1. Introduction
Researchers in the field of Plant Protection Products (PPPs) need to bridge the gap between evaluations from traditional human-based approaches and those enabled by Artificial Intelligence (AI) [
1]. Specifically, new PPPs undergo a rigorous safety screening before market entry. PPP developers must meticulously formulate and dose these PPPs to avoid harmful phytotoxic effects on crops, thus maintaining selectivity [
2]. Traditionally, experimenters assess the severity of phytotoxicity through visual observations. The reliability of these assessments depends on low variability among experimenters’ observations and proper rating scales [
3]. In Europe, technicians are required to operate according to Good Experimental Practice (GEP), which is based on international laws [
4]. GEP is a set of standards that ensures objectivity and precision in scientific experiments. The World Trade Organization Agreement on Sanitary and Phytosanitary Measures [
5] designates the International Plant Protection Convention (IPPC) as the authority for plant health standards [
6]. The European Union falls under the European and Mediterranean Plant Protection Organization (EPPO) within IPPC. EPPO is responsible for setting phytosanitary and PPP standards. EPPO standards address crop selectivity [
2] by providing evaluation methods involving both discrete and continuous values. However, experimenters often prefer using quantitative ordinal discrete scales due to their practicality [
7]. As observed by Chiang et al. [
3], percentage scales with intervals of 10% can reduce rater uncertainty. That is because 10% is commonly accepted as inter-rater error. This can potentially lead to inconsistencies with theoretical assumptions in variance analysis [
8,
9]. Nevertheless, the selectivity of PPPs is inherently a continuous variable, assumed to be inversely proportional to the percentage of phytotoxicity symptoms and their intensity. According to EPPO, phytotoxicity symptoms include (i) modifications in the development cycle, (ii) thinning, (iii) modifications in color, (iv) necrosis, (v) deformation, and (vi) effects on quantity and quality of the yield [
2]. General Phytotoxicity (PHYGEN) is an aggregate indicator that summarizes the above symptoms by defining the percentage of damage to a plant compared to a perfectly healthy reference plant [
10].
Imaging sensors have already been demonstrated to improve precision and objectivity in the detection of pathological symptoms [
7,
11]. Some spectral properties of plants, as recorded through multispectral sensors [
12], are recognized as indicators of photosynthetic efficiency [
13,
14]. Various methods, including multi-view approaches [
15,
16,
17], can be used to create 3D models of plants [
11]. Spectral and geometric features of plants can be used to virtually reproduce the plant appearance, as observed by an experimenter during assessment. When working with three-dimensional and multispectral data, a summary is necessary to obtain an accurate estimate of PHYGEN, like a direct human-based evaluation approach. Machine learning (ML) models from artificial intelligence (AI) can synthesize vast amounts of digital information in a robust and reasonable manner when guided by expert (low variation) experimenter annotations [
12]. Open platforms offer large labeled training datasets, allowing users to customize ML algorithms to their requirements [
18,
19]. Convolutional Neural Networks (CNNs) were found to be the most accurate method for symptom classification [
20,
21] while working with image-based data. CNNs were shown to be capable of rating EPPO symptoms, specifically “modifications in color”, at both leaf and canopy levels [
22]. Gómez-Zamanillo et al. [
23] proposed a method for assessing PHYGEN by classifying the most common symptoms. Their study demonstrated the effectiveness of CNNs as feature extractors for predicting PHYGEN rates or similar measures. The study utilized CNN to identify and classify color-related phytotoxicity symptoms from RGB images. Severity estimates were determined by assigning arbitrary weights to the detected symptoms. Rather, they relied on expert experimenters to quantify weights without optimizing scores. Currently, no CNN-based model has been proposed to generate a reasonable estimate of PHYGEN based on a comprehensive analysis of all symptoms. Weight optimization is highly appreciated as it is expected to enhance the accuracy of estimates and provide insights into the significance of each symptom in the toxicological mechanism of PPPs. Further challenges associated with the deployment of CNNs for plant disease detection and scoring are reported in Barbedo et al. [
24,
25]. In particular, these include (i) sensitivity of deductions to environmental and sensor-related issues, (ii) capability of generalization of the model, and (iii) training dataset quality. It is important to note that the quality of the training dataset is highly significant as it must be properly calibrated for the specific type of PPP being tested. Therefore, pre-trained networks relying on training datasets generated for different symptoms from different PPPs should not be used to test new PPPs. It is worth noting that, in order for CNN training to be robust and accurate enough, it requires huge training datasets consisting of thousands of images.
Table 1 shows some of the methods proposed in the literature for the estimation of PHYGEN, enhancing their suitability for new PPPs PHYGEN prediction.
Typical trials for new PPPs usually involve only a few hundred plants. This may not provide a sufficient dataset for robust training, testing, and deployment of a new CNN. It is noteworthy that CNNs maintain their efficacy when symptoms of phytotoxicity are well-documented and recognized within the training dataset. This specificity is a true challenge in ML optimization for the newer PPP-related trials since the explored symptomatology may not be cataloged.
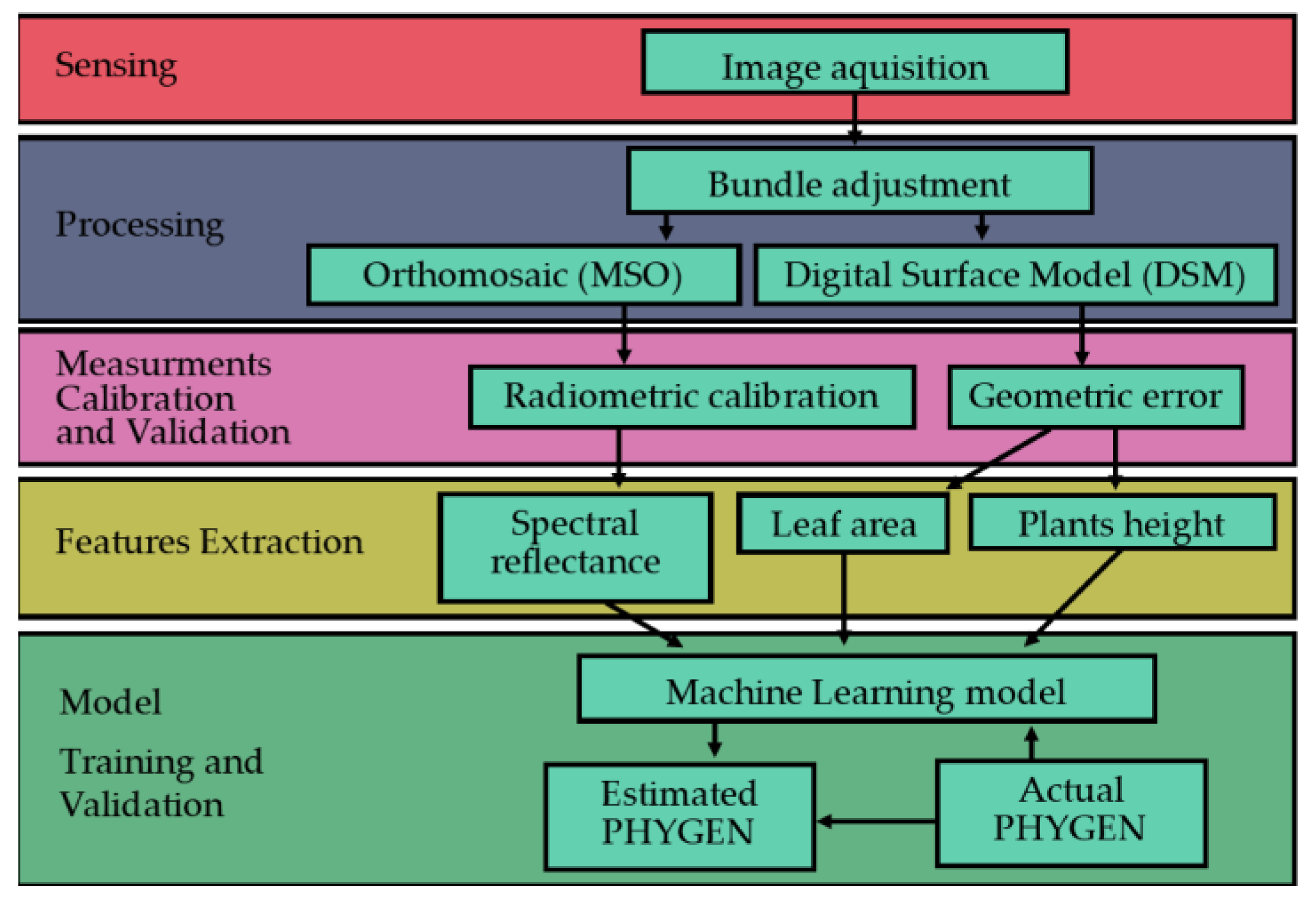
This work emphasizes that symptoms of phytotoxicity resulting from new PPPs can be unique due to their novelty, making them unpredictable. Therefore, screening trials are necessary. The proposed method involves a PHYGEN evaluation via a CV ML system for new PPPs operating in a greenhouse environment that overcome such limitations.
The system is specifically designed to address three key challenges in adopting AI, and specifically CV ML for new PPPs screening: small amount of training data, stability, and accuracy. Moreover, the model prediction suitability for ANOVA testing is also discussed.
The presented method requires only a small training sample with respect to CNN algorithms because it relies on a single linear regression and a logistic function. It takes a small training sample from the available study population, effectively addressing issues of under-representation of training datasets [
24], which is typical when testing new PPP phytotoxicity.
The system was found to reduce the impact of environmental and sensor-related factors on plant symptom detection, increasing the stability of plant pictures and measures. This is achieved through proper platform calibration techniques and a multi-view image capture approach that allows for the monitoring of errors of the geometrical and radiometric measures used to train and test the model. Model stability was tested using cross-validation. The results confirmed the robustness of the method regardless of the sample adopted. The accuracy of the model’s prediction was compared to the precision of human raters as described in the literature (10%) [
3] and to the state-of-the-art (SOTA) model for PHYGEN of non-new PPPs (6.74%) [
23]. It was not possible to find a direct comparison of a model predicting PHYGEN for new PPPs by CV ML in the literature. Therefore, the accuracy must be considered satisfactory if it is higher than the precision of human raters, and it is expected to be lower than that of CNN models with a greater amount of training data. The methodology also addresses the challenge of adopting discrete quantitative scales in the ML training step. It has been shown to improve the prediction of PHYGEN as a continuous scale variable, starting from quantitative ordinal discrete values, such as those obtained from ordinary approaches. Furthermore, as the PHYGEN estimates are now on a continuous scale, the ANOVA test can be more appropriately utilized, resolving the cumbersome lack of adaptation to the statistical theory that is often observed in the field of PPP screening.
4. Conclusions
The goal of this study was to test the operability and effectiveness of a controllable simple system based on multispectral digital photogrammetry and AI to support (and improve) current procedures for new PPP screening. This means that the system must be able to generate estimates of ordinarily recognized standard parameters (i.e., PHYGEN) and define the level of phytotoxicity of new PPPs before they enter the market. Basic requirements concern both compliance with accuracy standards and the robustness of the model output.
The proposed method can be made operational if proper Geomatics and AI skills are properly integrated. Geomatic skills are related to proper management of the acquisition system that involves both geometric (image block bundle adjustment) and radiometric-related operations needed to prepare the data that the predictors of the PHYGEN have to be extracted from. Hardware solutions proposed for the system exploit the abovementioned skills with the aim of reducing environmental and sensor-related issues. This makes acquired images more similar, partially overcoming one of the biggest problems recognized for the proper adoption of ML in phytopathometry: image features variability.
A strong constraint introduced by this specific field of study is the lack of a huge training dataset that cannot be reasonably supplied for new PPPs to be screened. In such situations, this type of screening is required.
The system operates in an effectively prepared greenhouse and requires significant infrastructure for the proper movement of the camera and lighting platform.
In this work, we present a simple solution to these requirements. In particular, after suggesting how to pre-process the data from a photogrammetric and radiometric point of view, we found some predictors for the model to be trained that are able to exploit both the geometric and spectral content of acquired data.
The predictors were analyzed and selected. They were used to train an ML algorithm integrating a LASSO and a logistic function to generate continuous estimates of PHYGEN. The robustness of the model was tested by conducting the training with a k-fold strategy and the correspondent statistics analyzed.
The proposed method/system showed stability (robustness), proving to be independent of the training sample. The accuracy of PHYGEN prediction from our model is consistent with the ones from traditional methods. Compared to other AI-based approaches (i.e., SOTA), it showed slightly higher performances in terms of correlation with expert scores applied for new PPPs (our model: R2 = 0.9, SOTA: R2 = 0.89).
In contrast, our model was not able to reach SOTA accuracy in PHYGEN scores prediction (our model: MAE = 10.66%, SOTA: MAE = 6.74%). However, it must be noted that SOTA is not intended for predictions concerning new PPPs, and the reference values we reported refer to previously tested PPPs (i.e., providing a huge amount of training data). A surprising capability of the model was to overcome the discrete nature of expert-based scores for PHYGEN. In fact, it is able to generate continuous scores of PHYGEN, even if trained on discrete ones. Their continuous nature provides a high added value since it makes it possible to test differences among groups using ordinary ANOVA-based methods.
However, some improvements are desirable, mostly in relation to a refinement of the hardware of the acquisition platform. A better-performing multispectral camera showing a higher spectral resolution and more rigorous calibration metadata is certainly a first step for future work. The active system providing controlled lighting can also be improved by using light sources that are able to generate a wider spectrum. Camera motion can be improved by using a stepper motor, allowing the possibility to stop the camera during image acquisition, thus avoiding blurring and reducing geometric deformations. Image processing could be also enhanced by strengthening automation in vegetation mask calculation from orthomosaic.
The most significant improvement of the model would be to train a CNN with such a small amount of data. The final activation layer of this CNN should be set to the logistic function proposed in this work. Further studies must test data augmentation techniques and such activation layers with MAE loss to predict PHYGEN in similar setups. Regardless of the solution, we maintain that the explicability of the model, where the physical meaning of predictors and their relationships can be somehow recognized, is an added value for those applications where precise decision making is involved.