Abstract
Seed reserve mobilization is a crucial physiological process during seed germination. Enhancing the reserve mobilization in sweet corn is vital for seed germination and seedling growth. In this study, a genome-wide association analysis (GWAS) was conducted to predict candidate genes for regulating the weight of mobilized reserved seeds (WMRS) and kernel weight (KW) in sweet corn. GWAS was performed using the BLINK model with the maize 56K SNP chip. The results indicated that there was a positive correlation between the WMRS and KW, with coefficients of variation of 68.18% and 44.63%. Association analysis identified thirteen SNPs associated with two traits, and linkage disequilibrium analysis revealed that eight of these SNPs were in strong linkage. A total of 298 candidate genes were identified within the confidence interval, of which 79 were annotated. About 20 candidate genes were identified through the comparison of homologous genes in Arabidopsis. These genes were enriched in regulating ribosome biogenesis, signal transduction, hormone synthesis, and RNA degradation processes. This study provides important insights into the genetic mechanisms governing germination traits in sweet corn, aiding further research into the localization and cloning of genes involved in the mobilization of reserve materials.
1. Introduction
Sweet corn (Zea mays saccharata) is a globally important economic crop, highly favored by consumers due to its high sugar content and rich nutritional profile [1,2]. In recent years, increasing market demand has led to a gradual expansion in the cultivation area of sweet corn, resulting in substantial economic benefits. Nonetheless, compared with field corn, sweet corn still faces challenges such as low germination rates, insufficient seed vigor, and poor seedling emergence [3,4]. Consequently, improving seed vigor and germination rates has become critical in contemporary production research.
Seed germination is a critical stage in the individual development of plants [5]. In this process, seedlings undergo a transition from heterotrophy to autotrophy [6]. In this phase, stored nutrients, including starch, proteins, and lipids, are mobilized to provide the necessary energy for seedling growth and development [7]. The mobilization and utilization of these reserve materials play a significant role in seedling growth. Healthy seedlings enhance plant tolerance and competitiveness, which are crucial for increasing crop yields [8]. The mobilization of reserve materials is one of the key focuses of research. Recent studies have reported on the utilization of phenotypes of seed storage materials in plants such as rice [9], wheat [10], and maize [7]. It has been discovered that both the seedling dry weight (SDW) and WMRS gradually increase during the sweet corn seed germination process [7]. Different seed sizes can also affect the utilization of seed storage material; large seeds have more storage material than small seeds [11]. It has been shown that under drought and salt-stress conditions, SDW and WMRS both exhibit decreasing trends as the severity of the conditions increases [12,13]. Researchers also found that low-temperature stress can reduce the utilization efficiency of stored materials [14,15].
Genome-wide association analysis (GWAS), also known as association mapping, is an effective method for studying complex traits based on linkage disequilibrium (LD) [16]. It combines bioinformatics and statistics using high-throughput sequencing technology and millions of SNP molecular markers [17]. GWAS is extensively used today for genetic screening in maize [18,19,20,21], rice [22], wheat [23], rape seed [24], etc. Through GWAS, 59 SNPs and 66 candidate genes associated with corn yield traits have been identified [25]. Ma et al. [26] identified 22 SNPs associated with grain weight. In addition, 48 SNPs and 37 genes related to maize quality traits were correlated by GWAS [27]. These findings indicate that GWAS can help in identifying important loci and genes for complex traits.
Despite the identification of several SNPs and candidate genes linked to germination, research on the mobilization of reserve materials in sweet corn is still limited, leaving the genetic mechanisms of seed germination unclear. In the present study, we assessed germination traits in 100 sweet corn inbred lines from various sources and utilized high-throughput SNP sequencing to identify critical genes associated with germination traits. The findings of the research assist in clarifying the genetic basis of germination traits and provide theoretical support for further studies on the localization and cloning of genes related to the mobilization of reserve materials.
2. Materials and Methods
2.1. Experimental Materials and Phenotype Assessment
One hundred sweet corn inbred lines were provided by the Anhui Research Institute of Maize Breeding Engineering and Technology (Chuzhou, China). The moisture content (MC) was calculated using the weight difference method. Three replications of the experiment were carried out, with 50 healthy sweet corn seeds selected for each replicate. Weights of dry kernels were measured separately using a balance; values for initial seed weight (ISW) were thereby obtained. The weighed seeds were placed in a germination chamber and incubated in the dark for 7 days at a controlled temperature of 28 ± 1 °C. The seedlings and seeds were separated and dried for constant weight at 105 ± 1 °C in an oven. Sample weights were measured, and values for seedling dry weight (SDW) and residual seed dry weight (RSDW) were obtained. Additionally, values for initial seed dry weight [ISDW = ISW × (1 − MC)] and weight of mobilized reserved seeds (WMRS = ISDW − RSDW) were calculated [12].
2.2. Phenotypic Data Analysis
The phenotypic data of 100 sweet corn inbred line materials were collected including WMRS and KW. Descriptive statistics, analysis of variance, and Pearson correlation coefficients were calculated for the phenotypic data using the IBM SPSS Statistics 24.0 and Microsoft Excel 2019 statistical analysis software. The RStudio 4.4.1 software was utilized to plot normal distributions. For each trait, the based broad-sense heritability was calculated as h2 = σ2g/(σ2g + σ2e/n), where σ2g is the genetic variance, σ2e is the residual error variance, and n represents the number of replications [28].
2.3. Genome-Wide Association Analysis
The software Plink 1.90 was used for data cleaning and filtering. A total of 36,747 SNPs without minor allele frequency (MAF) < 0.05 and a missing data rate > 0.2 were filtered out for subsequent analysis. GWAS of WMRS and KW was conducted using the BLINK model from the R GAPIT package. We used a threshold of −log10(p) ≥ 3.6 (p < 2.51 × 10−4) to identify significant SNPs. Using the PopLDdecay 3.42 software, we calculated the LD decay distance to be 200 kb [29]. The explained phenotypic variance of mutual SNPs was calculated using the R function lm().
2.4. Haplotype Analysis
Genotype data were extracted from 200 kb [29] upstream and downstream of the SNP loci for haplotype analysis. LD heatmaps were plotted using an online data analysis and visualization platform (https://www.bioinformatics.com.cn; accessed on 14 May 2024) [30]. To define the degree of LD block, a threshold value of r2 > 0.33 [31] was selected for this study. Allelic effect analysis was conducted by combining phenotypic and genotypic data, and the ggplot2 package was used for visualization.
2.5. Gene Function Annotation and Candidate Gene Prediction
Candidate genes within a 200 kb [32] range were identified using the B73 RefGen_v3 database from the Maize GDB (https://maizegdb.org/; accessed on 15 May 2024). Using the tools of the data analysis platform OmicShare (https://www.omicshare.com/; accessed on 19 May 2024), GO functional annotation of the candidate gene was performed, and KEGG enrichment analysis was carried out [33]. The RNA-seq data were obtained from the gene function and expression database in Maize GDB [34]. FPKM ≥ 1 [35] was used as the criterion for candidate gene expression, and the expression level of the target gene was standardized using the formula log2(FPKM + 1) [36]. Functional annotation of the candidate genes was performed using the UniProt database (https://www.uniprot.org/; accessed on 27 May 2024) and the NCBI database (https://www.ncbi.nlm.nih.gov/; accessed on 27 May 2024) [37].
3. Results
3.1. Phenotypic Analysis of Sweet Corn Inbred Lines
Statistical analysis of the WMRS and KW of 100 sweet corn inbred lines showed that WMRS ranged from 0.002 to 0.210 g/seed (Table 1). The average weight was 0.049 g/seed, with a standard deviation of 0.033, and a coefficient of variation of 68.18%. KW ranged from 0.039 to 0.298 g/seed. The average weight was 0.101 g, with a standard deviation of 0.045, and a coefficient of variation of 44.63%. WMRS and KW had a strong positive correlation. Both WMRS and KW exhibited a trend of normal distribution (Figure 1). The analysis of variance results showed that there was no significant difference in WMRS and KW between the groups (p > 0.05, Table 2). WMRS had a high heritability (h2 = 0.95), whereas KW had a moderate heritability (h2 = 0.87; Table 1). These findings indicated that the population’s broad phenotypic variations in WMRS and KW were largely a result of genetic factors and thus suitable for further association mapping.

Table 1.
Descriptive statistical analysis of WMRS and KW.
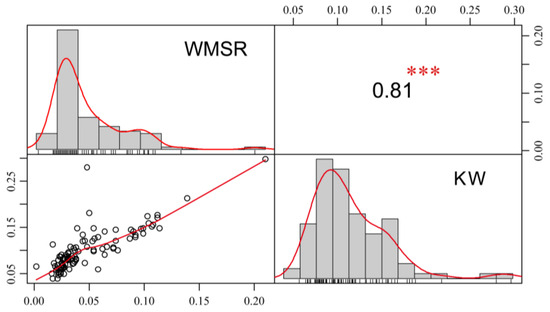
Figure 1.
Frequency distribution and correlation of WMRS and KW. The values are Pearson correlation coefficients between the two traits. *** indicates p < 0.001.

Table 2.
Analysis of variance for WMRS and KW.
3.2. Genome-Wide Association Analysis of WMRS and KW
GWAS results demonstrated that 13 SNPs were associated with these two traits, distributed across chromosomes 1, 3, 4, 6, 7, 8, and 10. Among these, eight SNPs were linked to WMRS, with SNP Affx-90065309 exhibiting the lowest p-value (Figure 2a,b). The PVE of these eight SNPs ranged from 13.2% to 24.6%. Additionally, five SNPs related to KW were located on chromosomes 1, 7, and 8. Specifically, three SNPs were located on chromosome 7 (Figure 2c,d). The locus with the lowest p-value was Affx-90522187 and the PVE of these seven SNPs ranged from 21.8% to 26.0% (Supplementary Table S1).
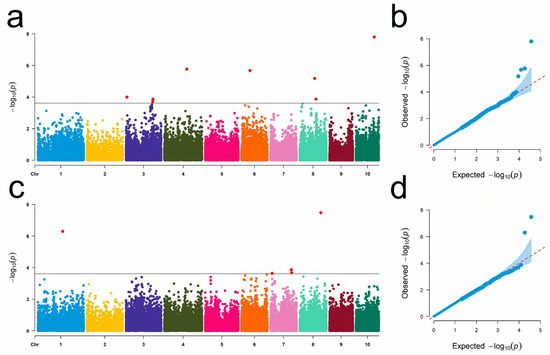
Figure 2.
Genome-wide association mapping of WMRS and KW. (a,b) represent the Manhattan and QQ plots for WMRS. (c,d) represent the Manhattan and QQ plots for KW. The black line indicates the genome-wide significance threshold of 2.51 × 10−4.
3.3. Analysis of Haplotypes and Allelic Variation Effects
LD analysis results indicated that multiple SNPs were in strong linkage. Affx-90862117, Affx-91208890, Affx-90276180, Affx-90802176, and Affx-115331806 showed a great linkage. Additionally, Affx-90065309, Affx-90448237, and Affx-91208575 also showed a strong linkage (Figure 3a,c). The effects of allelic variation in the linked SNPs were analyzed along with the phenotypic data. The loci Affx-90862117, Affx-90276180, Affx-90065309, and Affx-90522187 were responsible for phenotypic variations. The differences caused by Affx-91208890, Affx-90802176, and Affx-115331806 were not significant (Figure 3b,d, Supplementary Figure S1). At the Affx-90862117 locus, three alleles were present (T/T, C/T, C/C). There were 49 samples containing the T/T allele with an average WMRS of 0.059 g/seed, while 47 samples with the C/C allele had an average of 0.041 g/seed. The WMRS for the T/T alleles were 0.018 g/seed higher than that for the C/C alleles. Moreover, the Affx-90522187 locus also contained three alleles. The phenotypic variation range for samples with the T/T alleles were 0.04–0.18 g, while the range for those with the C/C alleles were 0.05–0.30 g. The KW for samples with the T/T alleles were 0.07 g higher compared to the C/C alleles, indicating T/T as a superior allele. We selected samples containing the T/T allele from these two loci and found that 13 materials fell within the intersection of both traits. Among these, materials such as FY39, YNT8-1, and FT4-1 showed high performance in both WMRS and KW (Supplementary Table S2).
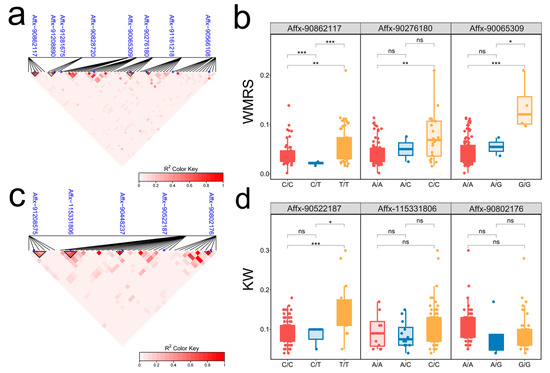
Figure 3.
LD heatmap of significant SNP loci and analysis of allelic variation effects. (a) WMRS LD heatmap. (c) KW LD heatmap. The triangular boxes indicate formed LD blocks exceeding the defined thresholds. The blue dots indicate where the loci were located. (b) Box plot of allelic effects of SNPs in WMRS. (d) Box plot of allelic effects of SNPs in KW. * p < 0.05; ** p < 0.01; *** p < 0.001; ns—not significant.
3.4. Candidate Gene Analysis
To further explore the candidate genes influencing WMRS and KW, 298 candidate genes were identified in the maize GDB based on significant SNPs. The results of GO functional annotation showed that 79 genes were annotated. These genes were enriched in 30 important GO terms. There were 60 genes involved in biological processes, 52 genes in cellular components and 62 genes in molecular functions (Figure 4a). Further analysis revealed that candidate genes in biological processes were associated with metabolism and cellular processes. These genes play a role in the synthesis of molecular complexes and internal organs within cellular components. In molecular functions, the candidate genes were focused on molecular interactions and catalytic activities (Supplementary Table S3).
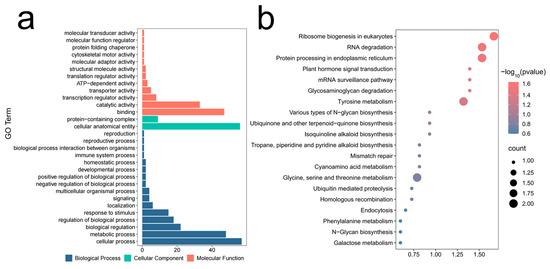
Figure 4.
Functional annotation and KEGG enrichment analysis of candidate genes. (a) GO function annotation bar chart. The blue blocks are biological processes, the green blocks are cellular components, and the orange blocks are molecular functions. (b) Bubble plot of KEGG enrichment analysis. Circles indicate numbers of genes, and p-values indicate the significance of enrichment.
The KEGG enrichment analysis revealed that 28 genes were enriched in 42 pathways. The candidate genes were enriched in pathways related to ribosome biogenesis in eukaryotes, RNA degradation, protein processing in the endoplasmic reticulum, and plant hormone signal transduction (Figure 4b, Supplementary Table S4). Further analysis showed that the gene GRMZM2G346839 encodes a crystalline protein. This maintains normal cellular function and survival by participating in protein synthesis, processing, and transport. The gene GRMZM2G132069 encodes a pectin phosphate kinase, which contributes to the cellular maintenance of energy homeostasis, cell proliferation, and metabolic stability. In addition, the gene GRMZM2G161035 encodes an endoplasmic reticulum chaperone that helps maintain cellular health by assisting in protein export and processing.
To determine which candidate is more likely related to the significant SNP, the expression patterns of these genes across different developmental stages were analyzed. The findings revealed that 63 annotated genes were expressed during both seedling and seed development. Among these, 41 of candidate genes were linked to WMRS, while 22 were associated with the regulation of KW (Figure 5). The analysis of the functional expression of homologous genes in Arabidopsis thaliana identified 20 candidate genes (Table 3, Supplementary Table S5). The twelve candidate genes were associated with WMRS, while eight were linked to KW. These candidate genes primarily participate in processes such as hormone synthesis, cell division, signal transduction, and energy metabolism.
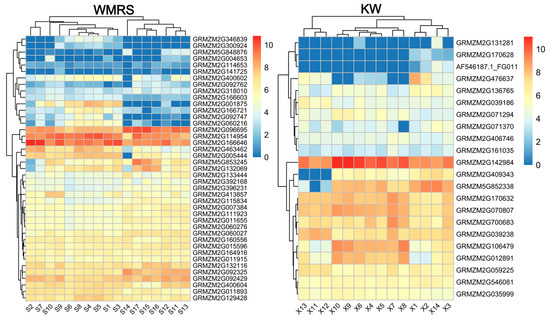
Figure 5.
Dynamic expression pattern analysis of candidate genes. The scale bars indicate the normalized gene expression levels. S1–S17 indicate the root system 3–7 days after sowing, and seed development 2–24 days after pollination. X1–X14 indicate the development of seeds, endosperm, and embryos 2–24 days after pollination.

Table 3.
Candidate genes for WMRS and KW in sweet corn, with functional annotations.
3.5. Regulatory Network of Candidate Genes
Based on the STRING database, protein interaction network analysis was conducted on 20 candidate genes. A total of 15 genes were obtained using the regulated network; these interacted with various functional proteins by forming a standalone group and by participating in the regulation of a variety of biological networks (Table 3, Figure 6). For example, the gene GRMZM2G300924 (M1) encodes the AP2/ERF protein that regulates the expression of key transcription factors. These genes were involved in ABA, GA metabolism, and the ABA downstream signaling pathways. The gene GRMZM2G096695 (M12) encodes the TRR14 protein. Overexpression of this gene reduces the growth-inhibiting and starch-accumulating effects of T6P, increasing fresh and dry weights and leaf areas. Furthermore, the candidate gene GRMZM2G161035 (M18) was annotated as a BIP protein. This can participate in the membrane fusion step of polar nuclei to ensure the correct shape and function of endosperm cells. This might result in the aberrant proliferation of endosperm nuclei.
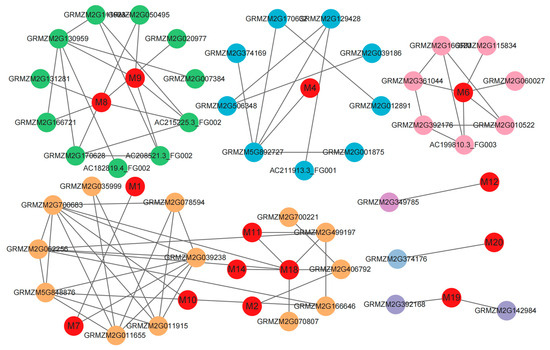
Figure 6.
Protein–protein interaction network analysis of 20 candidate genes. The nodes indicate proteins, and the lines indicate interactions between proteins. The red color indicates the protein encoded by the candidate gene. M1–M20 are the encoded proteins of the candidate genes.
These findings indicate that traits related to WMRS and KW are regulated by plant hormones, TRR, T6P, BIP, and other complex factors.
4. Discussion
4.1. Analysis of the Positioning Results for WMRS and KW
With the development of biotechnology, GWAS has been known as an important tool for the analysis of quantitative traits with large populations and high-throughput molecular markers; it has also identified important candidate genes [26,38]. GWAS was employed to identify 16 SNPs associated with panicle weight, and four candidate genes related to this characteristic were predicted [39]. Chen et al. [40] used GWAS to detect 13 SNPs linked to kernel color and identify 95 potential genes. In another study, 29 SNPs related to stem traits were identified from 377 inbred lines by association mapping, and 12 candidate genes were discovered [41]. Li et al. [42] detected 58 SNPs linked to germination characteristics and identified 36 candidate genes implicated in seed germination. Because the detected loci are affected by population structure, allele frequency, marker density, and modeling, this can lead to different results [43,44]. Based on the results of previous population genetic diversity analyses, 100 sweet corn inbred lines were classified into two subgroups of tropical and subtropical taxa [45]. Based on the origin of the material, the tropical subgroup contains eight materials and the subtropical subgroup contains 92 materials. The genetic distances of inbred lines within the same subgroups are small, while the genetic distances between different subgroups are large. Research utilizing PCA analysis revealed differences in genetic structure among various subgroups [46]. Chen et al. [47] analyzed the genetic diversity and population genetic structure of 250 sweet corn inbred lines using maize SNP chips, which showed that the experimental materials were divided into four subgroups. Wu et al. [48] analyzed the population structure of 100 sweet corn inbred lines and classified them into four subgroups of tropical, subtropical, temperate, and American hybrid inbred lines. In addition, principal component analysis and constructed kinship matrices were also carried out. Dang et al. [49] classified 190 sweet corn inbred lines and 287 glutinous maize inbred lines into three subgroups based on population structure and PCA analysis. The results of the present study are similar to those of previous national and international studies regarding population structure and PCA. In addition, false associations were effectively reduced by utilizing population structure, PCA, kinship, and combining appropriate models for GWAS.
To date, GWAS of WMRS and KW sweet corn has rarely been conducted. In the present study, a total of 13 SNPs regulating WMRS and KW were identified on chromosomes 1, 3, 4, 6, 7, 8, and 10. For the WMRS trait, the locus Affx-91208890 was identified on chromosome 6; this was situated within the interval of the previously localized QTL (qWMRS6) [45]. Regarding KW traits, it was discovered that the SNP Affx-115331806 was 5.6 Mb distant from the Affx-88993646 on chromosome 7 [50]. The Affx-90802176 on chromosome 1 was located 7.5 Mb away from the previously reported association at 1_154306450 [51]. In addition, Affx-115331806 on chromosome 7 was located within the interval of the QTL (KW-qCL7-6) identified by Wang et al. [52]. The related traits reported here are in line with the findings of previous national and international research, and they provide valuable insights for a deeper understanding of the genetic mechanisms underlying germination traits in sweet corn.
4.2. Candidate Gene Analysis for WMRS and KW
In the present study, a total of 20 candidate genes were found after 79 annotated genes had been put through further screening. Among these, twelve important genes regulating WMRS and eight important genes regulating KW were identified. The candidate gene GRMZM2G300924 at the Affx-90276180 locus was found to encode the AP2/ERF protein, which plays a key role in the regulatory network controlling the completion of seed germination [53,54]. This protein interacts with the photosensitive pigments (phyA and phyB), preventing them from binding to the promoters of genes that inhibit the completion of sprouting. In addition, phyA and phyB reduce the expression of ERF55 and ERF58 under light conditions, which further promotes seed germination. Seeds of ERF55 and ERF58 double mutants successfully complete germination even in the absence of light [55]. In addition, the Affx-91161218 locus gene GRMZM2G400604 was found to encode the ASL protein ASL9, a transcription factor linked with cytokinin signaling. Studies of mutants have shown that short-term accumulation and overexpression of ASL9 inhibit the cytokinin response, leading to developmental defects [56,57]. Knockdown of the cytokinin signaling activators ARR10 and ARR12 partially restores these defects [58]. In addition, the candidate gene GRMZM2G096695 was found to encode a teicoplanin-resistant glycoprotein (TRR14). This gene has been shown to confer resistance against the growth inhibition and starch accumulation effects of trehalose-6-phosphate (T6P) [59,60]. Compared with the wild type, the overexpression of TRR14 results in larger leaf areas and increased fresh and dry weights [61]. Therefore, it is hypothesized that this gene can enhance seedling growth.
Among the candidate genes regulating KW, the gene at the Affx-90802176 locus was found to encode mannosyltransferase (LEW). The LEW3 mutant exhibits abnormal development, reducing fertility and causing a lower seed set rate [62,63]. Therefore, it is hypothesized that this gene regulates seed sets and spike sizes, and also affects seed development. In the present study, the candidate genes GRMZM2G409343 and GRMZM5G852338 at Affx-90522187 were found to encode myosin XI, which plays a central role in cell expansion and organelle transport. Changes in these genes result in lower numbers of seeds per plant and reduced setting rates, compared with the wild type [64,65]. In conclusion, the results of the present study demonstrate that 12 candidate genes have an influence on WMRS through the regulation of root development, seed germination, cell differentiation, embryonic development, and seedling growth. The regulation of ovule, seed setting rates, as well as endosperm development, is controlled by eight candidate genes that affect KW. The findings presented in this paper provide important insights for a deeper understanding of the genetic mechanisms underlying germination traits in sweet corn.
5. Conclusions
The results of the present study reveal a strong positive correlation between WMRS and KW. Using GWAS, eight SNPs controlling WMRS and five SNPs controlling KW were identified. A total of 298 candidate genes were identified within the 200 kb confidence interval of significant SNPs. A total of 20 candidate genes were identified through functional annotation analysis. We found that twelve candidate genes regulate WMRS, and eight important candidate genes regulate KW. The main functions of these genes are related to processes such as transcription factor control, energy metabolism, hormone synthesis, and signaling. However, due to the limited number of groups in the present study, the accuracy and reliability of our GWAS results should be improved by increasing the number of associated groups in future studies. In the present study, we then sought to screen out the important genetic loci that regulate the seed germination of sweet corn. The results presented here provide theoretical support for analyzing the genetic mechanisms of seed reserve material and the production of seeds.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy14112648/s1, Figure S1: Boxplot of allele effects for significant SNPs; Table S1: results of the genome-wide association analysis for two traits; Table S2: phenotypic data of 100 sweet corn inbred lines; Table S3: GO functional annotation results of candidate genes; Table S4: enrichment analysis results of candidate genes; Table S5: summary of candidate genes within the confidence interval.
Author Contributions
Conceptualization, X.C. and L.C.; methodology, X.C.; software, Y.Y. and N.C.; validation, A.R., D.W. and T.S.; formal analysis, Y.Y.; investigation, N.C.; resources, X.C. and H.Y.; data curation, Y.Y. and L.C.; writing—original draft preparation, Y.Y., N.C., T.S., D.W., L.C. and X.C.; writing—review and editing, X.C., Y.Y., N.C., A.R., L.C. and H.Y.; visualization, Y.Y.; supervision, X.C. and L.C.; project administration, X.C.; funding acquisition, X.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Emergency Management of the National Natural Science Foundation of China (31440066), Key Discipline Construction Funds for Crop Science of Anhui Sciences and Technology University (No. XK-XJGF001), the Natural Science Research Project of Anhui University (KJ2019ZD57), and the Research Development Fund of Anhui Science and Technology University (FZ230126).
Data Availability Statement
Available on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ruanjaichon, V.; Khammona, K.; Thunnom, B.; Suriharn, K.; Kerdsri, C.; Aesomnuk, W.; Yongsuwan, A.; Chaomueang, N.; Thammapichai, P.; Arikit, S.; et al. Identification of Gene Associated with Sweetness in Corn (Zea mays L.) by Genome-Wide Association Study (GWAS) and Development of a Functional SNP Marker for Predicting Sweet Corn. Plants 2021, 10, 1239. [Google Scholar] [CrossRef] [PubMed]
- Tracy, W.F.; Shuler, S.L.; Dodson-Swenson, H. The Use of Endosperm Genes for Sweet Corn Improvement. In Plant Breeding Reviews; Wiley: Hoboken, NJ, USA, 2019; pp. 215–241. [Google Scholar]
- Huang, Y.; Lin, C.; He, F.; Li, Z.; Guan, Y.; Hu, Q.; Hu, J. Exogenous spermidine improves seed germination of sweet corn via involvement in phytohormone interactions, H2O2 and relevant gene expression. BMC Plant Biol. 2017, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-S.; Yun, H.S.; Seo, J.M. Optimum Harvest Time for High Quality Seed Production of Sweet and Super Sweet Corn Hybrids. Korean J. Crop Sci. 2004, 49, 373–380. [Google Scholar]
- Jiménez-López, S.; Mancera-Martínez, E.; Donayre-Torres, A.; Rangel, C.; Uribe, L.; March, S.; Jiménez-Sánchez, G.; Sánchez de Jiménez, E. Expression profile of maize (Zea mays L.) embryonic axes during germination: Translational regulation of ribosomal protein mRNAs. Plant Cell Physiol. 2011, 52, 1719–1733. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, S.L.; Charlton, W.L.; Baker, A.; Graham, I.A. Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J. 2002, 31, 639–647. [Google Scholar] [CrossRef]
- Cheng, X.X.; He, S.; Geng, G.H. Dynamic QTL analysis of seed reserve utilization in sh(2) sweet corn germination stages. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, J.; Miao, X.; Lin, X.; Liu, W.; Ren, H. Effects of exogenous silicon on maize seed germination and seedling growth. Sci. Rep. 2021, 11, 1014. [Google Scholar] [CrossRef]
- Zhao, Y.; Yin, T.; Ran, X.; Liu, W.; Shen, Y.; Guo, H.; Peng, Y.; Zhang, C.; Ding, Y.; Tang, S. Stimulus-responsive proteins involved in multi-process regulation of storage substance accumulation during rice grain filling under elevated temperature. BMC Plant Biol. 2023, 23, 547. [Google Scholar] [CrossRef]
- Li, B.B.; Zhang, S.B.; Lv, Y.Y.; Wei, S.; Hu, Y.S. Reactive oxygen species-induced protein carbonylation promotes deterioration of physiological ac-tivity of wheat seeds. PLoS ONE 2022, 17, e0263553. [Google Scholar] [CrossRef]
- Ambika, S.; Manonmani, V.; Somasundar, G. Review on Effect of Seed Size on Seedling Vigour and Seed Yield. Res. J. Seed Sci. 2014, 7, 31–38. [Google Scholar] [CrossRef]
- Soltani, A.; Gholipoor, M.; Zeinali, E. Seed reserve utilization and seedling growth of wheat as affected by drought and salinity. Environ. Exp. Bot. 2006, 55, 195–200. [Google Scholar] [CrossRef]
- Garg, G. Response in germination and seedling growth in Phaseolus mungo under salt and drought stress. J. Environ. Biol. 2010, 31, 261–264. [Google Scholar] [PubMed]
- Steckel, L.E.; Sprague, C.L.; Stoller, E.W.; Wax, L.M. Temperature effects on germination of nine Amaranthus species. Weed Sci. 2004, 52, 217–221. [Google Scholar] [CrossRef]
- Hussain, S.; Yin, H.; Peng, S.; Khan, F.A.; Khan, F.; Sameeullah, M.; Hussain, H.A.; Huang, J.; Cui, K.; Nie, L. Comparative Transcriptional Profiling of Primed and Non-primed Rice Seedlings under Sub-mergence Stress. Front Plant Sci. 2016, 7, 1125. [Google Scholar] [CrossRef]
- Jin, Y.; Li, D.; Liu, M.; Cui, Z.; Sun, D.; Li, C.; Zhang, A.; Cao, H.; Ruan, Y. Genome-Wide Association Study Identified Novel SNPs Associated with Chlorophyll Content in Maize. Genes 2023, 14, 1010. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Xi, N.; Liu, H.; Liu, P.; Xiang, C.; Zhang, C.; Zou, C.; Cheng, X.; Yu, H.; Zhang, M.; et al. An Integration of Linkage Mapping and GWAS Reveals the Key Genes for Ear Shank Length in Maize. Int. J. Mol. Sci. 2022, 23, 15073. [Google Scholar] [CrossRef]
- Ramekar, R.V.; Sa, K.J.; Park, K.-C.; Park, J.Y.; Park, K.J.; Lee, J.K. Genetic differentiation of Mutator insertion polymorphisms and association with agronomic traits in waxy and common maize. Genes. Genom. 2020, 42, 631–638. [Google Scholar] [CrossRef]
- Baseggio, M.; Murray, M.; Wu, D.; Ziegler, G.; Kaczmar, N.; Chamness, J.; Hamilton, J.P.; Buell, C.R.; Vatamaniuk, O.K.; Buckler, E.S.; et al. Genome-wide association study suggests an independent genetic basis of zinc and cadmium concentrations in fresh sweet corn kernels. G3 Genes|Genomes|Genet. 2021, 11, jkab186. [Google Scholar] [CrossRef]
- Yang, L.; Li, T.; Tian, X.; Yang, B.; Lao, Y.; Wang, Y.; Zhang, X.; Xue, J.; Xu, S. Genome-wide association study (GWAS) reveals genetic basis of ear-related traits in maize. Euphytica 2020, 216, 172. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, X.; Lu, C.; Chang, H.; Chachar, Z.; Fan, L.; An, Y.; Li, X.; Qi, Y. Genome-wide association study of carotenoids in maize kernel. Plant Genome 2024, 17, e20495. [Google Scholar] [CrossRef]
- Yang, T.; Dong, J.; Zhao, J.; Zhang, L.; Zhou, L.; Yang, W.; Ma, Y.; Wang, J.; Fu, H.; Chen, J.; et al. Genome-wide association mapping combined with gene-based haplotype analysis identify a novel gene for shoot length in rice (Oryza sativa L.). Theor. Appl. Genet. 2023, 136, 251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Peng, C.; Xu, W.; Li, Y.; Qi, X.; Zhao, M. Genome-wide association study of agronomic traits related to nitrogen use efficiency in Henan wheat. BMC Genom. 2024, 25, 7. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Ibrahim, S.; Kuang, L.; Ze, T.; Wang, X.; Wang, H.; Dun, X. Integrating genome-wide association study with transcriptomic data to predict candidate genes influencing Brassica napus root and biomass-related traits under low phosphorus conditions. Biotechnol. Biofuels Bioprod. 2023, 16, 149. [Google Scholar] [CrossRef]
- Zeng, T.; Meng, Z.; Yue, R.; Lu, S.; Li, W.; Li, W.; Meng, H.; Sun, Q. Genome wide association analysis for yield related traits in maize. BMC Plant Biol. 2022, 22, 449. [Google Scholar] [CrossRef]
- Ma, J.; Cao, Y. Genetic Dissection of Grain Yield of Maize and Yield-Related Traits Through Association Map-ping and Genomic Prediction. Front Plant Sci. 2021, 12, 690059. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ge, Z.; Wang, M.; Zhao, M.; Pei, Y.; Song, X. Genome-wide association study of quality traits and starch pasting properties of maize kernels. BMC Genom. 2023, 24, 59. [Google Scholar] [CrossRef]
- Coles, N.D.; McMullen, M.D.; Balint-Kurti, P.J.; Pratt, R.C.; Holland, J.B. Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics 2010, 184, 799–812. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Alemu, A.; Batista, L.; Singh, P.K.; Ceplitis, A.; Chawade, A. Haplotype-tagged SNPs improve genomic prediction accuracy for Fusarium head blight resistance and yield-related traits in wheat. Theor. Appl. Genet. 2023, 136, 92. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, M.; Chen, J.; Qing, C.; He, S.; Zou, C.; Yuan, G.; Yang, C.; Peng, H.; Pan, G.; et al. GWAS and WGCNA uncover hub genes controlling salt tolerance in maize (Zea mays L.) seedlings. Theor. Appl. Genet. 2021, 134, 3305–3318. [Google Scholar] [CrossRef]
- Bai, L.; Wu, C.; Lei, S.; Zou, M.; Wang, S.; Zhang, Z.; Bao, Z.; Ren, Z.; Liu, K.; Ma, Q.; et al. Potential anti-gout properties of Wuwei Shexiang pills based on network pharmacology and pharmacological verification. J. Ethnopharmacol. 2023, 305, 116147. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; De Leon, N.; Kaeppler, S.M. An Expanded Maize Gene Expression Atlas based on RNA Sequencing and its Use to Explore Root Development. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Sharifi Alishah, M.; Darvishzadeh, R.; Ahmadabadi, M.; Piri Kashtiban, Y.; Hasanpur, K. Identification of differentially expressed genes in salt-tolerant oilseed sunflower (Helianthus annuus L.) genotype by RNA sequencing. Mol. Biol. Rep. 2022, 49, 3583–3596. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Gao, K.; Yang, H.; Ju, T.; Zhu, J.; Tang, Z.; Zhao, L.; Chen, Q. Genome-wide analysis of metallothionein gene family in maize to reveal its role in development and stress resistance to heavy metal. Biol. Res. 2022, 55, 1. [Google Scholar] [CrossRef]
- Wan, W.; Wu, Y.; Hu, D.; Ye, F.; Wu, X.; Qi, X.; Liang, H.; Zhou, H.; Xue, J.; Xu, S.; et al. Genome-wide association analysis of kernel nutritional quality in two natural maize populations. Mol. Breed. 2023, 43, 18. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Guo, W.; Le, L.; Yu, J.; Wu, Y.; Li, D.; Wang, Y.; Wang, H.; Lu, X.; Qiao, H.; et al. Integration of high-throughput phenotyping, GWAS, and predictive models reveals the genetic architecture of plant height in maize. Mol. Plant 2023, 16, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Y.; Shaw, R.K.; Zhang, X.; Li, J.; Li, L.; Li, S.; Adnan, M.; Jiang, F.; Bi, Y.; et al. Genome-Wide Association Study and Prediction of Tassel Weight of Tropical Maize Germplasm in Multi-Parent Population. Int. J. Mol. Sci. 2024, 25, 1756. [Google Scholar] [CrossRef]
- Chen, W.; Cui, F.; Zhu, H.; Zhang, X.; Lu, S.; Lu, C.; Chang, H.; Fan, L.; Lin, H.; Fang, J.; et al. Genome-wide association study of kernel colour traits and mining of elite alleles from the major loci in maize. BMC Plant Biol. 2024, 24, 25. [Google Scholar] [CrossRef]
- Wang, B.; Yang, M.; Guo, H.; Wang, J.; Wang, Z.; Lu, H.; Qin, G.; Chen, J. Genome-wide association study for stalk lodging resistance related traits in maize (Zea mays L.). BMC Genom. 2024, 25, 19. [Google Scholar] [CrossRef]
- Li, Y.; Liang, Y.; Liu, M.; Zhang, Q.; Wang, Z.; Fan, J.; Ruan, Y.; Zhang, A.; Dong, X.; Yue, J.; et al. Genome-Wide Association Studies Provide Insights into the Genetic Architecture of Seed Germination Traits in Maize. Front. Plant Sci. 2022, 13, 930438. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, Y.; Wang, L.; Luo, Y.; Peng, Y.; Xu, Y.; Liu, X.; Wu, S.; Jian, L.; Xu, J.; et al. The genetic architecture of the dynamic changes in grain moisture in maize. Plant Biotechnol. J. 2021, 19, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Adu, G.B.; Badu-Apraku, B.; Akromah, R.; Garcia-Oliveira, A.L.; Awuku, F.J.; Gedil, M. Genetic diversity and population structure of early-maturing tropical maize inbred lines using SNP markers. PLoS ONE 2019, 14, e0214810. [Google Scholar] [CrossRef]
- Xu, Y.; Fang, H.; Zhou, B.; Yi, Y.; Wang, C.; Cheng, X.; Yu, H. Genome-wide association study of grain traits of sweet maize germplasm resources. Jiangsu J. Agric. Sci. 2021, 37, 289–295. [Google Scholar]
- Wang, C.; Xu, Y.; Cheng, X.; Zhou, Y.; Yu, H. Genome-wide association study of seed nutritional quality in sweet corn. Acta Agric. Zhejiangensis 2020, 32, 383–389. [Google Scholar]
- Chen, J.; Zhang, H.; Wang, T.; Li, X.; Wu, Z.; Lu, G. Genetic Diversity and Population Genetic Structure Analysis of Sweet Corn Inbred Lines. Mol. Plant Breed. 2022, 20, 6559–6565. [Google Scholar]
- Wu, Z.; Wang, T.; Chen, J.; Zhang, Y.; Lv, G. Sweet corn association panel and genome-wide association analysis reveal loci for chilling-tolerant germination. Sci. Rep. 2024, 14, 10791. [Google Scholar] [CrossRef]
- Dang, D.; Guan, Y.; Zheng, H.; Zhang, X.; Zhang, A.; Wang, H.; Ruan, Y.; Qin, L. Genome-Wide Association Study and Genomic Prediction on Plant Architecture Traits in Sweet Corn and Waxy Corn. Plants 2023, 12, 303. [Google Scholar] [CrossRef]
- Qu, Z.; Wu, Y.; Hu, D.; Li, T.; Liang, H.; Ye, F.; Xue, J.; Xu, S. Genome-Wide Association Analysis for Candidate Genes Contributing to Kernel-Related Traits in Maize. Front. Plant Sci. 2022, 13, 872292. [Google Scholar] [CrossRef]
- Zhang, K.; Dong, C.; Chang, J. Genome-Wide Association Study of 100-kernel Weight in Maize. J. Maize Sci. 2022, 32, 39–47. [Google Scholar]
- Wang, C.; Li, H.; Long, Y.; Dong, Z.; Wang, J.; Liu, C.; Wei, X.; Wan, X. A Systemic Investigation of Genetic Architecture and Gene Resources Controlling Kernel Size-Related Traits in Maize. Int. J. Mol. Sci. 2023, 24, 1025. [Google Scholar] [CrossRef] [PubMed]
- Legris, M.; Ince, Y.; Fankhauser, C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 2019, 10, 5219. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Yamaguchi, S.; Kamiya, Y.; Bae, G.; Chung, W.; Choi, G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006, 47, 124–139. [Google Scholar] [CrossRef]
- Li, Z.; Sheerin, D.J.; von Roepenack-Lahaye, E.; Stahl, M.; Hiltbrunner, A. The phytochrome interacting proteins ERF55 and ERF58 repress light-induced seed germination in Arabidopsis thaliana. Nat. Commun. 2022, 13, 1656. [Google Scholar] [CrossRef]
- Naito, T.; Yamashino, T.; Kiba, T.; Koizumi, N.; Kojima, M.; Sakakibara, H.; Mizuno, T. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 1269–1278. [Google Scholar] [CrossRef]
- Laplaze, L.; Benkova, E.; Casimiro, I.; Maes, L.; Vanneste, S.; Swarup, R.; Weijers, D.; Calvo, V.; Parizot, B.; Herrera-Rodriguez, M.B.; et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 2007, 19, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Roux, M.E.; Chevalier, J.R.; Dagdas, Y.F.; Yamashino, T.; Højgaard, S.D.; Knight, E.; Østergaard, L.; Rodriguez, E.; Petersen, M. The mRNA decapping machinery targets LBD3/ASL9 to mediate apical hook and lateral root development. Life Sci. Alliance 2023, 6, e202302090. [Google Scholar] [CrossRef]
- Aghdasi, M. Analysis of Trehalose-6-Phosphate Control over Carbon Allocation and Growth in Plants; Utrecht University: Utrecht, The Netherlands, 2006. [Google Scholar]
- Wingler, A.; Delatte, T.L.; O’hara, L.E.; Primavesi, L.F.; Jhurreea, D.; Paul, M.J.; Schluepmann, H. Trehalose 6-phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol. 2012, 158, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Rezapoor, H.; Aghdasi, M.; Sadeghipoor, H.R. The Impacts of TRR14-Overexpression on Arabidopsis thaliana Growth and Photosynthetic Parameters. Iran J. Biotechnol. 2017, 15, 33–41. [Google Scholar] [CrossRef][Green Version]
- Zhang, M.; Henquet, M.; Chen, Z.; Zhang, H.; Zhang, Y.; Ren, X.; Van Der Krol, S.; Gonneau, M.; Bosch, D.; Gong, Z. LEW3, encoding a putative alpha-1,2-mannosyltransferase (ALG11) in N-linked glycoprotein, plays vital roles in cell-wall biosynthesis and the abiotic stress response in Arabidopsis thaliana. Plant J. 2009, 60, 983–999. [Google Scholar] [CrossRef]
- Manzano, C.; Pallero-Baena, M.; Silva-Navas, J.; Neila, S.N.; Casimiro, I.; Casero, P.; Garcia-Mina, J.M.; Baigorri, R.; Rubio, L.; A Fernandez, J.; et al. A light-sensitive mutation in Arabidopsis LEW3 reveals the important role of N-glycosylation in root growth and development. J. Exp. Bot. 2017, 68, 5103–5116. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Nebenführ, A. Myosin XIK of Arabidopsis thaliana accumulates at the root hair tip and is required for fast root hair growth. PLoS ONE 2013, 8, e76745. [Google Scholar] [CrossRef] [PubMed]
- Madison, S.L.; Nebenführ, A. Understanding myosin functions in plants: Are we there yet? Curr. Opin. Plant Biol. 2013, 16, 710–717. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).