies on sunflower seeds using near-infrared spectroscopy have mostly focused on fat and protein content, but systematic reports on sunflower origin identification models based on NIRS and NMRS are lacking. This study processes near-infrared and nuclear magnetic spectroscopic information of the same variety of sunflowers from different origins, using ELM and KELM combined with a multi-source information fusion strategy to establish sunflower origin prediction models. These models are compared with single-data ELM and KELM origin prediction models to identify sunflowers from three origins, selecting the most effective method for common sunflower identification and quality control (Figure 1).
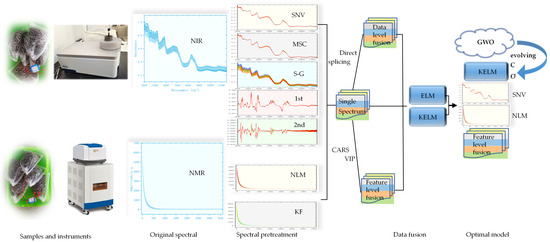
Figure 1.
Flowchart of this study.
2. Materials and Methods
2.1. Samples
The production of edible sunflowers in China is mainly concentrated in northern provinces such as Heilongjiang, Xinjiang, Inner Mongolia, Hebei, and Gansu, where the natural conditions are complex and the climate and ecological environments are diverse. According to research, the “LongshikuiNo.1” variety, bred by the Agricultural Sciences Institute of Heilongjiang, is an edible hybrid sunflower variety known for its high yield and good stress resistance. This variety is planted in major sunflower production areas in Xinjiang, Heilongjiang, Inner Mongolia, Shanxi, and Ningxia. Due to the unique geographical environments, the same variety of sunflowers grown in different locations exhibits variations in the content of dry matter, amylose, amylopectin, vitamin C, and monosaccharides [40,41,42,43,44,45,46,47]. For example, differences in soil nutrient content, water conditions, and especially the amount of sunlight in Heilongjiang, Xinjiang, and Inner Mongolia lead to variations in the growth and nutritional content of sunflower seeds, affecting the taste.
The edible sunflower seeds used in the experiments are from the most widely planted variety, LongshikuiNo.1 (Plant Inspection No. 6204232017001277). The geographic location of the 180 samples collected is shown in Figure 2 for a total of three provinces, as follows: 1-(60): sixty samples were collected from Altay Prefecture in Xinjiang; sixty from Heilongjiang Province (cities of 2-(20): Qiqihar, 3-(20): Daqing, 4-(20): Suihua); and sixty from Inner Mongolia (cities of 5-(20): Bayannur, 6-(20): Ordos, 7-(20): Chifeng). After removing damaged, shriveled, or insect-eaten seeds and shelling them, each sample was prepared by weighing approximately 5 ± 0.5 g, totaling 180 samples.
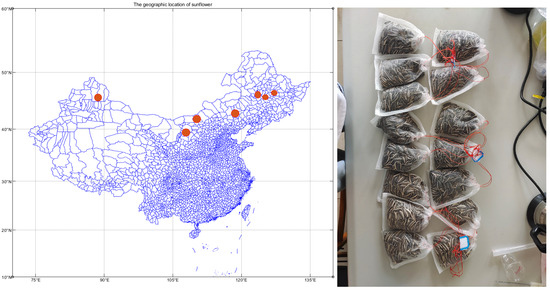
Figure 2.
Geographic location of edible sunflower.
The samples of the three provinces are labeled and geographically distributed as shown in Table 1, and the analysis found that the distribution of the dimensions of the samples in different regions did not vary much (about N 37°00′–48°03′). The 7-(20) samples belong to the Inner Mongolia region, but these samples and the three groups of samples labeled as belonging to the Heilongjiang region are geographically very close to each other, which may result in the fuzzy area of the classification of the sunflower origin identification model.

Table 1.
Specific geographical origin information of edible sunflower (Helianthus).
2.2. Instruments and Sampling
2.2.1. Near-Infrared Spectroscopy Collection and Spectral Analysis
The near-infrared (NIR) sampling instrument used was a small FT-NIR spectrometer produced by Bruker TANGO (The manufacturer of Bruker TANGO equipment is Bruker Optics GmbH & Co. KG, situated in Ettlingen, Germany), shown in Figure 3, measuring wavenumbers from 11,500 to 4000 cm⁻¹, with a resolution of 4 cm⁻¹ and a scanning speed of 8 times per second.
Figure 3.
TANGO FT-NIR spectrometer.
The chemometrics software used was OPUS software version 1.3.0. Following the one-hour preheating of the NIR spectrometer and the utilization of the built-in VU calibration unit for instrument testing, the sunflower samples were transferred from the incubator to a low-OH quartz cup tailored for solid sample analysis. These preparatory steps aimed to confirm that the spectrometer was operating at its optimal conditions. This setup ensured uniform coverage at the bottom of the cup and included a press to minimize disturbances caused by manual handling and external environmental factors. Along with a sample rotating stage, this increased the scanning area of the samples, enhancing their representativeness and eliminating heterogeneity. The transmittance of the samples was scanned, and after 32 scans per sample, the average spectrum was used as the research sample. The sample data were randomly split into a 70% training set and a 30% test set, resulting in 125 samples in the training set and 55 in the test set. The average original NIR spectra of the edible sunflower samples from three different origins are presented in Figure 4.
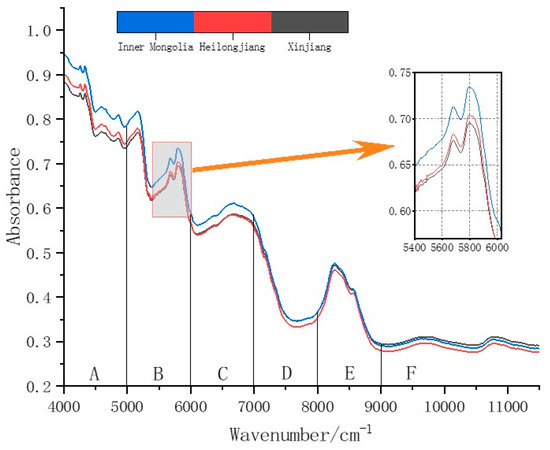
Figure 4.
Average original NIR spectra of edible sunflower samples from three different origins.
Infrared spectroscopy is a type of absorption spectroscopy that identifies functional groups by measuring the vibrational frequencies of chemical bonds within molecules. Near-infrared spectroscopy (NIRS) ranges from 0.75 to 2.5 μm (13,330–4000 cm−1), studying the combination and overtone absorption bands of chemical bonds connected to hydrogen in organic molecules. It is primarily used for quantitative analysis and identification of known substances. The NIR spectrum contains various levels of overtone bands and also includes many combination absorptions, making the spectral bands complex and rich in information. These spectral data provide a foundation for establishing models to identify the origins of samples [48,49,50]. As shown in Figure 4, the average NIR spectra of sunflower seeds from different origins are divided into six regions (A–F). Since about 50% of the oil in sunflower seeds is composed of fatty acids, the absorption observed in the NIR spectra is primarily due to the vibrational modes of C-H functional groups. The frequencies of the C-H groups can be attributed to three main functional groups: methylene, methyl, and vinyl groups [51]. These can be assigned to different regions in the NIR spectrum of sunflower seeds, as shown in Figure 4, with explanations of these regions’ functional group vibrations presented in Table 2.

Table 2.
Assignments of major NlR absorption bands for edible sunflower samples.
2.2.2. Nuclear Magnetic Resonance Collection and Spectral Analysis
The NMR instrument used was a two-dimensional NMR analyzer (MesoMR32-040V) produced by Shanghai Niumag Corporation (Shanghai, China), with a magnetic field strength of 0.50 ± 0.03 T and a longitudinal sample introduction direction, using a 1.5-inch temperature-controlled probe coil, shown in Figure 5. To ensure the reliability of the nuclear magnetic resonance (NMR) test results, the samples were placed in an incubator set to 26 °C for at least six hours to stabilize the sample temperature, thereby obtaining more representative test data. After turning on the power of the NMR equipment and setting the magnet temperature controller to 26 °C, the device was preheated for over 16 h to ensure that both the probe and the magnet were maintained at this constant temperature.

Figure 5.
MesoMR32-040V spectrometer by Shanghai Niumag.
The Niumag NMR analysis software version 4.0 was used to guide the calibration of the probe at the center of the magnetic field, followed by adjustments to the frequency offset (o1), 90° pulse width, and 180° pulse width. To minimize the effects of magnetic field inhomogeneity, which can cause dephasing of nuclear magnetism and inaccurate 180° pulses, the spin–spin interactions were characterized as much as possible, selecting the CPMG pulse sequence for collecting spin-echo spectra. The test samples were loaded into the non-magnetic, hydrogen-free test tube chamber of Niumag, ensuring the center of the sample was at the center of the magnetic field. After 16 cumulative sampling times per sample, the average value was used as the research sample. The sampling parameters were set as shown in Table 3.

Table 3.
Acquisition parameter values for Niumag magnetic resonance analysis software version 4.0.
The sample data were randomly split into a 70% training set and a 30% test set, resulting in 125 samples in the training set and 55 in the test set. The average original NMR spectra of the edible sunflower samples from three different origins are presented in Figure 6.
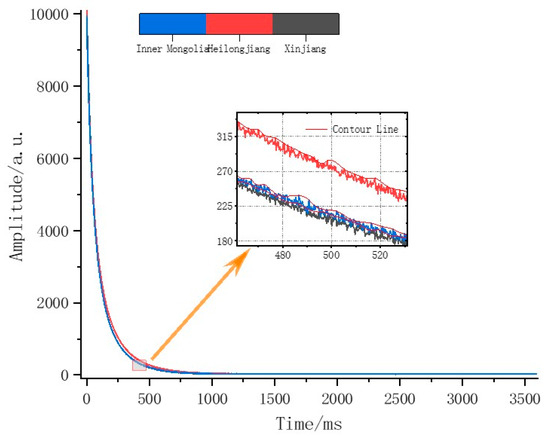
Figure 6.
Average original NMR spectra of edible sunflower samples from three different origins.
Nuclear magnetic resonance commonly occurs in materials containing specific atomic nuclei, such as hydrogen protons. When an external radio frequency pulse that matches the energy level splitting frequency is applied, hydrogen protons can absorb energy and resonate [52]. In this experiment, the spin-echo NMR spectra were collected using the Carr–Purcell–Meiboom–Gill (CPMG) pulse sequence. CPMG reduces the dephasing of the magnetization vector in a static magnetic field and enhances the detectability of the signal. After a 90° pulse, a subsequent 180° RF pulse applied at a time interval T induces dephasing; this pulse then inverts the magnetization by 180 degrees, effectively restoring it to its initial equilibrium orientation. At time 2 T, the first echo forms and then begins to dephase again; at time 3 T, a second 180° refocusing pulse is applied, and similarly, the second echo forms at time 4 T. This cycle repeats, with the number of applied 180° pulses being NECH, which depends on the interval time TE between the 180° pulses and the relaxation properties of the sample.
By observing the magnified section of the spin-echo NMR spectrum in Figure 6, variations can be seen in the spin-echo spectra of sunflowers from three origins, with significant differences in the curvature and signal intensity of the spin-echo peaks from Heilongjiang, Xinjiang, and Inner Mongolia. These differences are due to the varying accumulation of substances like water and oil in sunflowers, which are influenced by the local natural environment. The chemical structure and physical properties of water and oil determine their relaxation times. Typically, relaxation signals are divided into transverse relaxation (T2) and longitudinal relaxation (T1); generally, the larger the molecule size and the tighter the binding state, the smaller the T1 and T2 values. A single scanning process monitors the TE*NECH duration of transverse relaxation. The spin-echo NMR spectrum’s echo data consist of a binary data structure of each echo time and its corresponding peak value, as shown in Figure 6; the echo curve, which is the envelope formed by the echo peaks, is shown in the magnified part of Figure 6.
2.3. Establishment of the Sunflower Origin Identification Model
2.3.1. Principles and Evaluation of Extreme Learning Machine
To identify the origin of sunflowers from spectral data, a reliable and robust prediction model is often needed. Gradient-based learning methods, widely used for training neural networks, include Support Vector Machines (SVMs) and backpropagation (BP) neural networks [53,54]. These methods improve predictive performance through error minimization or backpropagation, requiring extensive parameter tuning. The complex model architectures and long iteration times of these models gradually become inadequate for complex problems [33]. However, the Extreme Learning Machine (ELM) and Kernel Extreme Learning Machine (KELM) proposed by Huang effectively overcome these issues [34,35]. Their key advantages include fast training speed and low complexity, overcoming the problems of local minima, overfitting, and inappropriate learning-rate selection associated with traditional gradient algorithms. KELM further utilizes kernel learning techniques, replacing random mapping with kernel mapping, which effectively improves the generalization and stability issues caused by the random assignment of hidden layer neurons, offering superior performance for nonlinear problems. ELM is a simple and efficient single-hidden-layer feedforward neural network learning algorithm. The input parameters are randomly generated and fixed, requiring no iterative solving. Only the output parameters between the output and hidden layers need to be processed, greatly accelerating the model’s learning speed. ELM algorithms have been developed for decades, and variants continue to be proposed. Numerous variants of ELM algorithms continue to improve their stability and generalization for specific applications [36,37,38,39].
A typical single-hidden-layer feedforward neural network (SLFN) structure is shown in Figure 7, consisting of an input layer with n neurons, a hidden layer with l neurons, and an output layer with m neurons.
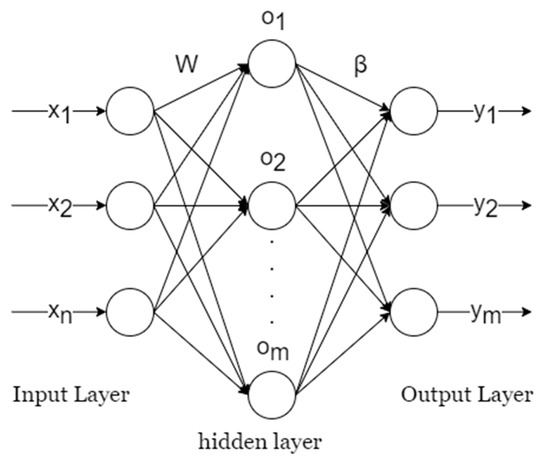
Figure 7.
Single-hidden-layer feedforward neural network architecture.
Before training, the Extreme Learning Machine (ELM) can randomly generate weights w and biases b. Determining the number of neurons in the hidden layer and the activation function for these neurons is only required to perform the calculation of β.
The Kernel Extreme Learning Machine (KELM) is an improved algorithm based on ELM that integrates a kernel function. KELM retains the advantages of ELM while enhancing the model’s predictive performance. The learning target matrix for ELM is shown as Equation (1):
The training of the network is transformed into solving a linear system problem; β is determined from , where is the generalized inverse matrix of . To enhance the stability of the neural network, a regularization coefficient C and the identity matrix I are introduced. Consequently, the least-squares solution for the output weights is shown as Equation (2):
By incorporating a kernel function into ELM, the kernel matrix is represented as Equation (3):
Incorporating Equation (3) into Equation (1), as shown in Equation (4), where is the given training samples, n is the number of samples, and represents the kernel function, we obtain:
Huang [55] first proposed a Kernel ELM 12 years ago, empirically specifying Gaussian and polynomial kernels. KELM has been developed in recent years. Common functions used as kernels are the Gaussian kernel, RBF kernel, polynomial kernel, Laplacian kernel, inverse square distance kernel, and inverse distance kernel [32]. It has been shown that extreme learning machines with “sigmoid” nodes usually require a large number of hidden layer nodes to achieve good generalization [56,57]. In addition, it has also been shown that KELM can map the data vector space to a higher-dimensional space for computation by introducing kernel functions while retaining the advantages of ELM [58].To a certain extent, the kernel solves the problem of large fluctuations in prediction due to the large number of nodes in the implicit layer of random weights and biases [57,59].The kernelized ELM has better generalization performance than ELM [60]. X. Liu [55] analyzed the general consistency of limit learning machines in training radial-base generative networks and which kernel functions should be chosen by kernel limit learning machines depending on the situation. S. Mojrian [61] used the radial-base-function (RBF) kernel of the Extreme Learning Machine (ELM) classification model with the Online Support Vector Machine and other baseline models. The Extreme Learning Machine with RBF kernel performs well under the evaluation metrics of accuracy, precision, mindfulness, and specificity; meanwhile, the algorithm of RBF-ELM proposed by Y. Qin [62] is validated on a low-carbon engineering problem, which further illustrates the computational classification advantages brought by RBF in mapping input vectors to a high-dimensional special space. The RBF formulation is shown in Equation (5), where σ is the kernel parameter:
Therefore, -RBF will be used to optimize the ELM, which becomes the KELM algorithm, and the optimized KELM model will be used for multispectral fusion data to identify the sunflower origin.
There are two parameters to be optimized (C and σ) for the KELM in the origin identification model. The regularization parameter C and the parameter σ of the RBF kernel function play a decisive role in determining the stability of the KELM for the identification model. Too large a value of C can lead to overfitting of the model, and vice versa, it can lead to underfitting due to insufficient learning. A variation in the kernel parameter σ affects the mapping from the input space to the feature space, which in turn has an impact on the nature of the feature space. In this study, the Gray Wolf Swarm Intelligence Optimization Algorithm is used to learn the optimal parameter values of C and σ in the KELM model. The Gray Wolf Optimization Algorithm is a pack-wise optimization algorithm that attempts to model the social hierarchy and hunting behavior of gray wolves from nature in order to find the most appropriate solution to the problem [63]. Since its proposal, it has performed well in all kinds of optimization problems [64], especially in the parameter optimization of the nuclear limit learning machine. Using GWO as a parameter optimization treatment works well [65]. The optimization flow of the GWO optimization algorithm is shown in Figure 8. The convergence curves of the two parametric models optimized by the GWO optimization algorithm are shown in Figure 9. The test shows that αWolf converges, on average, after 140 iterations of the model to find the global optimal solution of the regularization parameter C and the parameter σ of the RBF kernel function, which is in line with the results of the test by Wang [65] on the model performance dataset. This indicates that GWO can be used for KELM parameter seeking in the provenance identification model.
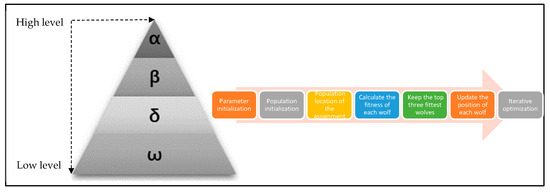
Figure 8.
Gray Wolf Optimizer Algorithm.
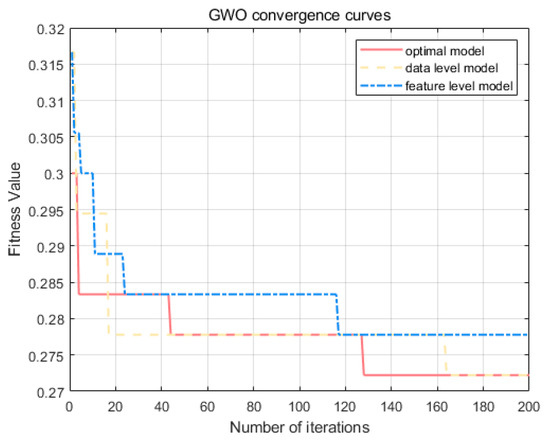
Figure 9.
Gray Wolf Optimizer convergence curves.
In the context of classifying sunflower origins, assessing the accuracy of the classification within each model is a necessary step. Considering the initial uniform distribution of samples from three origins, accuracy and the F1 score are used as evaluation criteria. The F1 score comprehensively considers precision and recall, both of which crucially affect the F1 score. A low value in either precision or recall will decrease the F1 score, as illustrated in Equation (6):
2.3.2. Multi-Source Information Fusion Techniques
Multi-source information fusion technology employs two fusion methods: data layer fusion and feature layer fusion [49,66]. The challenge in data layer fusion lies in directly connecting raw or preprocessed data and preserving the original information from different sources, which is often filled with noise and redundancy. This demands higher requirements for the applicability of the developed model. Prior to data fusion, normalization of multi-source datasets is essential. Variations among datasets collected by disparate instruments, manifesting as dimensional discrepancies or spectral intensity disparities, may compromise the fusion process and result in failure. Feature layer fusion involves extracting features from data from different sources and vectorizing the selected feature variables in a specific order. The challenge here is the selection of the feature matrix. To find better feature extraction algorithms for sunflower data, this study employs Competitive Adaptive Reweighted Sampling (CARS) and Variable Importance Projection (VIP) algorithms to eliminate irrelevant variables and generate a feature layer prediction matrix. The CARS algorithm initially uses a Monte Carlo model for sampling, randomly dividing the dataset for modeling analysis with a division ratio of 75%, and employs Partial Least Squares (PLS) for analysis and modeling. The absolute value percentage of regression coefficients is used to determine the importance of variables or their explanatory power toward the target variable. The CARS [25] algorithm begins with a preliminary modeling using all variables, followed by a gradual elimination of variables. As the algorithm iterates, the number of sampled variables decreases based on dynamically adjusted variable weights, retaining only those variables that contribute most to model performance. This weighted design allows the CARS algorithm to converge and effectively reflect the relationship between inputs and outputs. This efficacy is evaluated by calculating its RMSE and employing cross-validation root mean square error to assess the feature vector model, specifically the best set of variables. VIP [27] is a commonly used method in multivariate statistical analysis to assess the explanatory degree of different independent variables in the dependent variable model. The VIP method calculates the contribution of each independent variable in the PLS model to reflect its importance; the higher the VIP value, the greater the contribution of the data value to the label data. Therefore, based on VIP, a five-fold cross-validation is used to screen variables, with the experiment prioritizing variables with VIP values greater than 1 for constructing the sunflower feature layer fusion model.
3. Results and Discussion
3.1. Establishment and Verification of the Single-Spectrum Identification Model
In the experiments involving nuclear magnetic resonance (NMR) technology, the main factors affecting the signal-to-noise ratio (SNR) include the strength of the magnetic field and the choice of radiofrequency (RF) pulse sequence. The Carr–Purcell–Meiboom–Gill (CPMG) sequence, a commonly used sequence in low-field detection, was selected. Due to the low main magnetic field strength, the received spin-echo NMR signals are weak, and real echo signals can easily be drowned out by background noise, significantly impacting the accuracy and precision of subsequent sunflower origin identification models. Therefore, the experiment utilized multiple sampling averages, followed by the application of non-local means (NLMs) and Kalman Filter (KF) algorithms to suppress noise in the spin-echo NMR signals to enhance the SNR [67,68]. In the near-infrared spectroscopy (NIR) experiments, the heterogeneity in particle size and uniformity of samples led to the raw spectra containing disturbances such as fluorescence background, detector noise, and laser power fluctuations. Five preprocessing methods were employed to remove interference from the raw NIR data: standard normal variate (SNV), Multiplicative Scatter Correction (MSC), Savitzky–Golay smoothing filter (SG), first derivative, and second derivative [25]. In NIRS modeling, the SNV-ELM model sets the main parameter Hidden to 59 for the ELM, i.e., the number of nodes in the implicit layer is 59. In NMRS modeling, the NLM-ELM model sets the main parameter Hidden to 59 for the ELM, i.e., the number of nodes in the implicit layer is 59. In NMRS modeling, the NLM-KELM model, after optimization by the GWO optimization algorithm, adjusts the regularization parameter C to 179.39 and the kernel parameter σ to 93.65. In the origin identification model, the more optimal models for sunflower origin identification using these two data types are shown in Table 4. The experimental results indicate that combining data from two sources with data preprocessing techniques can effectively analyze and identify sunflower origins, with preprocessing methods significantly enhancing model recognition performance. Different preprocessing algorithms yielded varying results, with NIR spectroscopy combined with standard normal variate (SNV) preprocessing providing the best results, with recognition accuracies of 98.7% and 97.2% in the calibration and validation sets, respectively. The preprocessing can further enhance the predictive performance of the model, which is consistent with numerous results reported in the literature. However, the optimal preprocessing method is not the same for different NIR spectral prediction tasks. The same conclusion was reached in [25,27], while spin-echo spectroscopy combined with local linear embedding (NLM) preprocessing achieved the best results, with recognition accuracies of 97.6% and 96.4% in the calibration and validation sets, respectively. This is because the NLM preprocessing method uses the similarity between the neighborhood blocks of the current filter point and the neighborhood blocks of other points in the rectangular window to calculate the weights, which is more consistent with the preprocessing of NMR spectral data [67].

Table 4.
Optimal identification results of models using one type of spectral data.
Further analyses were conducted to determine the primary causes of misidentification in the sunflower identification models. As shown in Figure 10a, the optimal NIR model incorrectly identified two Inner Mongolian sunflower samples as originating from Heilongjiang while accurately classifying sunflowers from Xinjiang and Heilongjiang. As shown in Figure 10b, the optimal spin-echo NMR model incorrectly identified one Inner Mongolian sunflower sample as originating from Xinjiang and one Heilongjiang sample as originating from Inner Mongolia, achieving accurate classification for sunflowers from Xinjiang. The accuracy of the validation set for the spin-echo NMR model was 0.8% lower than that of the optimal NIR model, and the types of misidentification differed. This is because the NIR model focuses on capturing overtone and combination signals generated by vibrations of hydrogen-containing groups, whereas the spin-echo NMR spectroscopy model [48,49,50] measures the Larmor frequency (or phase change) of hydrogen protons in different chemical environments [51,52]. The focus of the spectral information obtained from the sunflower samples differs, thus affecting the modeling outcomes.
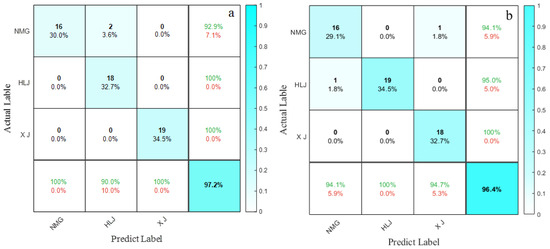
Figure 10.
(a) Confusion matrix for the optimal NIR spectroscopy model test dataset; (b) confusion matrix for the spin-echo NMR spectroscopy model test dataset.
These experimental results demonstrate the feasibility of combining NIR and NMR technologies with machine learning to construct models for identifying the origins of sunflowers. Both data sources are capable of being used to construct sunflower origin identification models, and in a comparison of the two spectroscopic technologies, NIR spectroscopy is more suitable for sunflower origin identification tasks [25,27], with models employing the KELM algorithm showing superior average identification accuracy. However, models relying solely on a single data source are unable to completely differentiate all sunflower samples by origin in the validation set, necessitating further exploration of multi-source data fusion modeling methods.
3.2. Feature Extraction
In the origin identification models that utilize data layer and feature layer fusion, the optimal preprocessing method is applied to the spectral data matrices individually, linking these matrices for data layer fusion to form a matrix of 180 samples and 13,845 variables. Of these, 1845 variables are provided by near-infrared spectroscopy and 12,000 variables by spin-echo NMR spectroscopy. Two sunflower origin identification models are constructed using the ELM and KELM machine learning methods.
In the origin identification models that use feature layer fusion, the feature matrices of spectral data are extracted using the CARS and VIP algorithms from the models with optimal preprocessing methods. As shown in Figure 11, the near-infrared spectroscopy data preprocessed with SNV are subjected to feature extraction by CARS. Upon reaching the 12th iteration, the CARS model attains an RMSE of approximately 0.087. Consequently, the 864 data points acquired at this juncture are deemed to represent the characteristic features for model construction. The spin-echo NMR spectroscopy data preprocessed with NLMs undergo feature extraction by CARS. At the 39th iteration, the CARS model achieves an RMSE of about 0.109, with 426 feature values obtained for feature-layer data-fusion modeling. The optimal preprocessed data, after VIP feature extraction calculation, yields VIP values where values corresponding to a VIP greater than 1 are filtered out—340 VIP features from near-infrared spectroscopy and 1639 from spin-echo NMR spectroscopy with VIP values greater than 1. The VIP value results are shown in Figure 12. After feature matrix extraction using the CARS algorithm, the near-infrared spectroscopy data form a matrix of 180 samples with 864 features, and the spin-echo NMR spectroscopy data form a matrix of 180 samples with 426 features. After feature matrix extraction using the VIP algorithm, the near-infrared spectroscopy data form a matrix of 180 samples with 340 features, and the spin-echo NMR spectroscopy data form a matrix of 180 samples with 1639 features. The feature matrices undergo two types of feature fusion strategies. The first strategy involves synthesizing the near-infrared feature matrix with the spin-echo NMR spectroscopy feature matrix by interleaving and merging the two feature matrices to form a combined-feature matrix—for example, merging the 864 features from the near-infrared spectroscopy and the 426 features from the spin-echo NMR spectroscopy processed by CARS into a 1290-dimensional combined-feature matrix. The second strategy simply involves concatenating the feature matrices of the two spectroscopies.
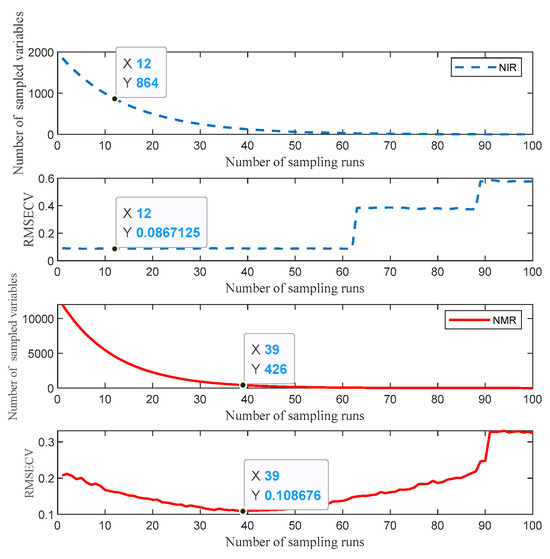
Figure 11.
Parameter diagram of CARS feature selection: near-infrared spectroscopy (blue), spin-echo NMR spectroscopy (red).
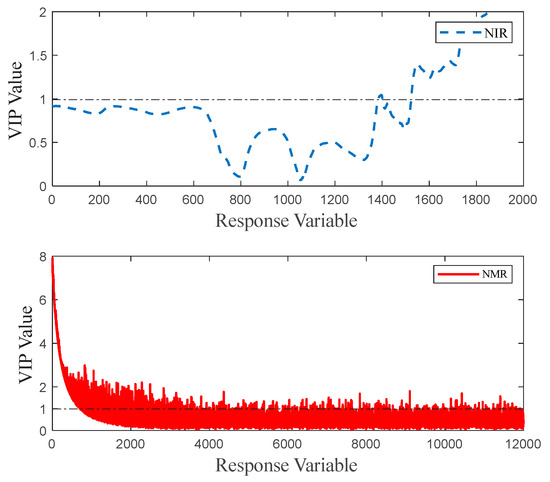
Figure 12.
Parameter diagram of VIP feature selection: near-infrared spectroscopy (blue), spin-echo NMR spectroscopy (red).
The aforementioned feature matrices, combined with joint feature and simple merging feature strategies, establish four types of feature-layer information fusion models: CARS–Simple Merge, VIP–Simple Merge, CARS–Joint Feature, and VIP–Joint Feature.
3.3. Establishment and Verification of the Data Fusion Identification Model
The KELM algorithm, when applied to the near-infrared spectroscopy with SNV preprocessing and the spin-echo NMR spectroscopy with NLM preprocessing as single-data-source prediction models, can achieve 100% identification of sunflowers from Xinjiang. However, both are prone to misclassification of sunflowers from Heilongjiang and Inner Mongolia. Following the method described in Section 2.3, these are fused at the data layer to establish ELM and KELM origin prediction models. The experimental results show that the optimal origin identification model with information fusion is KELM. The Kernel Extreme Learning Machine (KELM) model exhibits enhanced precision and F1 scores on the test set when contrasted with the top-performing single-spectrum predictor. Moreover, it manifests superior robustness relative to individual data source configurations, as evidenced by the data presented in Table 5.

Table 5.
Main parameters of the optimal model for origin identification of single data, data-level fusion data, and feature-level fusion data.
The data layer fusion model that utilizes the KELM algorithm combined with the SNV and NLM preprocessing methods achieves an identification accuracy of 98.2% on the validation set. However, as shown in Figure 13c, this model incorrectly classified one sunflower sample from Inner Mongolia as being from Heilongjiang.
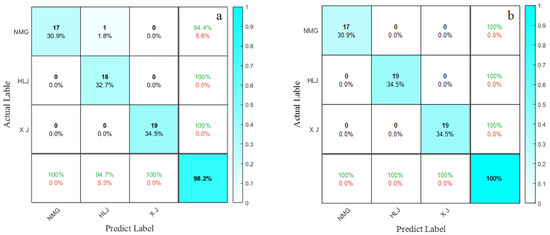
Figure 13.
(a) confusion matrix for the data-level optimization model test dataset; (b) confusion matrix for the feature-level optimization model test dataset.
Data layer fusion provides more comprehensive information about the initial sample compared to feature layer fusion, but directly linking the data may negatively affect the predictive performance of the recognition model. Although directly linked spectral data retain the most complete information, they also retain interfering cross-category information that is not useful for categorization, which may interfere with correct categorization. This was also demonstrated in a study by Zhou, Y. [27]. By integrating multiple data fusion strategies, the features of the data were further extracted to overcome the effect of simple data linkage on the model and further validate the above hypothesis. Table 6 demonstrates the experimental results of the feature layer fusion model.

Table 6.
Optimal identification results of the feature-level fusion strategy model.
Data fusion strategies can effectively enhance the accuracy of origin identification models. Studies [20,23,24,25,26,28] have also demonstrated that multi-source data fusion strategies at the feature level improve the prediction accuracy of provenance forecasting models. Further analysis reveals that raw information in low-level information fusion strategies hinders the synergy effect of spectral technology in multi-source spectrum fusion, resulting in unsatisfactory performance of low-level multi-source spectrum fusion.
Within the framework of the sunflower origin identification system, the integration of the KELM algorithm with the CARS–Joint Feature approach yields the most effective classifier. This model boasts 100% accuracy across both the training and validation datasets, and it secures the pinnacle F1 score on the test set, outperforming both standalone data sources and the finest data layer fusion alternatives.
The model can quickly and stably complete the task of identifying sunflower origins. The optimal feature layer fusion model improves the test set accuracy by 2.8% compared to the best single data source model and by 1.8% compared to the best data layer fusion model, meeting the needs for rapid and accurate detection of common sunflower origins.
4. Conclusions
This study innovatively established a multi-source data layer fusion and feature layer fusion model for sunflower origin identification, utilizing the information fusion techniques of near-infrared spectroscopy (NIRS) and spin-echo nuclear magnetic resonance (NMRS). Data collection was achieved through low-field nuclear magnetic resonance equipment and near-infrared spectrometers; various preprocessing algorithms were applied to the original data, and ELM and KELM models were separately constructed for origin identification, yielding predictions from single-source models. On this basis, four types of feature-layer information-fusion identification models were established using two feature extraction algorithms, CARS and VIP, combined with two feature fusion strategies: joint features and simple concatenation. The experimental results indicate that NIRS and NMRS are feasible for sunflower seed origin identification. However, the conclusion that the KELM algorithm with SNV preprocessing is the best single-spectrum model contradicts the findings in [49], where the first derivative (1st) was deemed superior. Yet, the results align with those in [27], possibly due to differences in sample attributes and acquisition environments across datasets, suggesting that optimal preprocessing methods should be determined experimentally for each sample [69]. Both data layer and feature layer fusion models significantly improved identification accuracy, with the CARS-KELM model achieving 100% accuracy on both the training and test sets using joint features. Under both fusion strategies, sunflower seed origin identification models showed significantly higher accuracy than single-spectrum models, consistent with the findings in [26] that fusion strategies significantly enhance origin identification accuracy, regardless of modeling methods. This suggests that fusing multispectral information at the feature level using joint features is a modeling approach that can improve prediction performance across different machine learning models. Sunflower seed origin identification models under both fusion strategies exhibited higher accuracy than single-spectrum models [25,28]. Regarding the influence of the number of hidden layers in extreme learning machines [39], the application of kernel functions has been found to effectively address this issue. It is recommended to further explore the specific parameters of single and combined kernel functions for origin prediction models in kernel extreme learning machines.
Although the KELM spectral fusion model achieved 100% accuracy on the test and prediction sets for LongshikuiNo.1 sunflower datasets from different origins, successfully resolving the issue of sunflower seed origin prediction, this study also verified that the feature layer fusion method combining NIRS and NMRS data is suitable for sunflower seed origin identification tasks, providing an application case for subsequent studies on origin modeling using multi-source spectral data. However, there may be potential limitations due to the design of single-layer neural networks with limited ability to analyze complex problems. The geographical range (in longitude and latitude) of the sunflower samples collected in this study was restricted to a similar longitude of 45° E (with a fluctuation of 2 degrees) and a latitude interclass fluctuation variation of 10°, indicating a lack of extensive geographic scope for the experimental samples. Future research will focus on evaluating the stability and robustness of the proposed method for sunflower seed origin prediction in a broader region and even for different crops. The study plans to incorporate more varieties and a larger geographic range of sunflower samples, extending the origin prediction model framework with multi-source information fusion strategies proposed in this study to traceability research on the origin of more crop varieties and expanding the application scope of the algorithm.
Author Contributions
Conceptualization, L.S. and H.L.; Methodology, H.L.; Software, J.N. and Z.W.; Validation, L.S., H.L., Z.W. and J.N.; Formal analysis, R.Z.; Investigation, R.Z.; Resources, L.S. and R.Z.; Data curation, L.S.; Writing—original draft, H.L and Z.W.; Writing—review & editing, L.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant number: 31772789).
Data Availability Statement
The original contributions presented in this study are included in this article; further inquiries can be directed to the corresponding author.
Acknowledgments
The authors thank the reviewers and editor for their insightful comments and constructive suggestions.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Marchioni, I.; Gabriele, M.; Carmassi, G.; Ruffoni, B.; Pistelli, L.; Pistelli, L.; Najar, B. Phytochemical, Nutritional and Mineral Content of Four Edible Flowers. Foods 2024, 13, 939. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, T.; Tabrizi, L.; Nazeri, V.; Etemadi, M. Nutrient distribution in various tissues of Licorice (Glycyrrhiza glabra L.) and the influence of soil fertility on the levels of its bioactive compounds. Ind. Crops Prod. 2024, 209, 118073. [Google Scholar] [CrossRef]
- Domaratskiy, E.O.; Bazaliy, V.V.; Domaratskiy, O.O.; Dobrovol, A.V.; Kyrychenko, N.V.; Kozlova, O.P. Influence of mineral nutrition and combined growth regulating chemical on nutrient status of sunflower. Indian J. Ecol. 2018, 45, 126–129. [Google Scholar]
- Yildiz, S.; Erdoğan, S. Quality Traits of the Nutrient Matter Compositions and Yield parameters of planted Silage Corn Zea mays L. and Sunflower Helianthus annuus L. at Conditions of Van. Türkiye Tarımsal Araștırmalar Derg. 2018, 5, 280–285. [Google Scholar] [CrossRef]
- Mrdja, J.; Crnobarac, J.; Radić, V.; Miklič, V. Sunflower seed quality and yield in relation to environmental conditions of production region. Helia 2012, 35, 123–134. [Google Scholar]
- Lagiso, T.M.; Singh, B.C.S.; Weyessa, B. Evaluation of sunflower (Helianthus annuus L.) genotypes for quantitative traits and character association of seed yield and yield components at Oromia region, Ethiopia. Euphytica 2021, 217, 27. [Google Scholar] [CrossRef]
- Varalakshmi, K.; Neelima, S.; Reddy, R.N.; Sreenivasulu, K. Genetic variability studies for yield and its component traits in newly developed sunflower (Helianthus annuus L.) hybrids. Electron. J. Plant Breed. 2020, 11, 301–305. [Google Scholar]
- Khan, F.A.; Iqbal, A.; Saeed, A.; Naeem, A.; Mehmood, M.A. Combining ability studies for yield and others quality traits of sunflower (Helianthus annuus L.) by using line× tester analysis. J. Agric. Res. 2021, 59, 7. [Google Scholar]
- Chen, X.; Zhang, H.; Teng, A.; Zhang, C.; Lei, L.; Ba, Y.; Wang, Z. Photosynthetic characteristics, yield and quality of sunflower response to deficit irrigation in a cold and arid environment. Front. Plant Sci. 2023, 14, 1280347. [Google Scholar] [CrossRef]
- Castillo-Lorenzo, E.; Pritchard, H.W.; Finch-Savage, W.E.; Seal, C.E. Comparison of seed and seedling functional traits in native Helianthus species and the crop H. annuus (sunflower). Plant Biol. 2019, 21, 533–543. [Google Scholar] [CrossRef]
- Heubl, G. New aspects of DNA-based authentication of Chinese medicinal plants by molecular biological techniques. Planta Med. 2010, 76, 1963–1974. [Google Scholar] [CrossRef] [PubMed]
- Timme, R.E.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am. J. Bot. 2007, 94, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, C. Boys and girls come out to play: The molecular biology of dioecious plants. Ann. Bot. 2000, 86, 211–221. [Google Scholar] [CrossRef]
- Sneh, B.; Jabaji-Hare, S.; Neate, S.; Dijst, G. Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control; Springer Science & Business Media: New York, NY, USA, 2013. [Google Scholar]
- Wang, H.; Li, J.; Tao, W.; Zhang, X.; Gao, X.; Yong, J.; Zhao, J.; Zhang, L.; Li, Y.; Duan, J. Lycium ruthenicum studies: Molecular biology, phytochemistry and pharmacology. Food Chem. 2018, 240, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Dai, X.; Guo, Z.; Wang, C. Identification of Rice from Different Origins by Two-dimensional Correlation Near Infrared Spectroscopy. J. Chin. Inst. Food Sci. Technol. 2023, 23, 331–338. [Google Scholar]
- Tong, P.; Lim, L.J.; Wei, T.; Elejalde, U.; Zhang, H.; Jiang, Y.; Cao, W. Rapid identification of the variety and geographical origin of Wuyou No. 4 rice by fourier transform near-infrared spectroscopy coupled with chemometrics. J. Cereal Sci. 2021, 102, 103322. [Google Scholar]
- Liu, X.; Bai, B.; Rogers, K.M.; Wu, D.; Qian, Q.; Qi, F.; Zhou, J.; Yao, C.; Song, W. Determining the geographical origin and cultivation methods of Shanghai special rice using NIR and IRMS. Food Chem. 2022, 394, 133425. [Google Scholar] [CrossRef]
- Hu, Y.-R.; Li, J.-Q.; Liu, H.-G.; Fan, M.-P.; Wang, Y.-Z. The Origin Identification Study of Boletus Edulis Based on the Infrared Spctrum Data Fusion Strategy. Spectrosc. Spectr. Anal. 2020, 40, 1276–1282. [Google Scholar]
- Hu, Y.-R.; Li, J.-Q.; Liu, H.-G.; Fan, M.-P.; Wang, Y.-Z. Infrared spectral study on the origin identification of Boletus Tomentipes based on the random forest algorithm and data fusion strategy. Spectrosc. Spectr. Anal. 2020, 40, 1495–1502. [Google Scholar]
- Xu, Z.-W.; Feng, C.-P.; Fan, S.; Zhang, B.-L.; Zhao, H.-F.; Liu, Y.-N.; Yue, H.-W.; Ji, X.; Zhang, S.-X.; Lu, W. Rapid determination of alcohol content in alcoholic beverages by low-field NMR. Food Ferment. Ind. 2022, 48, 254–261. [Google Scholar]
- Ackermann, S.M.; Dolsophon, K.; Monakhova, Y.B.; Kuballa, T.; Reusch, H.; Thongpanchang, T.; Bunzel, M.; Lachenmeier, D.W. Automated multicomponent analysis of soft drinks using 1D 1 H and 2D 1 H-1 H J-resolved NMR spectroscopy. Food Anal. Methods 2017, 10, 827–836. [Google Scholar] [CrossRef]
- Godinho, M.S.; Blanco, M.R.; Neto, F.F.G.; Lião, L.M.; Sena, M.M.; Tauler, R.; de Oliveira, A.E. Evaluation of transformer insulating oil quality using NIR, fluorescence, and NMR spectroscopic data fusion. Talanta 2014, 129, 143–149. [Google Scholar] [CrossRef]
- Li, Q.; Huang, Y.; Zhang, J.; Min, S. A fast determination of insecticide deltamethrin by spectral data fusion of UV–vis and NIR based on extreme learning machine. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 247, 119119. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Qu, C.; An, T.; Qian, J.; Shi, M.; Wang, X.; Hu, M. Applications of Molecular Spectral information Fusion to Distinguish the Rice From Different Growing Regions. Spectrosc. Spectr. Anal. 2023, 43, 2818–2824. [Google Scholar]
- Dai, Y.; Dai, Z.; Guo, G.; Wang, B. Nondestructive identification of rice varieties by the data fusion of Raman and near-infrared (NIR) spectroscopies. Anal. Lett. 2023, 56, 730–743. [Google Scholar] [CrossRef]
- Zhou, Y.; Zuo, Z.; Xu, F.; Wang, Y. Origin identification of Panax notoginseng by multi-sensor information fusion strategy of infrared spectra combined with random forest. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 226, 117619. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.-Y.; Wang, Y.-Z. FT-MIR and NIR spectral data fusion: A synergetic strategy for the geographical traceability of Panax notoginseng. Anal. Bioanal. Chem. 2018, 410, 91–103. [Google Scholar] [CrossRef]
- Mutyala, V.; Anaparthi, V.; Mundru, Y.; Yogi, M.K. Frontiers of AI beyond 2030: Novel Perspectives. J. Artif. Intell. 2022, 4, 246–262. [Google Scholar]
- Maragathavalli, P.; Thanushri, A.; Gayathri, S.N.L.; Asok, H. A Survey on Cyberbullying Predictive Model using Deep Learning Techniques. J. Trends Comput. Sci. Smart Technol. 2024, 6, 99–111. [Google Scholar] [CrossRef]
- Huang, G.-B.; Zhu, Q.-Y.; Siew, C.-K. Extreme learning machine: A new learning scheme of feedforward neural networks. In Proceedings of the 2004 IEEE International Joint Conference on Neural Networks (IEEE Cat. No. 04CH37541), Budapest, Hungary, 25–29 July 2004; pp. 985–990. [Google Scholar]
- Huang, G.-B.; Zhou, H.; Ding, X.; Zhang, R. Extreme learning machine for regression and multiclass classification. IEEE Trans. Syst. Man Cybern. Part B Cybern. 2011, 42, 513–529. [Google Scholar] [CrossRef]
- Li, H.; Chou, C.; Chen, Y.; Wang, S.; Wu, A. Robust and lightweight ensemble extreme learning machine engine based on eigenspace domain for compressed learning. IEEE Trans. Circuits Syst. I Regul. Pap. 2019, 66, 4699–4712. [Google Scholar] [CrossRef]
- Huang, G.-B.; Ding, X.; Zhou, H. Optimization method based extreme learning machine for classification. Neurocomputing 2010, 74, 155–163. [Google Scholar] [CrossRef]
- Leung, H.C.; Leung, C.S.; Wong, E.W.M. Fault and noise tolerance in the incremental extreme learning machine. IEEE Access 2019, 7, 155171–155183. [Google Scholar] [CrossRef]
- Dai, H.; Cao, J.; Wang, T.; Deng, M.; Yang, Z. Multilayer one-class extreme learning machine. Neural Netw. 2019, 115, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xie, X.; Zhang, T.; Bai, J.; Hou, M. A deep residual compensation extreme learning machine and applications. J. Forecast. 2020, 39, 986–999. [Google Scholar] [CrossRef]
- Jin, Z.; Zhou, G.; Gao, D.; Zhang, Y. EEG classification using sparse Bayesian extreme learning machine for brain–computer interface. Neural Comput. Appl. 2020, 32, 6601–6609. [Google Scholar] [CrossRef]
- Diker, A.; Avci, D.; Avci, E.; Gedikpinar, M. A new technique for ECG signal classification genetic algorithm Wavelet Kernel extreme learning machine. Optik 2019, 180, 46–55. [Google Scholar] [CrossRef]
- Ayaz, S.; Ahmad, M.; Zafar, M.; Ali, M.I.; Sultana, S.; Mustafa, M.R.U.; Kilic, O.; Çobanoğlu, D.N.; Demirpolat, A.; Ghani, A. Taxonomic significance of cypsela morphology in tribe Cichoreae (Asteraceae) using light microscopy and scanning electron microscopy. Microsc. Res. Tech. 2020, 83, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Carrijo, T.T.; Garbin, M.L.; Picanço Leite, W.; Mendonça, C.B.F.; Esteves, R.L.; Gonçalves-Esteves, V. Pollen morphology of some related genera of Vernonieae (Asteraceae) and its taxonomic significance. Plant Syst. Evol. 2013, 299, 1275–1283. [Google Scholar] [CrossRef]
- Ayaz, A.; Gu, Y. Macromorphological and foliar epidermal anatomical characteristics of Lilium rosthornii (Liliaceae): Implications for morphological adaptations and taxonomic significance. Microsc. Res. Tech. 2024, 1–7. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, C. Leaf morphological characteristics and taxonomic significance of eight Allium species. J. Northwest A F Univ.-Nat. Sci. Ed. 2018, 46, 107–116. [Google Scholar]
- Grace, O.M.; Simmonds, M.S.; Smith, G.F.; Van Wyk, A.E. Taxonomic significance of leaf surface morphology in Aloe section Pictae (Xanthorrhoeaceae). Bot. J. Linn. Soc. 2009, 160, 418–428. [Google Scholar] [CrossRef][Green Version]
- Freitas, F.S.; De-Paula, O.C.; Nakajima, J.N.; Marzinek, J. Fruits of Heterocoma (Vernonieae-Lychnophorinae): Taxonomic significance and a new pattern of phytomelanin deposition in Asteraceae. Bot. J. Linn. Soc. 2015, 179, 255–265. [Google Scholar] [CrossRef]
- Ranjbar, M.; Karami, S. Pollen morphology of some selected species of the tribes Brassiceae, Conringieae, Isatideae, and Plagiolobeae (Brassicaceae) in Iran, and its taxonomic significance. Palynology 2023, 47, 2138606. [Google Scholar] [CrossRef]
- Emlee, A.M.; Amri, C.N.A.C.; Midin, M.R. Taxonomic Significance of Leaf Micromorphology in Selected Garcinia from Peninsular Malaysia. Revel. Sci. 2023, 1. [Google Scholar] [CrossRef]
- Liu, Y.-D.; Ye, L.-Y.; Sun, X.-D.; Han, R.-B.; Xiao, H.-C.; Ma, K.-R.; Zhu, D.-N.; Wu, M.-M. Maturity evaluation model of tangerine based on spectral index. Chin. Opt. 2018, 11, 83–91. [Google Scholar]
- Pan, S.; Zhang, X.; Xu, W.; Yin, J.; Gu, H.; Yu, X. Rapid On-site identification of geographical origin and storage age of tangerine peel by Near-infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 271, 120936. [Google Scholar] [CrossRef]
- Jin, W.-L.; Cao, N.-L. Nondestructive grading test of rice seed activity using near infrared super-continuum laser spectrum. Chin. Opt. 2020, 13, 1032–1043. [Google Scholar]
- Westad, F.; Schmidt, A.; Kermit, M. Incorporating chemical band-assignment in near infrared spectroscopy regression models. J. Near Infrared Spectrosc. 2008, 16, 265–273. [Google Scholar] [CrossRef]
- Choi, H.; Vinograd, I.; Chaffey, C.; Curro, N. Inverse Laplace transformation analysis of stretched exponential relaxation. J. Magn. Reson. 2021, 331, 107050. [Google Scholar] [CrossRef]
- Werbos, P. Beyond Regression: New Tools for Prediction and Analysis in the Behavioral Sciences. Ph.D. Thesis, Committee on Applied Mathematics, Harvard University, Cambridge, MA, USA, 1974. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Liu, X.; Wang, L.; Huang, G.-B.; Zhang, J.; Yin, J. Multiple kernel extreme learning machine. Neurocomputing 2015, 149, 253–264. [Google Scholar] [CrossRef]
- Zhao, Y.-P. Parsimonious kernel extreme learning machine in primal via Cholesky factorization. Neural Netw. 2016, 80, 95–109. [Google Scholar] [CrossRef] [PubMed]
- You, C.-X.; Huang, J.-Q.; Lu, F. Recursive reduced kernel based extreme learning machine for aero-engine fault pattern recognition. Neurocomputing 2016, 214, 1038–1045. [Google Scholar] [CrossRef]
- Iosifidis, A.; Tefas, A.; Pitas, I. On the kernel extreme learning machine classifier. Pattern Recognit. Lett. 2015, 54, 11–17. [Google Scholar] [CrossRef]
- Luo, F.; Guo, W.; Yu, Y.; Chen, G. A multi-label classification algorithm based on kernel extreme learning machine. Neurocomputing 2017, 260, 313–320. [Google Scholar] [CrossRef]
- Iosifidis, A.; Gabbouj, M. On the kernel extreme learning machine speedup. Pattern Recognit. Lett. 2015, 68, 205–210. [Google Scholar] [CrossRef]
- Mojrian, S.; Pinter, G.; Joloudari, J.H.; Felde, I.; Szabo-Gali, A.; Nadai, L.; Mosavi, A. Hybrid machine learning model of extreme learning machine radial basis function for breast cancer detection and diagnosis; a multilayer fuzzy expert system. In Proceedings of the 2020 RIVF International Conference on Computing and Communication Technologies (RIVF), Ho Chi Minh City, Vietnam, 14–15 October 2020; pp. 1–7. [Google Scholar]
- Qin, Y.; Li, M.; De, G.; Huang, L.; Yang, S.; Tan, Q.; Tan, Z.; Zhou, F. Research on green management effect evaluation of power generation enterprises in China based on dynamic hesitation and improved extreme learning machine. Processes 2019, 7, 474. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey wolf optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Makhadmeh, S.N.; Al-Betar, M.A.; Doush, I.A.; Awadallah, M.A.; Kassaymeh, S.; Mirjalili, S.; Zitar, R.A. Recent advances in Grey Wolf Optimizer, its versions and applications. IEEE Access 2023, 12, 22991–23028. [Google Scholar] [CrossRef]
- Wang, M.; Chen, H.; Li, H.; Cai, Z.; Zhao, X.; Tong, C.; Li, J.; Xu, X. Grey wolf optimization evolving kernel extreme learning machine: Application to bankruptcy prediction. Eng. Appl. Artif. Intell. 2017, 63, 54–68. [Google Scholar] [CrossRef]
- Feng, L.; Wu, B.; Zhu, S.; Wang, J.; Su, Z.; Liu, F.; He, Y.; Zhang, C. Investigation on data fusion of multisource spectral data for rice leaf diseases identification using machine learning methods. Front. Plant Sci. 2020, 11, 577063. [Google Scholar] [CrossRef] [PubMed]
- Bouhrara, M.; Maring, M.C.; Spencer, R.G. A simple and fast adaptive nonlocal multispectral filtering algorithm for efficient noise reduction in magnetic resonance imaging. Magn. Reson. Imaging 2019, 55, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Huang, Z.; Ge, P.; Gao, F.; Gao, F. Adaptive denoising of photoacoustic signal and image based on modified Kalman filter. J. Biophotonics 2023, 16, e202200362. [Google Scholar] [CrossRef]
- Mokari, A.; Guo, S.; Bocklitz, T. Exploring the steps of infrared (IR) spectral analysis: Pre-processing,(classical) data modelling, and deep learning. Molecules 2023, 28, 6886. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).