Abstract
The WD40 gene family is a highly conserved protein family in plants that plays a crucial role in various life activities. Although eggplant (Solanum melongena L.) genome sequencing has been completed, there is limited research on the WD40 family in eggplant, and the regulatory mechanism of its involvement in anthocyanin synthesis remains poorly understood. The research identified the eggplant WD40 gene family, comprising 187 SmWD40 members that are unevenly distributed across 12 chromosomes of the eggplant. Phylogenetic analysis classified them into 11 subgroups, with members within the same subgroup having similar motifs and gene structures. The promoter of the SmWD40 genes contains a high number of light, stress, and hormone response elements. The expression patterns of 20 SmWD40 members of the S5 subgroup were analyzed during the formation of fruit color in long purple eggplant. Subsequently, we used virus-induced gene silencing (VIGS) to confirm the significance of the TTG1 (SmWD40-56) gene in subgroup S5 for anthocyanin synthesis in eggplant fruit. To investigate the molecular mechanism of SmWD40-56 in eggplant fruit color formation, we analyzed the expression patterns of structural genes for anthocyanin synthesis in eggplant fruit silenced for SmWD40-56. Finally, we predicted the protein interaction network of the SmWD40-56 gene to understand its potential regulatory mechanisms. The result showed that SmWD40-56 may regulate the structural genes involved in anthocyanin biosynthesis and plays an important role in eggplant fruit color formation. This study provides some basis for studying the mechanism of eggplant fruit color formation.
1. Introduction
Anthocyanin is a natural water-soluble pigment that belongs to a class of flavonoids [,,]. It is widely distributed in most plant organs and plays a crucial role in the development of blue, purple, and red traits in plant tissues [,]. Anthocyanins are important indicators for evaluating fruit quality and contribute significantly to plant resistance against various stresses [,,]. The regulation of anthocyanin synthesis is complex and involves both structural genes and transcription factors [].
The MBW complex is a complex formed by the MYB transcription factor, helix-loop-helix (bHLH) transcription factor and a WD40-repeat protein [,], which is widely involved in flavonoid synthesis, morphogenesis, and resistance physiology in plants [,,]. Anthocyanin regulation by MBW complexes is complex and sophisticated []. MBW complexes can regulate the structural genes of anthocyanin synthesis such as dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), anthocyanidin synthase (ANS), flavanone 3-hydroxylase (F3H), and so on, and thus play an important role in regulating anthocyanin synthesis []. In addition, the MBW complex can activate anthocyanin accumulation induced by jasmonic acid (JA) []. WD40 proteins are a type of transcription factor with unique structures []. They are one of the most diverse protein families in plants [,]. The WD40 domain is unique and folds into a β-propeller structure, making it one of the most diverse structures in the eukaryotic genome. This domain serves as a platform for protein interaction and assembly [,,]. WD40 proteins play an active role in regulating anthocyanin synthesis by enhancing and activating gene expression. Transparent testa glabra 1 (TTG1) and AN11 belong to the WD40 gene family and are frequently reported as anthocyanin-regulated genes [,]. TTG1 and AN11 can bind to R2R3-MYB transcription factors such as MYB113/114 and polynucleotide adenylyltransferase 1/2 (PAP1/2), thereby regulating the expression levels of anthocyanin synthesizing structural genes such as ANS, F3H, and DFR, which ultimately have an impact on anthocyanin biosynthesis [,]. In Petunia hybrida Vilm, the PhAN11 mutants showed the reduced expression of DFR. Further studies in Arabidopsis have revealed that the homologous gene of AN11, AtTTG1, functions similarly to AN11 [,]. Several studies have highlighted the regulatory function of WD40 genes in the coloration of flowers and fruits. For example, the expression of the cotton TTG1 gene in the MiTTG1 mutant of Matthiola incana L. can partially restore some of the anthocyanin phenotypes []. Similarly, the apple MdTTG1 gene can form a complex with bHLH to initiate the transcription of downstream genes involved in anthocyanin synthesis, such as MdDFR and MdUFGT [].
Eggplant (Solanum melongena L.) stands as a vital vegetable with global cultivation potential. The peel color of eggplant, a crucial quality trait, greatly influences the evaluation of fruit maturity and consumer preferences [,]. The peel color of eggplant exhibits a diverse range, including common purple, white, green, and other colors. Research indicates a direct correlation between eggplant fruit color and the contents of anthocyanin and chlorophyll. A higher anthocyanin content results in a darker peel color, and the fruit is green when anthocyanin content is low, or white when chlorophyll content is also low [,]. The molecular mechanism of eggplant fruit color formation has been a hot topic of research. Expression analyses of structural (CHS, DFR and ANS) and regulatory genes (Myc, MYbB and MYbC) for anthocyanin synthesis in eggplants of different fruit colors revealed that the expression levels of anthocyanin biosynthesis genes were higher in purple fruits than in white fruits, with the upregulation of Myc and MYbC genes showing a trend consistent with that of anthocyanin biosynthesis genes []. Expression profiling of the structural genes for anthocyanin synthesis in eggplants (SmCHS, SmCHI, SmF3H, SmDFR, and SmANS) for different tissues, light conditions, and temperatures revealed that all genes were expressed at higher levels in the pericarp tissues except SmF3H; the expressions of SmCHI and SmANS were completely dependent on light. At low temperatures, all the structural genes were upregulated to a certain extent []. The MBW complex also plays an important role in fruit coloration in eggplant, with the MYB transcription factor being the most intensively studied. MYB transcription factors such as SmMYB1, SmMYB2, SmMYB75, SmMYB44, SmMYB86, and SmMYB35 have been identified to contribute to anthocyanin synthesis and accumulation in eggplant [,,,]. Some MYB transcription factors, like SmMYB86, inhibited anthocyanin synthesis by binding to the promoters of SmCHS, SmF3H, and SmANS. Silencing SmMYB86 can significantly increase the accumulation level of anthocyanin accumulation []. In eggplant, the bHLH regulation of anthocyanin synthesis involves two main branches. One branch involves AN1, which activates anthocyanin biosynthesis through the MYB-AN1-WD40 (MBW) complex, while the other branch involves JAF13, which forms the MYB-JAF13-WD40 (MBW) complex to regulate downstream AN1, thereby activating anthocyanin biosynthesis []. WD40 provides a stable platform for MYB and bHLH proteins in the MBW complex. The overexpression of the WD40 protein STAN11 in potato upregulates the STDFR gene and increases anthocyanin accumulation []. In tomato, SlAN11 can interact with bHLH transcription factors (SLT8 and SLGL3) to form an MBW complex with SLANT or SLGL3, thereby regulating SlDFR expression and anthocyanin synthesis [].
In recent years, with the continuous increase in people’s demand for healthy food and product quality, more and more attention has been paid to the influence of anthocyanin content on the quality of eggplant fruit, and the research on the coloring mechanism of eggplant fruit has been deepened [,]. Currently, research on the mechanisms of color formation in eggplant is primarily focused on the structural genes that encode synthases and transcription factors within the MBW complex []. While studies on anthocyanin synthetase and the MBW complex involving the MYB and bHLH gene families have reached a certain level of maturity, so far there have been fewer studies on WD40 in eggplant [,]. Further exploration is warranted on the intricate interactions between WD40, MYB, and bHLH in regulating anthocyanin synthesis and influencing eggplant fruit color. Therefore, the comprehensive analysis of the WD40 family in eggplant assumes great importance in unraveling the mechanisms behind anthocyanin synthesis in eggplant fruit and enhancing the quality of eggplant. In this research, 187 members of the eggplant WD40 family were identified based on genome-wide data. The physicochemical properties analysis, phylogenetic analysis, chromosome mapping, cis-acting element analysis, and collinearity analysis were performed. This research examined the expression pattern of WD40 in different growth stages of long purple eggplant fruits and validated the function of the SmTTG1 (SmWD56) gene through virus-induced gene silencing (VIGS). Additionally, expression levels of several primary structural genes in anthocyanin synthesis were measured after SmTTG1 (SmWD40-56) silencing, and the interaction protein of the SmWD40-56 gene was predicted to delve into its role in anthocyanin synthesis and fruit color formation in eggplant. In conclusion, this study provides a thorough analysis of the eggplant WD40 family. The results are significant for the investigation of eggplant fruit color formation and the cultivation of superior varieties.
2. Materials and Methods
2.1. Identification and Physicochemical Property of SmWD40 Gene Family
Genomic data for eggplant were sourced from the eggplant genome database (http://www.eggplant-hq.cn/Eggplant) (accessed on 8 August 2023). The Hidden Markov Model (HMM) file (PF00400) of the WD40 conserved domain was obtained by downloading from Pfam (http://pfam-legacy.xfam.org) (accessed on 8 August 2023) to characterize the SmWD40 members. Protein sequence analysis of candidate genes was conducted using NCBI’s CD-Search functional (https://www.ncbi.nlm.nih.gov) (accessed on 8 August 2023) analysis tool to screen for SmWD40 members with complete WD40 structures. The final set of SmWD40 members was derived by excluding structurally incomplete genes. Subsequently, SmWD40 protein sequences were submitted for an analysis of the physicochemical properties of SmWD40 members to ExpasyProteparam (https://web.expasy.org/proteparam/) (accessed on 8 August 2023).
2.2. Chromosomal Location and Phylogenetic Analysis of SmWD40
The chromosome localization of SmWD40 members was performed by TBtools v2.052 software []. The Arabidopsis WD40 member protein sequence was obtained from TAIR (https://www.arabidopsis.org/) (accessed on 10 August 2023). The WD40 gene family protein sequences of eggplant and Arabidopsis were aligned by MEGA11 v11.0.10 software. The phylogenetic tree was constructed using the NJ method, and the bootstrap value was set at 1000.
2.3. Conserved Motif Analysis and Cis-Acting Element Analysis of SmWD40
MEME (https://meme-suite.org/meme/) (accessed on 14 August 2023) was utilized to analyze the motif characteristics of SmWD40 members, while their conserved domain information underwent analysis through the CD-Search function of NCBI (https://www.ncbi.nlm.nih.gov) (accessed on 14 August 2023). Upstream 2000 bp nucleic acid sequences from each SmWD40 member were extracted and uploaded to PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on 14 August 2023) for cis-acting element analysis in promoter regions. The conserved motifs and cis-acting elements, combined with phylogenetic relationships, were visually presented by the TBtools v2.052 software [].
2.4. Collinearity Analysis of SmWD40
SmWD40 collinearity analysis was performed by the MCScanX plug-in in TBtools v2.052 software, and the collinearity relationship was visualized by TBtools v2.052 software [].
2.5. Plant Materials and Treatments
The eggplant used in this experiment was the long purple eggplant (9211: A strain from Solanum melongena cv. Kmzc-7), the seeds of which were saved by our research group. At the beginning of the experiment, the seeds were soaked in distilled water for 24 h, and then the seeds were germinated in 15 mm glass petri dishes lined with moistened filter paper. Subsequently, they were sown in peat and vermiculite mixed medium (3:1), and then transplanted to a 15 cm diameter planting pot where they grew to 2 pairs of true leaves. Next, they were cultivated in the planting base of Southwest Forestry University (102.454308° E, 25.035229° N), with a temperature of 25 °C, relative humidity maintained at 60–70%, and a light–dark cycle of 14 h to 10 h. After the flower buds appeared, they were utilized for subsequent experiments.
To investigate the potential mechanism of the S5 subgroup in eggplant fruit color formation, gene expression levels in eggplant peel (0.5–0.8 mm) were analyzed by qRT-PCR at 5, 15, and 25 d after the end of flowering. The stage I (5 d) represents the early phase of eggplant fruit development, when the fruit color was full green. The stage II (15 d) refers to the middle stage of eggplant fruit development, when the fruit color began to appear purple, and there were still large areas of green. In stage III (25 d), the eggplant fruit became mature, and the fruit was covered with purple. The surface of the fruit was washed with ddH2O and frozen in liquid nitrogen, and then stored at −80 °C.
2.6. Virus-Induced Gene Silencing
The experimental treatments were slightly adapted based on previous research on eggplant []. When the eggplant material grew to the early stage of flower bud development, two pairs of mature leaves below the flower bud were infected by injection. The bacterial solution had a concentration of OD600 = 0.8, with equal proportions of TRV1 and TRV2 bacterial solutions mixed in equal proportions 1 h prior injection. To control the experimental variables, two control groups were established, consisting of empty TRV2 and H2O. When the eggplant fruit reached maturity, TRV2 detection primers were adopted to assess the virus transmission within the fruit, and the amplified fragment length was determined using the 5000 bp marker (Biomed Biotech Co., Ltd., Beijing, China). The expression level of SmTTG1 was detected by qRT-PCR.
2.7. Analysis of Anthocyanin Synthesis Structural Gene Expression Level
The anthocyanin synthesis structural genes in eggplant were acquired from the eggplant genome database (http://www.eggplant-hq.cn/Eggplant) (accessed on 12 September 2023). Expression levels of structural genes were measured by qRT-PCR, including CHS (Smechr0500387), CHI (Smechr0500261), F3H (Smechr0101736), DFR (Smechr0202337), flavonol synthase (FLS, Smechr0401078), and PAL (Smechr0400589).
2.8. qRT-PCR and Viral Vector Construction
RNA was extracted using FastPure Universal Plant Total RNA Isolation Kit (Vazyme Biotech Co., Ltd., Nanjing, China). RNA integrity was detected using 1% agarose gel electrophoresis, and the OD260/280 values of RNA were detected using a nucleic acid proteometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) to ensure that the quality of RNA met the requirements of subsequent experiments. Subsequently, RNA was reverse-transcribed into cDNA using All-In-One 5X RT MasterMix (Applied Biological Materials Inc., Shanghai, China). The qRT-PCR primers were designed using the primer-Blast function of NCBI database (Table A1). To prevent the presence of gDNA contamination in RNA, negative control experiments were performed during qRT-PCR using unreversed transcribed RNA as a template, and no contamination was considered to have occurred with no significant amplification of the negative control. qRT-PCR was performed using ArtiCanCEO SYBR qPCR Mix (Tsingke Biotechnology Co., Ltd., Beijing, China). The qRT-PCR reaction process was as follows: pre-denaturation at 95 °C for 1 min, followed by cycling for 10 s at 95 °C and 20 s at 60 °C for 40 cycles. Fluorescence was collected at the 60 °C reaction step. The Actin gene (Smechr1001310.1) served as the internal reference for a calculation of the gene relative expression using 2−ΔΔCt. The primer amplification efficiencies for qRT-PCR are shown in Table A1. The VIGS experiment was performed using tobacco rattle virus vectors. The 300 bp length fragment was identified as the silencing fragment using the VIGS online prediction tool (https://vigs.solgenomics.net/) (accessed on 16 August 2023), and the target fragment was amplified by MegaFi TM Fidelity 2X PCR MasterMix (Applied Biological Materials Inc., Shanghai, China). TRV2 was linearized using KpnI endonuclease (New England Biolabs Inc., Beijing, China). The ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, China) was employed to homologously recombine the ligated fragment and linearized vector. Leveraging recombinant plasmids, E. coli (Top 10) was transformed, and the bacterial solution was sequenced at Tsingke Biotech Co., Ltd. (Tsingke Biotechnology Co., Ltd., Beijing, China). Using the Trelief® reg Plasmid Mini Kit Plus (Beijing Tsingke Biotech Co., Ltd.) kit, the correct bacterial liquid vector was extracted, and the agrobacterium (GV3101) was transformed by the thermal stimulation method.
2.9. Interaction Network Analysis of SmWD40-56
To investigate the potential regulatory mechanism of SmWD40-56 in eggplant, we performed protein sequencing of the eggplant whole genome interaction protein by SmWD40-56 via STRING (https://cn.string-db.org) (accessed on 3 October 2023).
3. Results
3.1. Identification of the WD40 Genes in Eggplant
WD40 members were systematically identified in the entire eggplant genome utilizing a Hidden Markov Model. After manually removing the ineligible genes, we obtained a total of 187 compliant WD40 genes. The genes were designated as SmWD40-1 through SmWD40-187 in the order of their arrangement on the chromosomes. An analysis of physicochemical properties revealed the diversity within the SmWD40 protein family. The length of SmWD40 proteins varied from 169 amino acids (SmWD40-164) to 3249 amino acids (SmWD40-83), while the molecular weight ranged from 18,088.63 Da (SmWD40-164) to 364,048.07 Da (SmWD40-83). The analysis of the theoretical isoelectric point (pI) revealed that 125 SmWD40 proteins have a pI value of less than 7, indicating a negative charge in an alkaline pH environment. One protein (SmWD40-143) had a pI of 7, indicating no charge in a neutral environment, and 61 proteins exhibited a pI value greater than 7, suggesting a positive charge in an acidic pH range. Instability index analysis categorized 81 SmWD40 proteins as stable, with an instability coefficient of below 40, while the remaining 106 SmWD40 proteins were deemed unstable, boasting an instability coefficient surpassing 40. Aliphatic Index predictions ranged from 54.81 (SmWD40-141) to 103.1 (SmWD40-168). The grand average of hydropathicity prediction revealed that with the exception of SmWD40-65, SmWD40-78, and SmWD40-168, the grand average of hydropathicity for the remaining 184 SmWD40 proteins was less than 1, indicating a hydrophilic nature (Table A2).
3.2. Chromosome Localization of SmWD40 Genes
The analysis of chromosomal localization for the 187 identified WD40 genes indicated that the SmWD40 gene family members are distributed across 12 chromosomes, primarily concentrated at the chromosome ends. The analysis of gene density further indicated that the SmWD40 genes are mainly located in regions with high gene density on the chromosomes. Chromosomes 01, 02, and 03 have a higher abundance of SmWD40 genes, with 29, 19, and 27 members, respectively. In contrast, the numbers of SmWD40 genes on chromosomes 09, 11, and 12 were relatively low, with 10, 10, and 6 genes, respectively (Figure 1).
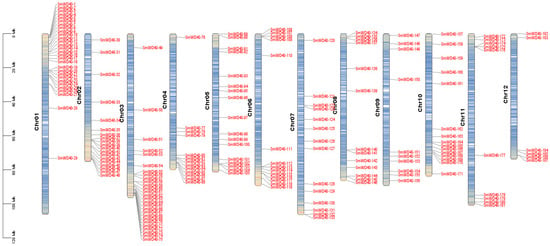
Figure 1.
Chromosomal localization of the SmWD40 genes. Each bar corresponds to the length of a chromosome, with red and blue indicating regions with high and low gene density, respectively.
3.3. Phylogenetic Analysis of SmWD40 Proteins
To investigate the phylogenetic relationships within the SmWD40 gene family, the phylogenetic tree of 187 WD40 members in eggplant and 162 in Arabidopsis was constructed using the neighbor-joining (NJ) method (Figure 2). The study revealed that the phylogenetic relationship can be divided into 11 distinct subgroups. Subgroups S1 and S9 had the largest numbers of members (44), while subgroup S8 had the least (20). Notably, each subgroup included both eggplant and Arabidopsis proteins; this result indicates that WD40 is highly conserved in eggplant and Arabidopsis.
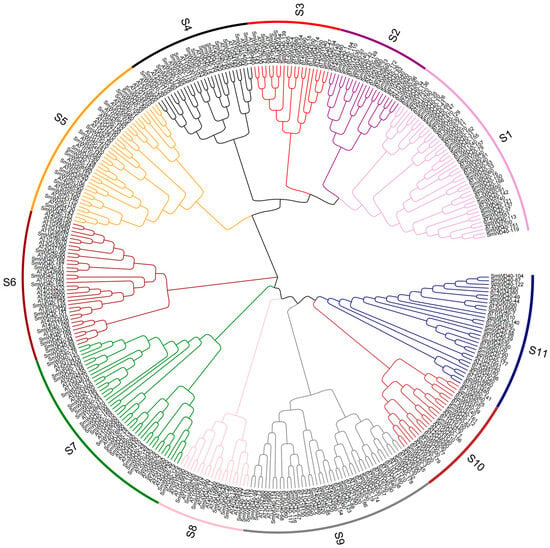
Figure 2.
Phylogenetic trees for WD40 proteins in eggplant and Arabidopsis, with different colors representing various subgroups. AT: Arabidopsis thaliana. Sm: Solanum melongena.
3.4. Conserved Domains and Cis-Acting Elements Prediction of SmWD40s
Motifs and conserved domains in SmWD40 were integrated based on phylogenetic relationships for further analysis. The results reveal a distribution of similarities in motifs and domains among members of the same subgroup in phylogenetic development, with motif 1 being the most prevalent across the majority of members (Figure 3A). The conserved domain analysis (Figure 3B) demonstrated that all members exhibited the WD40-specific domain, confirming the accurate identification of SmWD40 members. In addition, a parallel distribution of motif 10 and the CTLH domain regions was observed in the subgroups S3 and S5, suggesting that there may be functional similarity related to the structure.
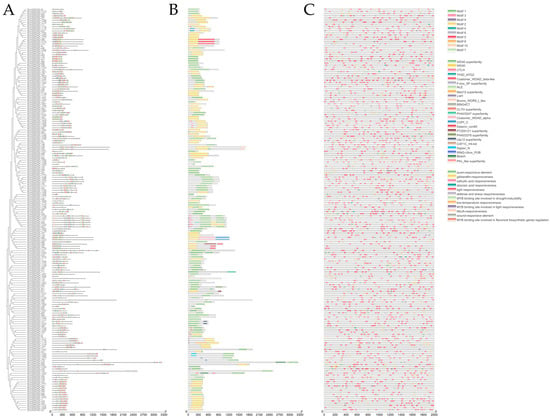
Figure 3.
SmWD40 gene conserved domain, motif, and the cis-acting elements analysis. (A) Motif compositions. (B) Conserved domains. (C) Cis-acting elements.
Analyzing cis-acting elements in the upstream 2000 bp promoter region of the SmWD40 genes (Figure 3C) showed that light response elements were the most abundant among SmWD40 promoters. Among the hormone response elements, MeJA response elements were the most prevalent, with SmWD40-135 having the largest number of members (20). SmWD40-50 exhibited the largest number of auxin response elements (four members), while SmWD40-168 and SmW40-162 had the largest numbers of gibberellin response elements (five members). Among the salicylic acid response elements, SmWD40-3, SmWD40-20, and SmWD40-177 had the highest counts, with three members each. SmWD40-41 had the largest number of abscisic acid response elements, with nine members.
3.5. Collinearity Analysis of SmWD40 Genes
In order to understand the amplification mechanism of SmWD40 genes, the collinearities of SmWD40 genes were analyzed (Figure 4). The results show that there were 16 pairs of fragment duplication genes in SmWD40. Collinear gene pairs were distributed across most chromosomes, except Chr10 and Chr11, with a predominant concentration in high-gene-density regions at both ends of the chromosomes. Notably, SmWD40-121 showed a collinear relationship with SmWD40-112 and SmWD40-43, and SmWD40-94 displayed collinear relationships with SmWD40-81 and SmWD40-58. Meanwhile, SmWD40-71 was collinearly associated with SmWD40-7, SmWD40-33, and SmWD40-84, and SmWD40-43 exhibited a collinear correlation with both SmWD40-121 and SmWD40-90.
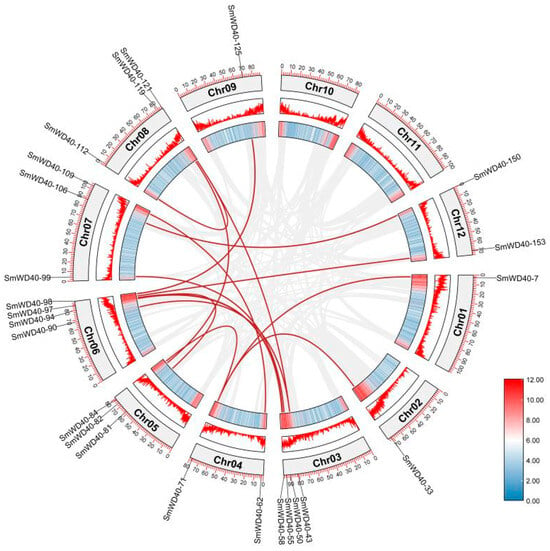
Figure 4.
Collinearity analysis of SmWD40s. The red line represents collinear genes. The colors of the bars represent the gene density on the chromosome, with red and blue signifying high and low density, respectively.
3.6. Expression Pattern Analysis of S5 Subgroup Genes in Fruit Color Development
Previous studies underscored the pivotal role of WD40 in the synthesis and accumulation of anthocyanins in plants. Current studies in Arabidopsis have shown that several genes in the S5 subgroup, such as AT5G24520 (AtTTG1) and AT1G12910 (AtAN11), have regulatory effects on anthocyanins. There may be similarities in gene function in the same subgroups [], so we hypothesize that the SmWD40 genes in the S5 subgroup may be involved in the regulation of anthocyanin synthesis in eggplant. To explore the potential influence mechanism of the SmWD40 family in eggplant fruit color formation, we analyzed the expression patterns of S5 subgroup genes in eggplant fruits at different growth times (5 d, 15 d, 25 d) after the end of flowering by qRT-PCR (Figure 5A). The results show that these SmWD40 members exhibited different expression patterns (Figure 5B). To be specific, two distinct categories emerged: Firstly, genes exhibiting high levels in stage I and subsequently decreasing expression in stages II and III, such as SmWD40-21, SmWD40-27, SmWD40-43, SmWD40-56, SmWD40-58, SmWD40-70, SmWD40-128, SmWD40-145, SmWD40-153, SmWD40-160 and SmWD40-182. It is suggested that these genes have a potential regulatory role in the initiation of anthocyanin synthesis in eggplant fruits. On the other hand, genes with lower expressions in stage I but elevated expressions in stages II and III were identified. Examples include SmWD40-30, SmWD40-92, SmWD40-105, SmWD40-106, SmWD40-132, SmWD40-152 and SmWD40-181. These genes may exert potential regulatory functions in pigment accumulation and transport in eggplant fruits.
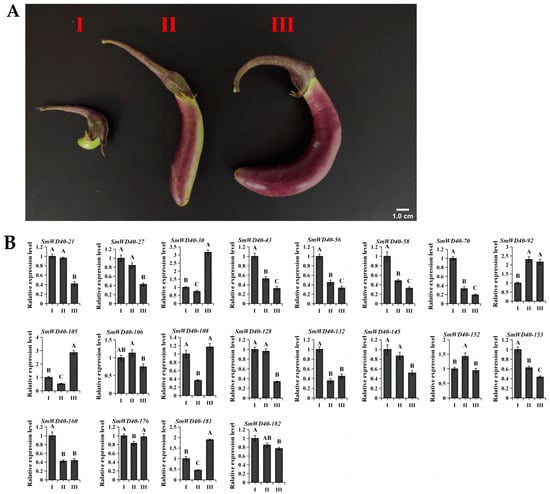
Figure 5.
Expression pattern analysis of S5 subgroup genes in fruit color development. (A) Eggplant fruits at different growth times after end of flowering. Stages I, II, and III represent 5, 15, and 25 d after the end of flowering, respectively. In stage I, the fruits were completely green; in stage II, the fruits began to show purple color, but were still partially green; and in stage III, the fruits were covered with purple coloring. (B) Expression patterns within the subgroup S5 at distinct stages of fruit color development. The y-axis depicts the relative gene expression, while the x-axis corresponds to various stages of fruit color development. The data, presented as mean ± standard deviation from three biological replicates, reveal remarkable differences between samples, as indicated by distinct capital letters (p < 0.01) based on one-way ANOVA and Tukey’s test. Error bars represent the standard deviation (SD).
3.7. Analysis of the Effects of Silencing SmWD40-56 on Fruit Color and the Expression Pattern of Structural Genes for Anthocyanin Synthesis after Silencing
TTG1 plays a crucial role in anthocyanin formation in various plant species. Based on its expression patterns, the eggplant TTG1 gene, known as SmWD40-56, may contribute to the early stages of anthocyanin formation in eggplant fruit. To investigate the regulatory role of the TTG1 gene in anthocyanin synthesis in eggplant fruits, we reduced the expression levels of SmWD40-56 genes using VIGS technology. Detection primers specific to tobacco rattle virus (TRV) were used to assess mature fruits of infected plants. The results show that both Empty TRV2-infected plants and TRV2-SmWD40-56 were able to detect the amplification bands of TRV. Noticeably, the amplified fragments of TRV2-SmWD40-56 were approximately 300 bp longer in comparison to those of Empty TRV2 (Figure 6A). This observation confirms that the TRV was successfully transmitted in eggplant fruits, and that the SmWD40-56 silenced fragment was successfully transmitted with viral vectors.
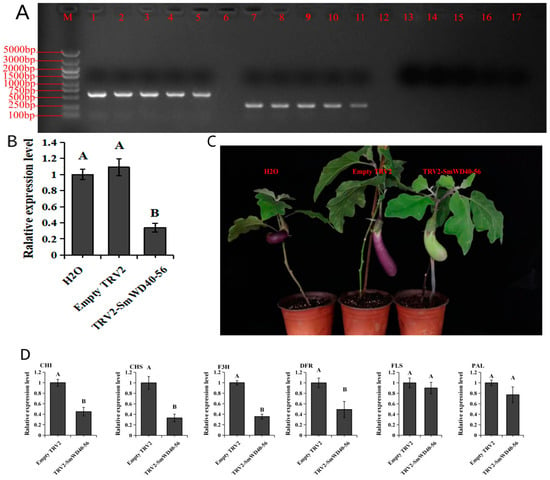
Figure 6.
Analysis of the effect of silencing SmWD40-56 on fruit color and the expression pattern of structural genes for anthocyanin synthesis after silencing. (A) Gel electrophoresis of different silencing treatment groups. M: 5000 bp marker. 1–5: TRV2-SmWD40-56. 7–11: Empty TRV2. 13–17: H2O. (B) The expression level of SmWD40-56 under distinct silencing treatments. (C) Fruit phenotype of long purple eggplant under varying silencing treatments. (D) Effect of silencing SmWD40-56 on anthocyanin synthesis structural genes in long purple eggplant fruit. The results in (B,D) are based on one-way ANOVA and Tukey’s test (p < 0.01), with capital letters showing significant differences and error bars showing standard deviation.
After the qRT-PCR assay, it was evident that VIGS significantly downregulated the expression of the SmWD40-56 gene in eggplant fruits (Figure 6B). The colors of the mature fruits in plants with silenced SmWD40-56 were significantly different from those of the control plants (Figure 6C). The analysis focused on the expression levels of structural genes responsible for anthocyanin synthesis in fruit that had been silenced for SmWD40-56. The results show that chalcone-flavanone isomerase (CHI), chalcone synthase (CHS), F3H, and DFR expression levels decreased significantly after SmWD40-56 silencing, indicating that SmWD40-56 has a potential regulatory effect on these anthocyanin synthases (Figure 6D).
3.8. Protein Interaction Network of SmWD40-56 in Eggplant
To investigate the regulation mechanism of SmWD40-56, we used STRING to predict the interaction network of the SmWD40-56 protein in eggplant (Figure 7). The results show that SmWD40-56 interacted with genes related to flavonoid synthesis (Smechr0501995.1, Smechr0202337.1, Smechr1000237.1, Smechr0500261.1, Smechr0202240.1, Smechr0302994.1 and Smechr0401078.1), the anthocyanin-containing compound metabolic process (Smechr0401078.1, Smechr1000237.1, Smechr0202337.1) and the jasmonic acid mediated signaling pathway (Smechr0802367.1, Smechr0101484.1). In addition, SmWD40-56 interacted with genes related to pathways such as cell differentiation, protein ubiquitination, and the regulation of biosynthesis processes. The KEGG pathway prediction for the interacting proteins indicates their potential involvement in flavonoid biosynthesis. Local cluster analysis highlighted a group of interacting genes including SmWD40-56 and Smechr0202337.1, Smechr0302994.1, Smechr0202240.1, Smechr0500261.1, as well as Smechr0401078.1. This group may regulate plant anthocyanin synthesis through its naringenin 3-dioxygenase activity, chalcone isomerase activity, L-ascorbic acid binding, and oxidoreductase activity.
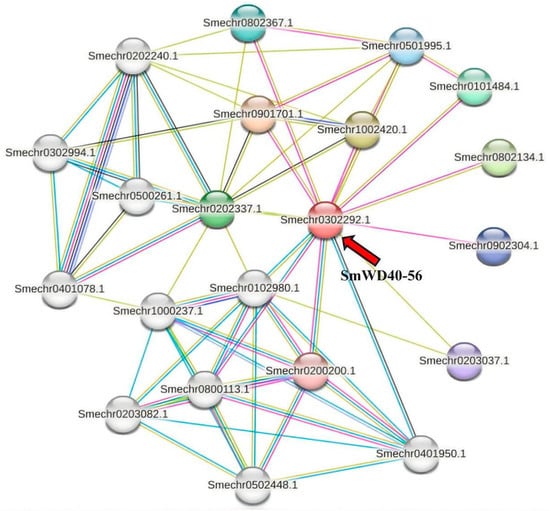
Figure 7.
Protein interaction network of SmWD40-56. Each node corresponds to a distinct protein. The colored and uncolored nodes signify proteins potentially forming the first and the second shell of interactions with SmWD40-56, respectively. Lines connecting nodes denote known or predicted protein–protein interactions. The red arrow identifies the SmWD40-56 protein.
4. Discussion
At present, with the advancement in sequencing technology, the genome-wide identification of numerous gene families has been successfully conducted in a variety of plants. However, the eggplant WD40 gene family is still unclear []. In this study, the WD40 gene family of eggplant was characterized at the genome-wide level and subjected to the analysis of predicted physicochemical properties of proteins, sequence characterization, chromosomal localization, cis-acting element analysis, and gene duplication events. Expression patterns for members in subgroup S5 of the long purple eggplant fruit were investigated at distinct stages of color formation using qRT-PCR. It was found that these genes have complex expression patterns during fruit development. The eggplant TTG1 gene (SmWD40-56) is highly expressed in the early stages of fruit color formation and has potential regulatory functions in the initiation of anthocyanin synthesis. Finally, we used VIGS technology to verify the function of SmWD40-56 gene. Expression levels of structural genes in anthocyanin synthesis in eggplant fruits after silencing were analyzed by qRT-PCR, and we analyzed the protein interaction network of the SmWD40-56 gene. The silencing of the SmWD40-56 gene resulted in significant changes in the color of long purple eggplant fruits and a significant decrease in the expression levels of CHS, CHI, F3H and DFR. Combined with protein interaction network analysis, it was found that the SmWD40-56 gene may affect eggplant fruit color by interacting with multiple genes and thus regulating the expression levels of structural genes for anthocyanin synthesis.
Anthocyanins hold great potential in many aspects, such as human health and improving the ornamental value of plants [,]. Meanwhile, they find widespread applications in horticultural traits, food production, and health care [,,]. In addition, anthocyanins are extremely critical in addressing adverse environmental conditions like drought and hypothermia [,]. Consequently, anthocyanin biosynthesis remains a focal point of research [,,]. The crucial involvement of R2R3-MYB, bHLH, and WD40 in anthocyanin synthesis is well-established [,,]. As transcription factors, they collectively form the MBW complex and regulate gene expression in anthocyanin biosynthetic pathways by binding to corresponding cis-acting elements in the promoters of structural gene [,]. The previous study identified the WD40 gene family in different types of plants. Specifically, in tomatoes, 207 WD40 members were identified and classified into 12 subgroups, and the analysis revealed that the WD40 protein may be involved in tomato fruit development []. A total of 258 WD40 genes were identified in Rhododendron simsii Planch, and transcriptome analysis revealed that these genes play potential regulatory roles in anthocyanin synthesis []. A total of 178 WD40 genes were identified in the potato genome, among which TTG1 and its interactions with proteins play an important role in potato anthocyanin synthesis []. Previous studies have highlighted the significance of TTG1, a member of the WD40 family, in anthocyanin biosynthesis in different plants, such as fabe bean [], pomegranate [], and Arabidopsis []. This research analyzed cis-acting elements and found that SmWD40 contains numerous light-, hormone-, and stress-response elements. This suggests that SmWD40 plays a significant role in eggplant photomorphogenesis, hormone response, and resistance to physiological processes. These findings are consistent with previous research on anthocyanin function [,,]. The examination of the SmWD40 gene at different stages of eggplant color formation highlighted the potential regulatory role of members from subgroup S5 in fruit anthocyanin formation. The significance of SmWD40-56 in the formation of anthocyanin in eggplants was subsequently confirmed by employing the VIGS of the eggplant TTG1 gene (SmWD40-56).
At present, the process of synthesizing anthocyanin in plants is well understood, and it is a branch of the flavonoid synthesis pathway. Phenylalanine serves as the precursor for anthocyanin synthesis [,]. Under the influence of enzymes like phenylalanine ammonia-lyase (PAL) and CHS, phenylalanine transforms into chalcone []. Chalcone, in turn, forms naringenin through the activity of CHI, and further converts to dihydrokaempferol under the action of F3H []. Eventually, dihydrokaempferol undergoes a series of reactions catalyzed by DFR, leading to the formation of leucoanthocyanidin []. In this study, the protein interaction network analysis of the SmWD40-56 gene showed that this gene was highly correlated with the synthesis of chalcone and naringenin. Moreover, gene expression levels of CHS, CHI, F3H and DFR were significantly reduced after the silencing of SmWD40-56. Therefore, it could be speculated that the SmWD40-56 gene may interact with the above proteins to regulate and activate anthocyanin synthesis-related genes, thereby affecting the color formation of eggplant fruits.
5. Conclusions
In this study, 187 WD40 genes were identified in the whole genome of eggplant from the HMM file, which were irregularly distributed on 12 chromosomes of eggplant. Phylogenetic analysis showed that these SmWD40 proteins could be categorized into 11 subgroups. Gene motif and conserved domain analyses indicated similarities in gene structure among the same subgroups of phylogeny, suggesting the possibility of similar gene functions. The analysis of cis-acting elements revealed that all SmWD40 genes contain light-responsive elements, indicating a strong correlation between their gene expression and light. In addition, a large number of hormone-responsive elements and resistance-responsive elements were identified, suggesting that SmWD40 is regulated by a complex signaling network involved in the physiological process of resistance in eggplant. The analysis of the expression pattern of the S5 subgroup of genes during eggplant fruit development suggests that this subgroup of genes may play a regulatory role in processes such as the initiation of eggplant fruit color formation and anthocyanin accumulation. After applying VIGS technology to decrease the expression levels of SmWD40-56 genes in long purple eggplant fruit, the intensity of purple color in the fruits decreased significantly. The qRT-PCR results indicate that silencing SmWD40-56 significantly reduced the expression levels of CHS, CHI, F3H, and DFR genes. Additionally, protein interaction network analysis revealed a strong correlation between the SmWD40-56 gene and anthocyanin synthesis. This study serves as a reference for comprehending the mechanism of eggplant fruit color formation and enhancing eggplant quality.
Author Contributions
Conceptualization, Y.Y., Q.C. and Z.S.; formal analysis, Y.Y. and Q.C.; investigation, Y.Y., Q.C. and Y.W.; methodology, Y.Y., Q.C., Y.W., Z.S. and L.L.; project administration, Y.Y. and Q.C.; resources, Z.S. and L.L.; supervision, Z.S. and L.L.; validation, Y.Y. and Q.C.; visualization, Y.Y. and Q.C.; writing—original draft, Y.Y.; writing—review and editing, Y.Y., Q.C., Z.S. and L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The Key Research and Development Program of Yunnan Province (202202AEO90012-04), International cooperation project of Yunnan Province (2019IB011).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
The primer sequences used in this research.
Table A1.
The primer sequences used in this research.
| Primer Name | Gene ID | Forward Primer (5′->3′) | Reverse Primer (5′->3′) | Function | Amplification Efficiency |
|---|---|---|---|---|---|
| SmWD40-21 | Smechr0102060.1 | GGATGGGGATGCAAGGACAA | CATGCATCAAGTTCTCGCCG | SmWD40-21 qRT-PCR | 0.9612 |
| SmWD40-27 | Smechr0102379.1 | TGGAATAGGTGTGGGGGACT | TGTGCATAACCACCAAACTGG | SmWD40-27 qRT-PCR | 0.9956 |
| SmWD40-30 | Smechr0200037.1 | AGAGGAATCTGGGGTTGGGA | AGCCAAACGGTAAGGGACAG | SmWD40-30 qRT-PCR | 0.9835 |
| SmWD40-43 | Smechr0202538.1 | ATGACTTCACCTACCGAGCC | AGAAGGGATCCATCGGAAACG | SmWD40-43 qRT-PCR | 0.9744 |
| SmWD40-56 | Smechr0302292.1 | TGAGTGCCGAAAATAACAATAAAAC | GGGCTATCAATTGGGTTTCGAC | SmWD40-56 qRT-PCR | 0.9725 |
| SmWD40-58 | Smechr0302644.1 | ACAGTCATGTTACCGCAGGTT | AGGTAGCCAGAGGTTGACCA | SmWD40-58 qRT-PCR | 0.9882 |
| SmWD40-70 | Smechr0303426.1 | GCTGGGACCTCTCGGTCA | ACCATCGGCATGTGTCTCTT | SmWD40-70 qRT-PCR | 0.9673 |
| SmWD40-92 | Smechr0500827.1 | AGCAGGTTCTAGCTGGGATG | AGGGCCTGTTCAACGAGATG | SmWD40-92 qRT-PCR | 0.9956 |
| SmWD40-105 | Smechr0502667.1 | TGATATTTCATCCCTGCACGA | AGGCCTTTTCCAGGTGAGTC | SmWD40-105 qRT-PCR | 1.0014 |
| SmWD40-106 | Smechr0600010.1 | CATCGGTCGACCATTCGAGA | TTTGGAGGGAGTGGTGCTTC | SmWD40-106 qRT-PCR | 0.9892 |
| SmWD40-108 | Smechr0600126.1 | TTTGCTCCTTGGCACAACAA | TGCAACTTTCTCGTCTGCCC | SmWD40-108 qRT-PCR | 0.9766 |
| SmWD40-128 | Smechr0701564.1 | TGCAAAAGATTGGTCTGGTGG | TGTTTATGTAAAGAGGCGCGA | SmWD40-128 qRT-PCR | 0.9883 |
| SmWD40-132 | Smechr0702707.1 | ATGTCGTGCTGCCATGAACA | GCTCTCAAAGCCAAATACACG | SmWD40-132 qRT-PCR | 0.9965 |
| SmWD40-145 | Smechr0802330.1 | AGAGTACGATCAACATCTCGGA | GGCGCAATAGATGGCATGG | SmWD40-145 qRT-PCR | 0.9746 |
| SmWD40-152 | Smechr0901596.1 | GTGCAGACCCTGAAAAAGGC | CACCACCTGTCCCCATGTTG | SmWD40-152 qRT-PCR | 1.0233 |
| SmWD40-153 | Smechr0901745.1 | CGGGGACATACTGGCTTTGT | TCACCGGCATAGAATGAGCC | SmWD40-153 qRT-PCR | 0.9516 |
| SmWD40-160 | Smechr1000926.1 | CCCCCTCTTATTGTCAGCGG | GAGGCGTCAGAGAAACAGACA | SmWD40-160 qRT-PCR | 0.9777 |
| SmWD40-176 | Smechr1100624.1 | ATGACCGCACAACTGCCAAA | TAGCCGGATGGTCCGATCTT | SmWD40-176 qRT-PCR | 0.1023 |
| SmWD40-181 | Smechr1102707.1 | AACAACCCTTTGGTCCCAGC | GTTAGAAGCAGCCGTTGTGC | SmWD40-181 qRT-PCR | 0.9761 |
| SmWD40-182 | Smechr1200022.1 | CAGGTCATAACGGCCCTGTG | TGGAGCCCATTCACACGAAA | SmWD40-182 qRT-PCR | 0.9552 |
| SmCHI | Smechr0500261.1 | CGATCGCTTAGCAGAAGCCG | GCGAACGATATCTTTGCACTACT | SmCHI qRT-PCR | 0.9765 |
| SmCHS | Smechr0500387.1 | GCTCGTCGTCTGTGCTGAG | ATCCCCGTTAGGCACGAATG | SmCHS qRT-PCR | 0.9838 |
| SmF3H | Smechr0101736.1 | AGCAATGGGATATACAAAGGCATC | CGCTAACGCAGGTCCAACT | SmF3H qRT-PCR | 0.9689 |
| SmDFR | Smechr0202337.1 | AGTCAGGGGAATGTTGAGCA | TCCAGCTAGTCTCGTCATAGA | SmDFR qRT-PCR | 1.0332 |
| SmFLS | Smechr0401078.1 | AAGGTCAGAGCACAACAGCC | GTCGATGACTGGCACCTCAA | SmFLS qRT-PCR | 0.9777 |
| SmPAL | Smechr0400589.1 | TGGAACTTTCGAGGGCGAAT | CGTCGGGCGATCGTACTTTT | SmPAL qRT-PCR | 0.9923 |
| SmActin | Smechr1001310.1 | GGTCGGAATGGGACAGAAGG | GGTGCCTCAGTCAGGAGAAC | Reference Gene | 0.9936 |
| SmWD40-56-VIGS | Smechr0302292.1 | ctccatggggatccgcCCAATGACATTCTTG | agacgcgtgagctcgATCAGCAGAAACGGAAG | SmWD40-56 amplification of silencing fragments | |
| TRV2 | none | TGGGAGATGATACGCTGTT | CCTAAAACTTCAGACACG | Detect infected plants |

Table A2.
Identification and physicochemical analysis of SmWD40 protein.
Table A2.
Identification and physicochemical analysis of SmWD40 protein.
| Gene Name | Gene ID | Number of Amino Acids | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|---|
| SmWD40-1 | Smechr0100054.1 | 538 | 60030.64 | 4.34 | 34.27 | 70.46 | −0.589 |
| SmWD40-2 | Smechr0100084.1 | 403 | 45571.6 | 4.69 | 40.3 | 77.59 | −0.516 |
| SmWD40-3 | Smechr0100138.1 | 440 | 48400.93 | 5.45 | 36.58 | 87.09 | −0.189 |
| SmWD40-4 | Smechr0100203.1 | 493 | 55068.17 | 9.04 | 48.05 | 72.17 | −0.398 |
| SmWD40-5 | Smechr0100250.1 | 377 | 40950.02 | 7.19 | 29.56 | 76.47 | −0.233 |
| SmWD40-6 | Smechr0100269.1 | 757 | 82965.53 | 5.51 | 32.25 | 94.66 | −0.151 |
| SmWD40-7 | Smechr0100290.1 | 469 | 51874.12 | 8.72 | 41.8 | 76.46 | −0.486 |
| SmWD40-8 | Smechr0100456.1 | 340 | 37891.44 | 8.51 | 35.37 | 68.5 | −0.521 |
| SmWD40-9 | Smechr0100664.1 | 424 | 48291.91 | 4.7 | 46.24 | 78.4 | −0.548 |
| SmWD40-10 | Smechr0100732.1 | 747 | 84865.95 | 5.11 | 51.97 | 73.31 | −0.732 |
| SmWD40-11 | Smechr0100835.1 | 1412 | 150253.47 | 6.92 | 39.42 | 88.53 | −0.101 |
| SmWD40-12 | Smechr0100970.1 | 1132 | 124636.89 | 6.6 | 42.72 | 84.89 | −0.214 |
| SmWD40-13 | Smechr0101114.1 | 982 | 106620.95 | 5.42 | 52.75 | 75.51 | −0.339 |
| SmWD40-14 | Smechr0101146.1 | 759 | 83050.86 | 6.88 | 34.38 | 82.71 | −0.321 |
| SmWD40-15 | Smechr0101232.1 | 526 | 57618.08 | 4.84 | 35.11 | 87.05 | −0.282 |
| SmWD40-16 | Smechr0101322.1 | 2513 | 276904.98 | 6.3 | 42.12 | 86.42 | −0.061 |
| SmWD40-17 | Smechr0101516.1 | 484 | 53493.77 | 9.17 | 39.41 | 72.77 | −0.477 |
| SmWD40-18 | Smechr0101622.1 | 441 | 47647.5 | 8.92 | 32.22 | 86.73 | −0.199 |
| SmWD40-19 | Smechr0102034.1 | 907 | 101447.44 | 8.01 | 42.57 | 93.84 | −0.141 |
| SmWD40-20 | Smechr0102054.1 | 442 | 49465.81 | 9.04 | 37.15 | 82.62 | −0.238 |
| SmWD40-21 | Smechr0102060.1 | 1126 | 121602.13 | 5.23 | 49.64 | 78.53 | −0.288 |
| SmWD40-22 | Smechr0102098.1 | 419 | 46256.05 | 9.03 | 40.99 | 84.39 | −0.19 |
| SmWD40-23 | Smechr0102184.1 | 1382 | 154085.28 | 6.18 | 35.94 | 86.11 | −0.15 |
| SmWD40-24 | Smechr0102204.1 | 1667 | 186041.55 | 6.55 | 46.43 | 73.11 | −0.597 |
| SmWD40-25 | Smechr0102232.1 | 353 | 39736.77 | 5.72 | 35.53 | 75.38 | −0.292 |
| SmWD40-26 | Smechr0102378.1 | 208 | 23235.25 | 6.85 | 52.26 | 73.94 | −0.32 |
| SmWD40-27 | Smechr0102379.1 | 528 | 59948.58 | 9.06 | 50.64 | 73.86 | −0.476 |
| SmWD40-28 | Smechr0102778.1 | 1698 | 190580.35 | 6.39 | 50.3 | 67.02 | −0.738 |
| SmWD40-29 | Smechr0103230.1 | 496 | 55498.41 | 8.63 | 47.27 | 76.05 | −0.406 |
| SmWD40-30 | Smechr0200037.1 | 462 | 52158.93 | 8.17 | 30.21 | 90.65 | −0.177 |
| SmWD40-31 | Smechr0200130.1 | 598 | 65181.09 | 6.65 | 41.09 | 91.79 | −0.08 |
| SmWD40-32 | Smechr0200257.1 | 820 | 90126.36 | 8.35 | 38.2 | 87.24 | −0.286 |
| SmWD40-33 | Smechr0200514.1 | 239 | 26251.65 | 4.69 | 24.93 | 101.17 | −0.029 |
| SmWD40-34 | Smechr0200897.1 | 773 | 86581.58 | 7.06 | 51.25 | 89.5 | −0.232 |
| SmWD40-35 | Smechr0201236.1 | 369 | 40099.45 | 7.04 | 24.63 | 79.7 | −0.189 |
| SmWD40-36 | Smechr0201464.1 | 516 | 58044.79 | 6.6 | 50.01 | 69.38 | −0.766 |
| SmWD40-37 | Smechr0201698.1 | 671 | 73205.32 | 5.75 | 44.53 | 79.66 | −0.261 |
| SmWD40-38 | Smechr0201774.1 | 1027 | 113880.83 | 6.41 | 55.77 | 80.68 | −0.452 |
| SmWD40-39 | Smechr0201777.1 | 336 | 36529.3 | 6.86 | 39.42 | 81.25 | −0.208 |
| SmWD40-40 | Smechr0201803.1 | 432 | 48118.81 | 4.86 | 47.8 | 58.5 | −0.526 |
| SmWD40-41 | Smechr0202123.1 | 1051 | 115331.91 | 6.84 | 42.3 | 92.12 | −0.186 |
| SmWD40-42 | Smechr0202247.1 | 332 | 36903.65 | 6.09 | 37.48 | 78.07 | −0.228 |
| SmWD40-43 | Smechr0202538.1 | 314 | 34577.84 | 6.36 | 22.95 | 80.99 | −0.291 |
| SmWD40-44 | Smechr0202756.1 | 471 | 52145.57 | 8.9 | 42.35 | 71 | −0.524 |
| SmWD40-45 | Smechr0202868.1 | 440 | 48281.7 | 5.53 | 36.32 | 87.55 | −0.189 |
| SmWD40-46 | Smechr0202967.1 | 499 | 54952.24 | 7.31 | 41.3 | 82.65 | −0.244 |
| SmWD40-47 | Smechr0203005.1 | 302 | 32550.54 | 5.84 | 22.74 | 79.11 | −0.216 |
| SmWD40-48 | Smechr0203082.1 | 514 | 56664.04 | 8.77 | 50.81 | 74.36 | −0.388 |
| SmWD40-49 | Smechr0300525.1 | 460 | 51307.37 | 8.6 | 40.56 | 70.93 | −0.491 |
| SmWD40-50 | Smechr0301092.1 | 432 | 47227.71 | 9.15 | 41.93 | 87.59 | −0.168 |
| SmWD40-51 | Smechr0301403.1 | 528 | 59266.6 | 9.56 | 36.37 | 78.52 | −0.52 |
| SmWD40-52 | Smechr0301606.1 | 434 | 47495.08 | 9.01 | 37.03 | 83.53 | −0.235 |
| SmWD40-53 | Smechr0301684.1 | 421 | 47215.62 | 7.94 | 42.57 | 82.16 | −0.144 |
| SmWD40-54 | Smechr0301928.1 | 430 | 47321.27 | 8.13 | 32.35 | 71.56 | −0.381 |
| SmWD40-55 | Smechr0302236.1 | 714 | 78581.52 | 6.43 | 46.39 | 75.84 | −0.369 |
| SmWD40-56 | Smechr0302292.1 | 417 | 47092.84 | 4.89 | 49.66 | 79.28 | −0.433 |
| SmWD40-57 | Smechr0302364.1 | 871 | 96184.7 | 6.05 | 41.83 | 87.3 | −0.181 |
| SmWD40-58 | Smechr0302644.1 | 385 | 42188.43 | 6.49 | 32.66 | 83.32 | −0.242 |
| SmWD40-59 | Smechr0302647.1 | 392 | 44101.14 | 7.17 | 43.02 | 89.97 | −0.243 |
| SmWD40-60 | Smechr0302855.1 | 404 | 44930.37 | 8.86 | 35.58 | 84.41 | −0.215 |
| SmWD40-61 | Smechr0302946.1 | 980 | 108146.29 | 5.79 | 47.3 | 79.45 | −0.408 |
| SmWD40-62 | Smechr0302999.1 | 387 | 42791.76 | 4.97 | 32.53 | 76.02 | −0.42 |
| SmWD40-63 | Smechr0303123.1 | 1132 | 124471.51 | 6.57 | 39.07 | 82.47 | −0.249 |
| SmWD40-64 | Smechr0303179.1 | 1130 | 124653.99 | 6.76 | 39.34 | 79.57 | −0.35 |
| SmWD40-65 | Smechr0303180.1 | 391 | 42538.27 | 5.22 | 52.86 | 89.05 | 0.017 |
| SmWD40-66 | Smechr0303262.1 | 564 | 63489.27 | 8.57 | 56 | 83.67 | −0.459 |
| SmWD40-67 | Smechr0303326.1 | 759 | 85606.12 | 6.63 | 39.59 | 94.18 | −0.162 |
| SmWD40-68 | Smechr0303331.1 | 609 | 66187.92 | 6.15 | 24.42 | 89.26 | −0.139 |
| SmWD40-69 | Smechr0303392.1 | 1821 | 202371.41 | 6.31 | 41.24 | 97.76 | −0.075 |
| SmWD40-70 | Smechr0303426.1 | 354 | 38347.79 | 5.4 | 38.02 | 74.92 | −0.329 |
| SmWD40-71 | Smechr0303432.1 | 903 | 101063.15 | 6.23 | 38.07 | 85.2 | −0.162 |
| SmWD40-72 | Smechr0303452.1 | 313 | 35957.79 | 5.13 | 42.84 | 85.59 | −0.286 |
| SmWD40-73 | Smechr0303453.1 | 916 | 103343.75 | 4.87 | 32.34 | 84.49 | −0.286 |
| SmWD40-74 | Smechr0303517.1 | 301 | 32399.3 | 5.09 | 32.56 | 81.96 | −0.158 |
| SmWD40-75 | Smechr0303588.1 | 459 | 50643.58 | 5.6 | 41.09 | 76.86 | −0.449 |
| SmWD40-76 | Smechr0400216.1 | 874 | 96741.3 | 5.58 | 47.68 | 70.55 | −0.581 |
| SmWD40-77 | Smechr0401256.1 | 326 | 35928.7 | 7.23 | 30.61 | 82.24 | −0.313 |
| SmWD40-78 | Smechr0401335.1 | 854 | 94768.78 | 5.31 | 39.39 | 94.67 | 0.025 |
| SmWD40-79 | Smechr0401384.1 | 1484 | 166473.85 | 5.68 | 51.06 | 89.97 | −0.285 |
| SmWD40-80 | Smechr0401851.1 | 450 | 50649.48 | 5 | 38.44 | 74.56 | −0.414 |
| SmWD40-81 | Smechr0401950.1 | 789 | 86855.86 | 6.27 | 51.99 | 79.15 | −0.397 |
| SmWD40-82 | Smechr0401952.1 | 665 | 73574.83 | 6.21 | 46.54 | 78.32 | −0.376 |
| SmWD40-83 | Smechr0401998.1 | 3249 | 364048.07 | 6.09 | 47.63 | 96 | −0.116 |
| SmWD40-84 | Smechr0402112.1 | 814 | 89327.63 | 6.31 | 39.76 | 84.9 | −0.191 |
| SmWD40-85 | Smechr0402166.1 | 580 | 65695.44 | 5.61 | 24.67 | 68.57 | −0.767 |
| SmWD40-86 | Smechr0402222.1 | 448 | 49231.7 | 6.03 | 48.3 | 89.44 | −0.212 |
| SmWD40-87 | Smechr0402308.1 | 447 | 49013.24 | 8.59 | 36.1 | 77.56 | −0.4 |
| SmWD40-88 | Smechr0402434.1 | 1218 | 137204.73 | 6.63 | 33.33 | 91.33 | −0.261 |
| SmWD40-89 | Smechr0500012.1 | 1895 | 211341.88 | 5.36 | 48.62 | 89.18 | −0.294 |
| SmWD40-90 | Smechr0500094.1 | 397 | 42640.62 | 4.3 | 47.4 | 75.42 | −0.271 |
| SmWD40-91 | Smechr0500782.1 | 438 | 49316.61 | 6.01 | 45 | 94.18 | −0.174 |
| SmWD40-92 | Smechr0500827.1 | 581 | 64645.73 | 5.75 | 42.19 | 93.87 | −0.199 |
| SmWD40-93 | Smechr0501136.1 | 348 | 38447.5 | 8.69 | 36.1 | 68.65 | −0.412 |
| SmWD40-94 | Smechr0501240.1 | 302 | 32668.9 | 5.9 | 29.18 | 83.54 | −0.135 |
| SmWD40-95 | Smechr0501281.1 | 623 | 69179.42 | 5.84 | 38.02 | 84.06 | −0.154 |
| SmWD40-96 | Smechr0501342.1 | 437 | 48304.69 | 8.37 | 35.31 | 82.59 | −0.288 |
| SmWD40-97 | Smechr0501500.1 | 334 | 36872.71 | 5.99 | 41.91 | 83.8 | −0.14 |
| SmWD40-98 | Smechr0501683.1 | 455 | 50098.93 | 8.06 | 44.26 | 82.68 | −0.168 |
| SmWD40-99 | Smechr0501744.1 | 929 | 104910.34 | 4.93 | 32.25 | 86.76 | −0.302 |
| SmWD40-100 | Smechr0501824.1 | 205 | 24016.19 | 8.46 | 49.64 | 62.73 | −1.172 |
| SmWD40-101 | Smechr0502137.1 | 486 | 53616.15 | 9.15 | 41.21 | 72.61 | −0.531 |
| SmWD40-102 | Smechr0502285.1 | 821 | 90061.98 | 7.33 | 38.18 | 85.7 | −0.162 |
| SmWD40-103 | Smechr0502290.1 | 453 | 49769.86 | 8.69 | 42.92 | 76.87 | −0.385 |
| SmWD40-104 | Smechr0502356.1 | 410 | 45405.3 | 8.34 | 30.5 | 74.85 | −0.383 |
| SmWD40-105 | Smechr0502667.1 | 1663 | 184193.33 | 6.19 | 51.02 | 89.99 | −0.146 |
| SmWD40-106 | Smechr0600010.1 | 487 | 55330.28 | 7.69 | 48.69 | 76.86 | −0.441 |
| SmWD40-107 | Smechr0600111.1 | 434 | 47259.87 | 5.42 | 41 | 91.68 | −0.058 |
| SmWD40-108 | Smechr0600126.1 | 826 | 91024.23 | 6.08 | 41.81 | 87.19 | −0.123 |
| SmWD40-109 | Smechr0600145.1 | 650 | 71617.27 | 5.45 | 47.01 | 70.52 | −0.661 |
| SmWD40-110 | Smechr0600493.1 | 473 | 53158.71 | 6.35 | 52.38 | 78.9 | −0.465 |
| SmWD40-111 | Smechr0601395.1 | 695 | 77092.81 | 5.98 | 44.39 | 83.41 | −0.31 |
| SmWD40-112 | Smechr0601940.1 | 673 | 74624.11 | 6.28 | 48.35 | 78.66 | −0.39 |
| SmWD40-113 | Smechr0601947.1 | 905 | 99310.05 | 6.42 | 54.43 | 65.31 | −0.713 |
| SmWD40-114 | Smechr0602039.1 | 514 | 58190.36 | 6.53 | 48.13 | 82.47 | −0.361 |
| SmWD40-115 | Smechr0602212.1 | 917 | 103580.79 | 4.89 | 31.65 | 86.65 | −0.284 |
| SmWD40-116 | Smechr0602338.1 | 323 | 35657.39 | 7.03 | 30.15 | 87.77 | −0.16 |
| SmWD40-117 | Smechr0602339.1 | 315 | 34997.6 | 6.65 | 33.71 | 86.6 | −0.194 |
| SmWD40-118 | Smechr0602689.1 | 416 | 47296.75 | 6.12 | 31.03 | 83.92 | −0.143 |
| SmWD40-119 | Smechr0603088.1 | 778 | 84819.8 | 6.43 | 49.78 | 67.98 | −0.539 |
| SmWD40-120 | Smechr0700242.1 | 1134 | 125041.54 | 6.46 | 37.69 | 75.56 | −0.343 |
| SmWD40-121 | Smechr0700811.1 | 1300 | 146022.8 | 5.8 | 43.64 | 99 | −0.109 |
| SmWD40-122 | Smechr0700862.1 | 443 | 48787.03 | 8.85 | 30.77 | 78.28 | −0.373 |
| SmWD40-123 | Smechr0700863.1 | 1219 | 136975.43 | 6.55 | 31.74 | 91.23 | −0.244 |
| SmWD40-124 | Smechr0700946.1 | 472 | 51755.65 | 5.08 | 48.5 | 75.4 | −0.434 |
| SmWD40-125 | Smechr0701007.1 | 507 | 55962.59 | 5.45 | 43.86 | 78.42 | −0.403 |
| SmWD40-126 | Smechr0701091.1 | 369 | 40180.43 | 5.37 | 40.88 | 83.79 | −0.132 |
| SmWD40-127 | Smechr0701168.1 | 323 | 34828.15 | 5.6 | 37.42 | 86.04 | −0.074 |
| SmWD40-128 | Smechr0701564.1 | 580 | 65174.62 | 6.28 | 39.93 | 95.5 | −0.206 |
| SmWD40-129 | Smechr0701747.1 | 391 | 42415.3 | 5.34 | 36.39 | 95.73 | −0.109 |
| SmWD40-130 | Smechr0702099.1 | 452 | 51980.04 | 9.59 | 43.59 | 69.87 | −0.662 |
| SmWD40-131 | Smechr0702444.1 | 931 | 102681.23 | 6.26 | 43.32 | 70 | −0.628 |
| SmWD40-132 | Smechr0702707.1 | 313 | 34969.47 | 6.5 | 24.56 | 78.21 | −0.377 |
| SmWD40-133 | Smechr0702744.1 | 816 | 89440.77 | 6.49 | 44.01 | 87.76 | −0.112 |
| SmWD40-134 | Smechr0800148.1 | 734 | 81426.56 | 5.72 | 39.17 | 73.27 | −0.401 |
| SmWD40-135 | Smechr0800182.1 | 422 | 47143.08 | 9.1 | 45.03 | 77.65 | −0.405 |
| SmWD40-136 | Smechr0800245.1 | 417 | 46766.29 | 8.84 | 46.53 | 81.77 | −0.252 |
| SmWD40-137 | Smechr0800403.1 | 656 | 72721.72 | 6.1 | 49.6 | 74.94 | −0.387 |
| SmWD40-138 | Smechr0800720.1 | 440 | 48200.45 | 5.83 | 40.79 | 85.11 | −0.081 |
| SmWD40-139 | Smechr0800881.1 | 1088 | 121328.9 | 5.85 | 43.4 | 89.59 | −0.173 |
| SmWD40-140 | Smechr0801440.1 | 473 | 53597.8 | 8.73 | 50.14 | 75.39 | −0.374 |
| SmWD40-141 | Smechr0801468.1 | 756 | 83339.82 | 8.48 | 55.39 | 54.81 | −0.728 |
| SmWD40-142 | Smechr0801696.1 | 1909 | 207813.61 | 5.27 | 48.43 | 84.89 | −0.365 |
| SmWD40-143 | Smechr0801889.1 | 1137 | 124958.9 | 7 | 44.38 | 77.12 | −0.319 |
| SmWD40-144 | Smechr0802284.1 | 484 | 53175.71 | 9.25 | 51.85 | 75.72 | −0.408 |
| SmWD40-145 | Smechr0802330.1 | 589 | 67263.63 | 6.31 | 45.83 | 62.43 | −0.787 |
| SmWD40-146 | Smechr0802536.1 | 728 | 81007.96 | 6.91 | 38.18 | 71.57 | −0.49 |
| SmWD40-147 | Smechr0900060.1 | 908 | 100784.21 | 8.76 | 50.2 | 72.28 | −0.565 |
| SmWD40-148 | Smechr0900415.1 | 911 | 102220.86 | 8.76 | 49.83 | 71.68 | −0.678 |
| SmWD40-149 | Smechr0900611.1 | 925 | 99984.42 | 8.06 | 38.82 | 88.01 | −0.001 |
| SmWD40-150 | Smechr0900982.1 | 1353 | 148539.39 | 5.95 | 46.24 | 90.78 | −0.088 |
| SmWD40-151 | Smechr0901524.1 | 761 | 83468.49 | 6.57 | 44.54 | 67.95 | −0.554 |
| SmWD40-152 | Smechr0901596.1 | 516 | 58319.57 | 5.52 | 32.1 | 92 | −0.233 |
| SmWD40-153 | Smechr0901745.1 | 318 | 34965.64 | 8.83 | 26.12 | 82.67 | −0.191 |
| SmWD40-154 | Smechr0902013.1 | 349 | 38922.34 | 5.03 | 25.92 | 72.84 | −0.378 |
| SmWD40-155 | Smechr0902163.1 | 599 | 64872.51 | 6.42 | 40.41 | 91.65 | −0.08 |
| SmWD40-156 | Smechr0902418.1 | 521 | 56569.94 | 6.18 | 29.8 | 84.09 | −0.254 |
| SmWD40-157 | Smechr1000020.1 | 300 | 33350.69 | 9.75 | 36.8 | 72.17 | −0.586 |
| SmWD40-158 | Smechr1000478.1 | 377 | 42581.03 | 9.12 | 41.57 | 78.7 | −0.528 |
| SmWD40-159 | Smechr1000759.1 | 212 | 24088.84 | 8.75 | 28.49 | 93.3 | −0.193 |
| SmWD40-160 | Smechr1000926.1 | 341 | 37563.78 | 7.11 | 36.75 | 72.02 | −0.358 |
| SmWD40-161 | Smechr1001004.1 | 315 | 35276.69 | 5.99 | 36.49 | 78 | −0.321 |
| SmWD40-162 | Smechr1001354.1 | 783 | 84904.31 | 6.5 | 47.52 | 76.87 | −0.368 |
| SmWD40-163 | Smechr1001468.1 | 391 | 43489.13 | 8.63 | 50.52 | 81.3 | −0.401 |
| SmWD40-164 | Smechr1001592.1 | 169 | 18088.63 | 9.1 | 42.47 | 92.9 | −0.034 |
| SmWD40-165 | Smechr1001624.1 | 924 | 102745.94 | 6.71 | 53.42 | 64.89 | −0.716 |
| SmWD40-166 | Smechr1001754.1 | 1345 | 148416.1 | 6.06 | 44.42 | 89.62 | −0.097 |
| SmWD40-167 | Smechr1001849.1 | 533 | 59967.06 | 9.58 | 42.66 | 83.32 | −0.38 |
| SmWD40-168 | Smechr1001886.1 | 187 | 20631.09 | 8.57 | 49.91 | 103.1 | 0.107 |
| SmWD40-169 | Smechr1001952.1 | 343 | 38023.91 | 5.38 | 45.56 | 86.91 | −0.145 |
| SmWD40-170 | Smechr1001992.1 | 325 | 35950.37 | 5.77 | 37.24 | 80.37 | −0.229 |
| SmWD40-171 | Smechr1002755.1 | 555 | 61974.65 | 6.07 | 40.04 | 80.22 | −0.611 |
| SmWD40-172 | Smechr1100180.1 | 380 | 41235.45 | 6.23 | 37.84 | 79.34 | −0.309 |
| SmWD40-173 | Smechr1100321.1 | 354 | 38754.14 | 4.72 | 33.59 | 88.95 | −0.342 |
| SmWD40-174 | Smechr1100584.1 | 443 | 48823.45 | 6.5 | 41.29 | 83.43 | −0.227 |
| SmWD40-175 | Smechr1100594.1 | 650 | 72901.54 | 6.33 | 44.64 | 66.37 | −0.547 |
| SmWD40-176 | Smechr1100624.1 | 298 | 33074.38 | 6.85 | 38.76 | 84.97 | −0.233 |
| SmWD40-177 | Smechr1101663.1 | 430 | 47675.99 | 9.16 | 42.56 | 80 | −0.29 |
| SmWD40-178 | Smechr1102321.1 | 821 | 89984.91 | 6.49 | 39.31 | 88.33 | −0.292 |
| SmWD40-179 | Smechr1102445.1 | 580 | 63841.26 | 5.43 | 49.8 | 82.79 | −0.37 |
| SmWD40-180 | Smechr1102608.1 | 493 | 53785.19 | 4.52 | 40.31 | 84.12 | −0.326 |
| SmWD40-181 | Smechr1102707.1 | 424 | 46942.13 | 8.38 | 41.49 | 84.03 | −0.262 |
| SmWD40-182 | Smechr1200022.1 | 584 | 65408.14 | 6.12 | 39.28 | 99.61 | −0.099 |
| SmWD40-183 | Smechr1200183.1 | 323 | 34904.16 | 5.64 | 38.71 | 83.62 | −0.071 |
| SmWD40-184 | Smechr1201542.1 | 476 | 53542.49 | 6.47 | 44.76 | 76.16 | −0.501 |
| SmWD40-185 | Smechr1201594.1 | 557 | 63080.15 | 8.55 | 38.78 | 86.62 | −0.32 |
| SmWD40-186 | Smechr1201735.1 | 1071 | 116791.17 | 6.24 | 38.87 | 73.52 | −0.419 |
| SmWD40-187 | Smechr1201743.1 | 672 | 76211.31 | 6.84 | 41.77 | 83.79 | −0.294 |
References
- Harborne, J.B.; Williams, C.A. Anthocyanins and other flavonoids. Nat. Prod. Rep. 2001, 18, 310–333. [Google Scholar] [CrossRef]
- Middleton, E.; Kandaswami, C.; Theoharides, T.C. The Effects of Plant Flavonoids on Mammalian Cells: Implications for Inflammation, Heart Disease, and Cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar] [CrossRef]
- Mohammed, H.A.; Khan, R.A. Anthocyanins: Traditional Uses, Structural and Functional Variations, Approaches to Increase Yields and Products’ Quality, Hepatoprotection, Liver Longevity, and Commercial Products. Int. J. Mol. Sci. 2022, 23, 2149. [Google Scholar] [CrossRef]
- Wu, X.; Beecher, G.R.; Holden, J.M.; Haytowitz, D.B.; Gebhardt, S.E.; Prior, R.L. Concentrations of Anthocyanins in Common Foods in the United States and Estimation of Normal Consumption. J. Agric. Food Chem. 2006, 54, 4069–4075. [Google Scholar] [CrossRef]
- An, J.; Zhang, X.; Bi, S.; You, C.; Wang, X.; Hao, Y. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, S.; Cheng, Y.; Peng, Z.; Han, J. Transcriptome profiling of anthocyanin-related genes reveals effects of light intensity on anthocyanin biosynthesis in red leaf lettuce. PeerJ 2018, 6, e4607. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Lim, S.; Lee, J.G.; Lee, E.J. VcBBX, VcMYB21, and VcR2R3MYB Transcription Factors Are Involved in UV–B-Induced Anthocyanin Biosynthesis in the Peel of Harvested Blueberry Fruit. J. Agric. Food Chem. 2017, 65, 2066–2073. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Zhao, M.; Morohashi, K.; Hatlestad, G.; Grotewold, E.; Lloyd, A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 2008, 135, 1991–1999. [Google Scholar] [CrossRef]
- Wei, Q.; Zhang, F.; Sun, F.; Luo, Q.; Wang, R.; Hu, R.; Chen, M.; Chang, J.; Yang, G.; He, G. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant Sci. 2017, 265, 112–123. [Google Scholar] [CrossRef]
- Gil-Muñoz, F.; Sánchez-Navarro, J.A.; Besada, C.; Salvador, A.; Badenes, M.L.; Naval, M.d.M.; Ríos, G. MBW complexes impinge on anthocyanidin reductase gene regulation for proanthocyanidin biosynthesis in persimmon fruit. Sci. Rep. 2020, 10, 3543. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–b HLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-Domain Proteins Interact with the WD-Repeat/bHLH/MYB Complexes to Regulate Jasmonate-Mediated Anthocyanin Accumulation and Trichome Initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Gaitatzes, C.; Saxena, K.; Neer, E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999, 24, 181–185. [Google Scholar] [CrossRef]
- Stirnimann, C.U.; Petsalaki, E.; Russell, R.B.; Müller, C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010, 35, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Fong, H.; Hurley, J.B.; Hopkins, R.S.; Miake-Lye, R.; Johnson, M.S.; Doolittle, R.F.; Simon, M.I. Repetitive segmental structure of the transducin beta subunit: Homology with the CDC4 gene and identification of related mRNAs. Proc. Natl. Acad. Sci. USA 1986, 83, 2162–2166. [Google Scholar] [CrossRef] [PubMed]
- Wall, M.A.; Coleman, D.E.; Lee, E.; Iñiguez-Lluhi, J.A.; Posner, B.A.; Gilman, A.G.; Sprang, S.R. The structure of the G protein heterotrimer Giα1β1γ2. Cell 1995, 83, 1047–1058. [Google Scholar] [CrossRef]
- Sondek, J.; Bohm, A.; Lambright, D.G.; Hamm, H.E.; Sigler, P.B. Crystal structure of a GA protein βγdimer at 2.1 Å resolution. Nature 1996, 379, 369–374. [Google Scholar] [CrossRef]
- Miller, J.C.; Chezem, W.R.; Clay, N.K. Ternary WD40 Repeat-Containing Protein Complexes: Evolution, Composition and Roles in Plant Immunity. Front. Plant Sci. 2016, 6, 1108. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 Locus, Which Regulates Trichome Differentiation and Anthocyanin Biosynthesis in Arabidopsis, Encodes a WD40 Repeat Protein. Plant Cell 1999, 11, 1337–1349. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.M.; de Vos, R.C.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-b HLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006, 46, 768–779. [Google Scholar] [CrossRef]
- de Vetten, N.; Quattrocchio, F.; Mol, J.; Koes, R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997, 11, 1422–1434. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.A.; Walker, A.R.; Timmis, J.N.; Orford, S.J. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 2005, 57, 67–81. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Tian, Y.; Chen, K.; Wang, X.; Hao, Y. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 2012, 169, 710–717. [Google Scholar] [CrossRef]
- Mat Sulaiman, N.N.; Rafii, M.Y.; Duangjit, J.; Ramlee, S.I.; Phumichai, C.; Oladosu, Y.; Datta, D.R.; Musa, I. Genetic Variability of Eggplant Germplasm Evaluated under Open Field and Glasshouse Cropping Conditions. Agronomy 2020, 10, 436. [Google Scholar] [CrossRef]
- Chapman, M.A. Introduction: The Importance of Eggplant; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–10. [Google Scholar] [CrossRef]
- Gisbert, C.; Dumm, J.M.; Prohens, J.; Vilanova, S.; Stommel, J.R. A Spontaneous Eggplant (Solanum melongena L.) Color Mutant Conditions Anthocyanin-free Fruit Pigmentation. HortScience 2016, 51, 793–798. [Google Scholar] [CrossRef]
- Nothmann, J.; Rylski, I.; Spigelman, M. Color and variations in color intensity of fruit of eggplant cultivars. Sci. Hortic. 1976, 4, 191–197. [Google Scholar] [CrossRef]
- Stommel, J.R.; Dumm, J.M. Coordinated Regulation of Biosynthetic and Regulatory Genes Coincides with Anthocyanin Accumulation in Developing Eggplant Fruit. J. Am. Soc. Hortic. Sci. 2015, 140, 129–135. [Google Scholar] [CrossRef]
- Jiang, M.; Liu, Y.; Ren, L.; Lian, H.; Chen, H. Molecular cloning and characterization of anthocyanin biosynthesis genes in eggplant (Solanum melongena L.). Acta Physiol. Plant. 2016, 38, 163. [Google Scholar] [CrossRef]
- Jiang, M.; Ren, L.; Lian, H.; Liu, Y.; Chen, H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016, 249, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chu, G.; Hu, Z.; Gao, Q.; Cui, B.; Tian, S.; Wang, B.; Chen, G. Genetically engineered anthocyanin pathway for high health-promoting pigment production in eggplant. Mol. Breed. 2016, 36, 54. [Google Scholar] [CrossRef]
- Babak, O.G.; Nekrashevich, N.A.; Nikitinskaya, T.V.; Yatsevich, K.K.; Kilchevsky, A.V. Study of the Myb-factor polymorphism based on comparative genomics of vegetable Solanaceae crops (tomato, pepper, eggplant) to search for DNA markers that differentiate samples by the anthocyans accumulation. Dokl. Natl. Acad. Sci. Belarus 2019, 63, 721–729. [Google Scholar] [CrossRef]
- Zhou, L.; He, Y.; Li, J.; Liu, Y.; Chen, H. CBFs Function in Anthocyanin Biosynthesis by Interacting with MYB113 in Eggplant (Solanum melongena L.). Plant Cell Physiol. 2020, 61, 416–426. [Google Scholar] [CrossRef]
- Li, L.; He, Y.; Ge, H.; Liu, Y.; Chen, H. Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Sci. 2021, 302, 110696. [Google Scholar] [CrossRef]
- Montefiori, M.; Brendolise, C.; Dare, A.P.; Lin-Wang, K.; Davies, K.M.; Hellens, R.P.; Allan, A.C. In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 2015, 66, 1427–1436. [Google Scholar] [CrossRef]
- Li, W.; Wang, B.; Wang, M.; Chen, M.; Yin, J.M.; Kaleri, G.M.; Zhang, R.J.; Zuo, T.N.; You, X.; Yang, Q. Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation. J. Integr. Plant Biol. 2014, 56, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, J.; Chen, Y.; Tang, H.; Wang, Y.; He, Y.; Ou, Y.; Sun, X.; Wang, S.; Yao, Y. Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic. Res. 2018, 5, 27. [Google Scholar] [CrossRef]
- Collonnier, C.; Fock, I.; Kashyap, V.; Rotino, G.; Daunay, M.; Lian, Y.; Mariska, I.; Rajam, M.; Servaes, A.; Ducreux, G. Applications of biotechnology in eggplant. Plant Cell Tissue Organ Cult. 2001, 65, 91–107. [Google Scholar] [CrossRef]
- Gürbüz, N.; Uluişik, S.; Frary, A.; Frary, A.; Doğanlar, S. Health benefits and bioactive compounds of eggplant. Food Chem. 2018, 268, 602–610. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, A.; Wu, X.; Zhu, Z.; Yang, Z.; Zhu, Y.; Zha, D. Transcriptome analysis revealed expression of genes related to anthocyanin biosynthesis in eggplant (Solanum melongena L.) under high-temperature stress. BMC Plant Biol. 2019, 19, 387. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Z.; Chu, G.; Huang, C.; Tian, S.; Zhao, Z.; Chen, G. Anthocyanin Accumulation and Molecular Analysis of Anthocyanin Biosynthesis-Associated Genes in Eggplant (Solanum melongena L.). J. Agric. Food Chem. 2014, 62, 2906–2912. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.-I.; Jung, H.-J.; Hur, Y.; Nou, I.-S. Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rap. Funct. Integr. Genom. 2015, 15, 383–394. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fu, D.; Zhu, B.; Yan, H.; Shen, X.; Zuo, J.; Zhu, Y.; Luo, Y. Virus-induced Gene Silencing in Eggplant (Solanum melongena). J. Integr. Plant Biol. 2012, 54, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Wang, J.; Wang, W.; Hu, T.; Hu, H.; Bao, C. A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 2020, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef]
- Casta eda-Ovando, A.; Pacheco-Hernández, M.d.L.; Páez-Hernández, M.E.; Rodríguez, J.A.; Galán-Vidal, C.A. Chemical studies of anthocyanins: A review. Food Chem. 2009, 113, 859–871. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071. [Google Scholar] [CrossRef]
- Passeri, V.; Koes, R.; Quattrocchio, F.M. New Challenges for the Design of High Value Plant Products: Stabilization of Anthocyanins in Plant Vacuoles. Front. Plant Sci. 2016, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.N.; Koski, M.H. The effects of climate change on floral anthocyanin polymorphisms. Proc. R. Soc. B 2021, 288, 20202693. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Sun, L.; Wei, L.; Yuan, J.; Kong, F.; Zhang, Y.; Miao, X.; Xia, G.; Liu, S. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress. Plant J. 2021, 105, 1010–1025. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.; Visser, R.G.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Giusti, M.M. Anthocyanins: Natural Colorants with Health-Promoting Properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.H.; Cho, M.; Choi, M.G.; Das, P.K.; Lee, S.-K.; Choi, S.-B.; Park, Y.-I. Identification of genes that may regulate the expression of the transcription factor production of anthocyanin pigment 1 (PAP1)/MYB75 involved in Arabidopsis anthocyanin biosynthesis. Plant Cell Rep. 2015, 34, 805–815. [Google Scholar] [CrossRef]
- Cominelli, E.; Gusmaroli, G.; Allegra, D.; Galbiati, M.; Wade, H.K.; Jenkins, G.I.; Tonelli, C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 2008, 165, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.A.; Glover, B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Tanaka, Y.; Ohmiya, A.J.C.o.i.b. Seeing is believing: Engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 2008, 19, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Yang, T.; Wang, B.; Yang, H.; Wang, J.; Yu, Q. Genome-Wide Identification of the WD40 Gene Family in Tomato (Solanum lycopersicum L.). Genes 2023, 14, 1273. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ye, D.; Li, Y.; Hu, P.; Xu, R.; Wang, X. Genome-wide identification and bioinformatics analysis of the WD40 transcription factor family and candidate gene screening for anthocyanin biosynthesis in Rhododendron simsii. Front. Genet. 2023, 14, 1172321. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, Y.; Coulter, J.A.; Shen, B.; Li, Y.; Li, C.; Cao, Z.; Zhang, J. The WD40 gene family in potato (Solanum tuberosum L.): Genome-wide analysis and identification of anthocyanin and drought-related WD40s. Agronomy 2020, 10, 401. [Google Scholar] [CrossRef]
- Gutierrez, N.; Torres, A.M. Characterization and diagnostic marker for TTG1 regulating tannin and anthocyanin biosynthesis in faba bean. Sci. Rep. 2019, 9, 16174. [Google Scholar] [CrossRef]
- Ben-Simhon, Z.; Judeinstein, S.; Nadler-Hassar, T.; Trainin, T.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 2011, 234, 865–881. [Google Scholar] [CrossRef]
- Song, S.; Liu, B.; Song, J.; Pang, S.; Song, T.; Gao, S.; Zhang, Y.; Huang, H.; Qi, T. A molecular framework for signaling crosstalk between jasmonate and ethylene in anthocyanin biosynthesis, trichome development, and defenses against insect herbivores in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 1770–1788. [Google Scholar] [CrossRef]
- Clifford, M.N. Anthocyanins–nature, occurrence and dietary burden. J. Sci. Food Agric. 2000, 80, 1063–1072. [Google Scholar] [CrossRef]
- Timbola, A.K.; Souza, C.D.d.; Giacomelli, C.; Spinelli, A. Electrochemical oxidation of quercetin in hydro-alcoholic solution. J. Braz. Chem. Soc. 2006, 17, 139–148. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).