Identification of AP2/ERF Transcription Factors and Characterization of AP2/ERF Genes Related to Low-Temperature Stress Response and Fruit Development in Luffa
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Growth Conditions, and Stress Treatment
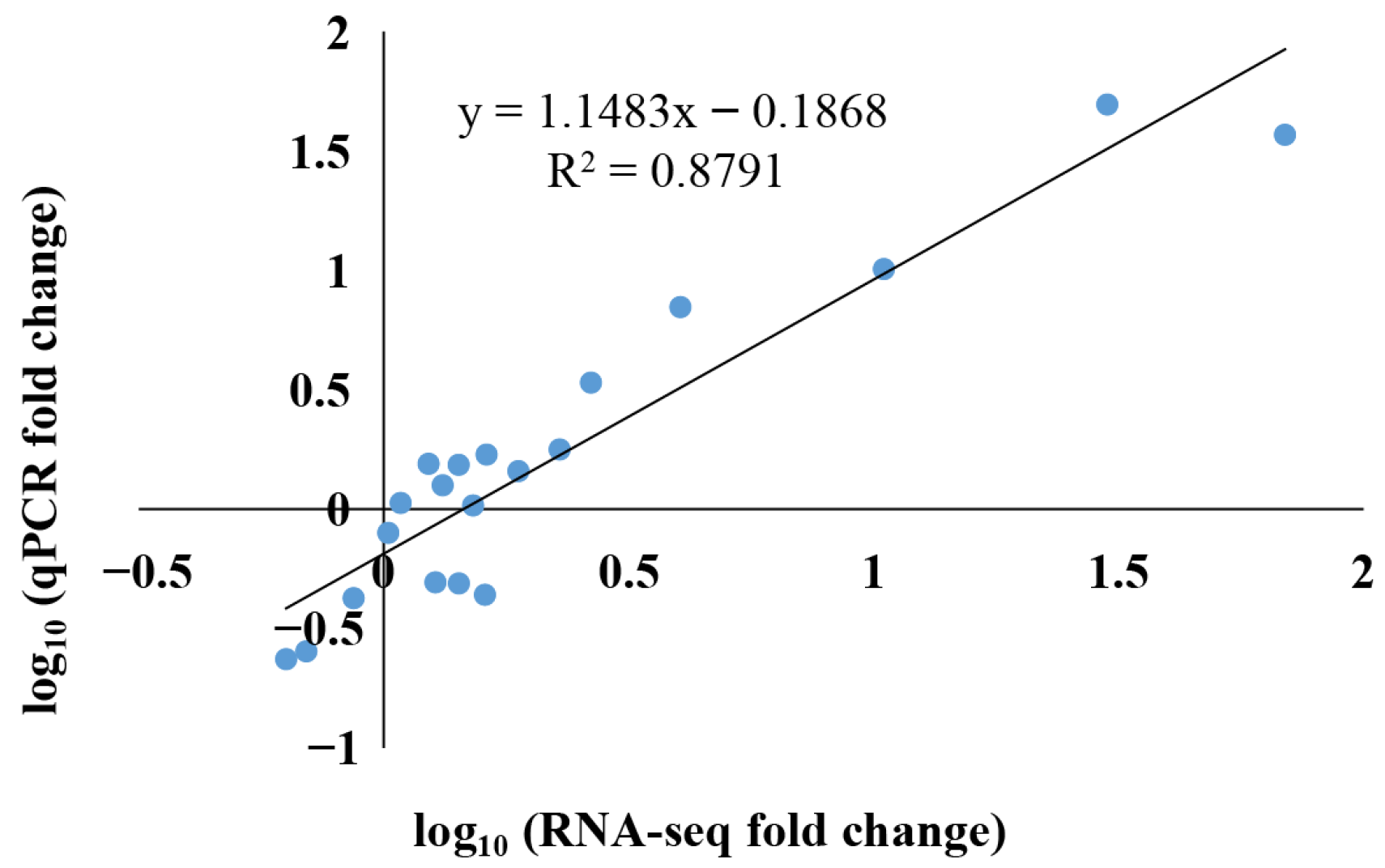
2.2. Total RNA Extraction, RNA Sequencing, and Gene Expression Analysis
2.3. The Identification of LcAP2/ERF Genes from Genome of the Luffa
2.4. Bioinformatics Analysis of the LcAP2/ERF Gene Family
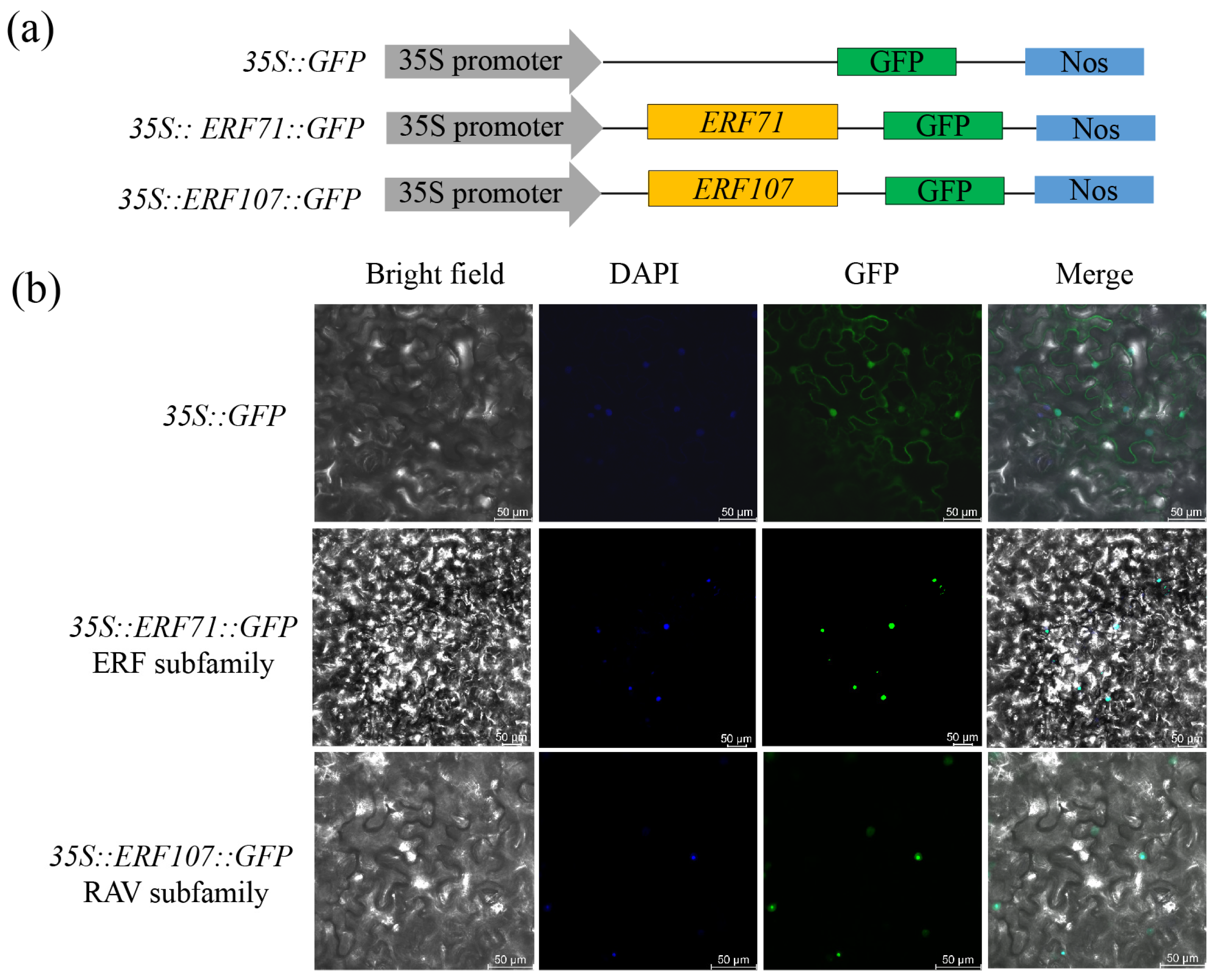
2.5. Subcellular Localization of LcAP2/ERF Transcription Factors
3. Results
3.1. Characterization of LcAP2/ERF GENES in Luffa
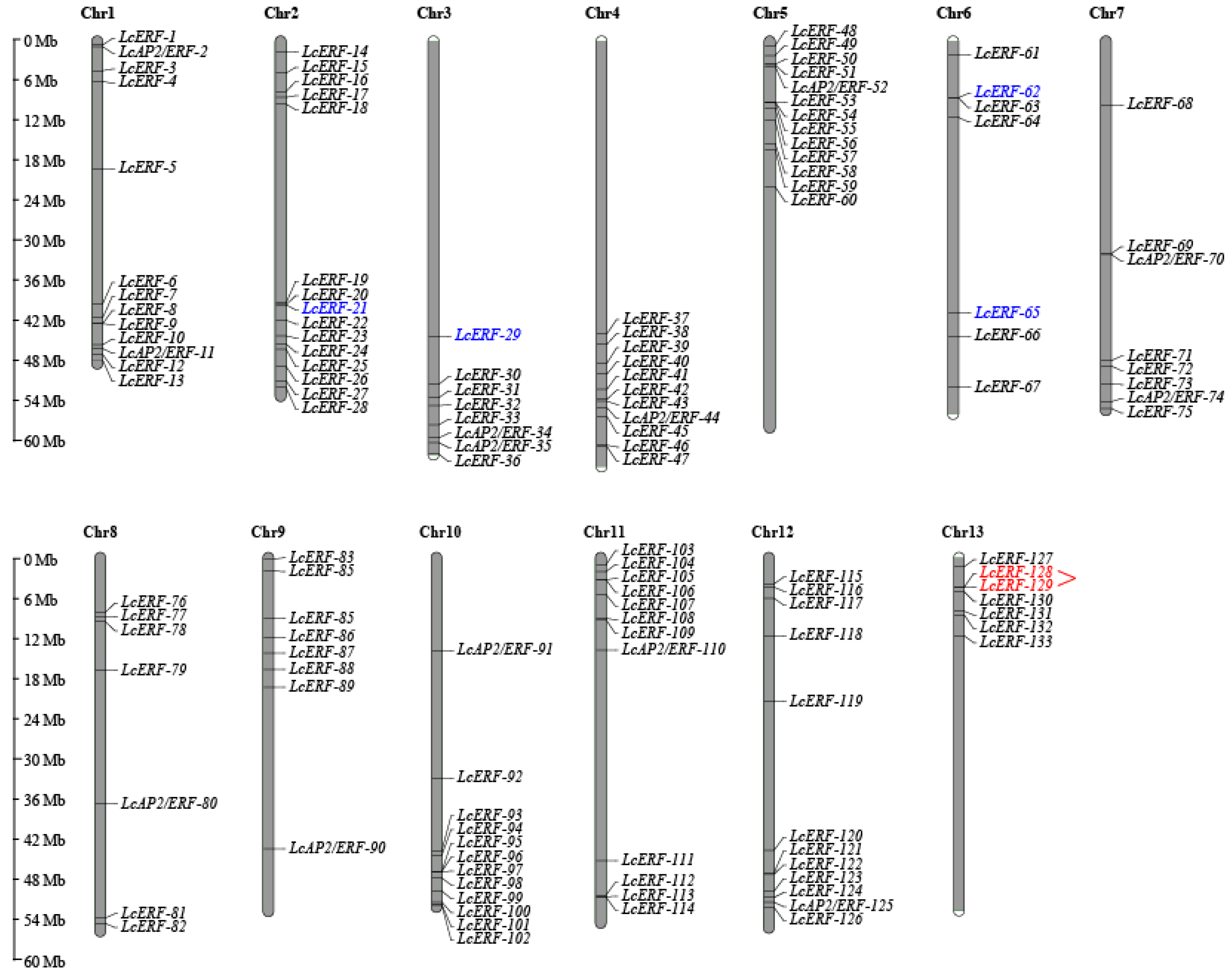
3.2. The Chromosome Localization and Replication of LcAP2/ERF Genes
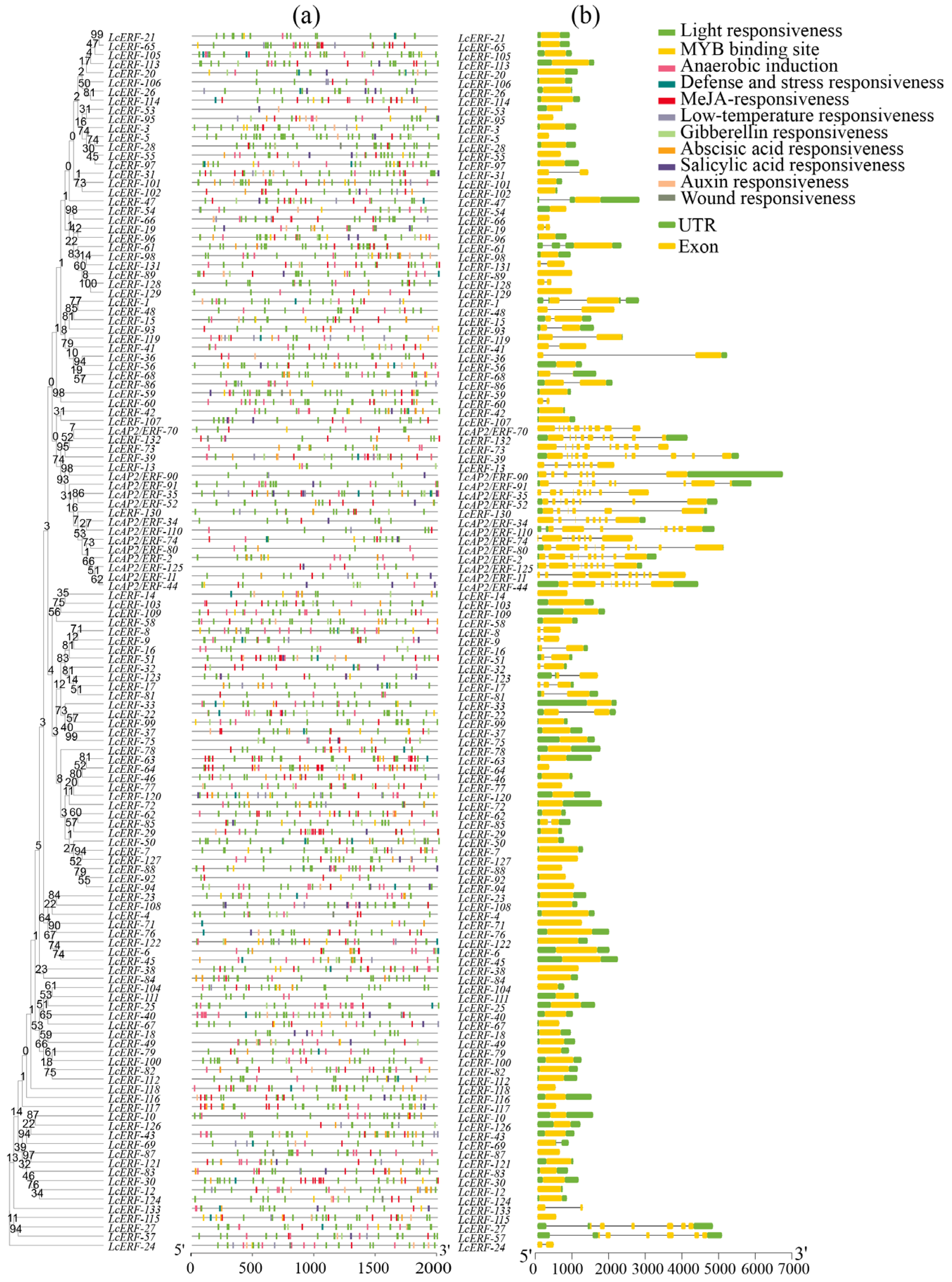
3.3. Promoter Cis-Acting Elements and Structural Analyses of LcAP2/ERF Genes
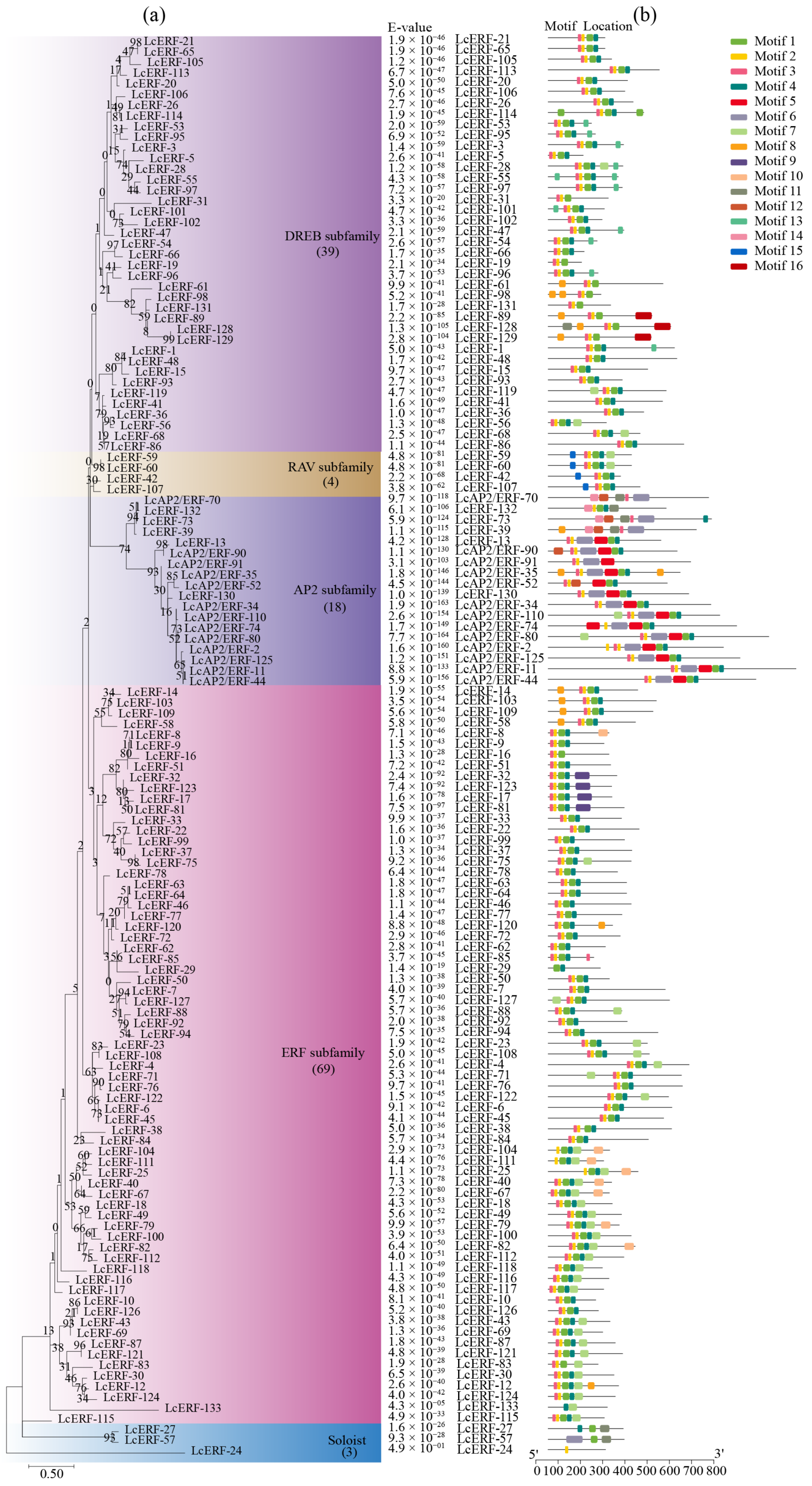
3.4. The Protein Phylogenetic Relationships and Conservation Patterns of LcAP2/ERF Transcription Factors
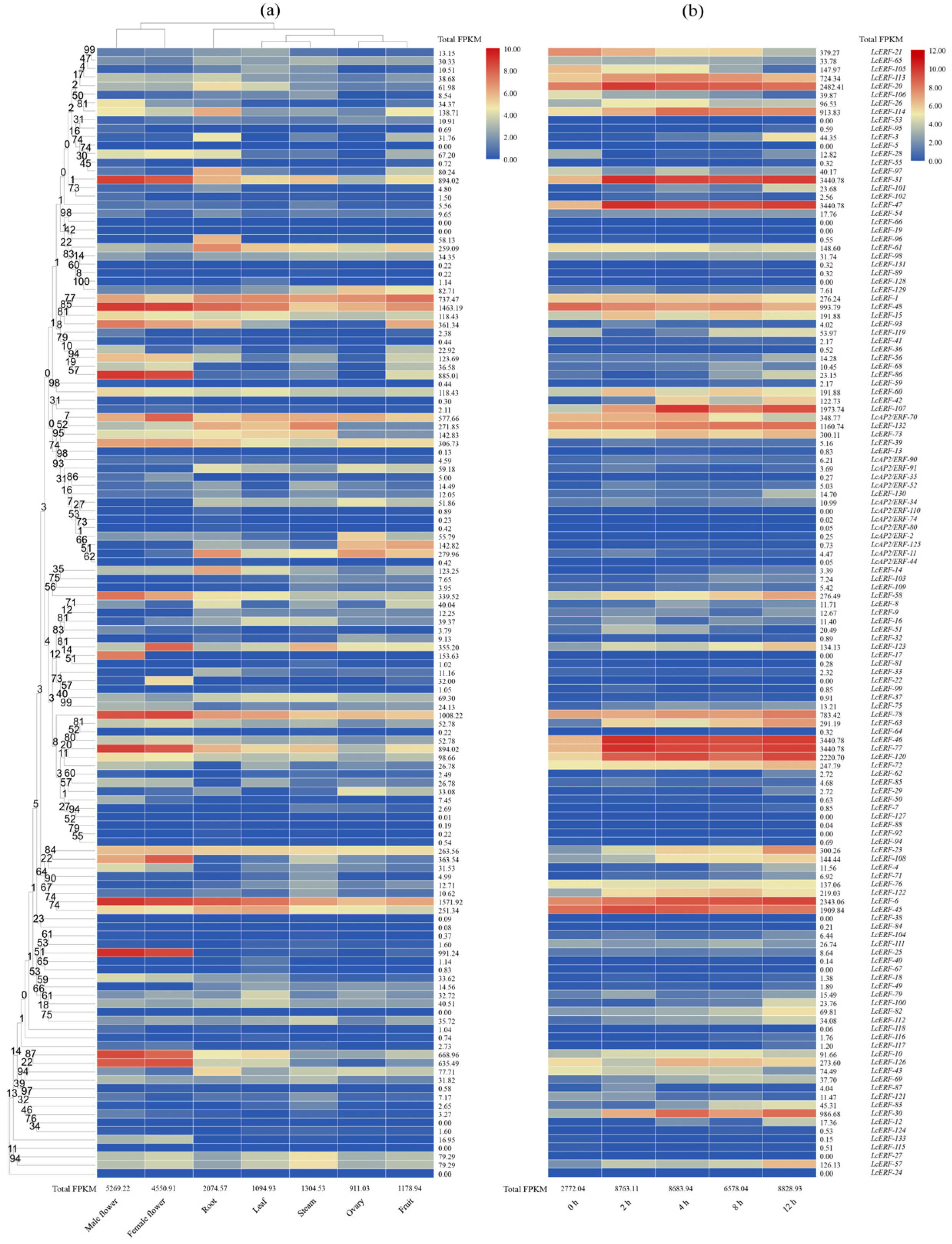
3.5. LcAP2/ERF Expression Profiles in Seven Tissues
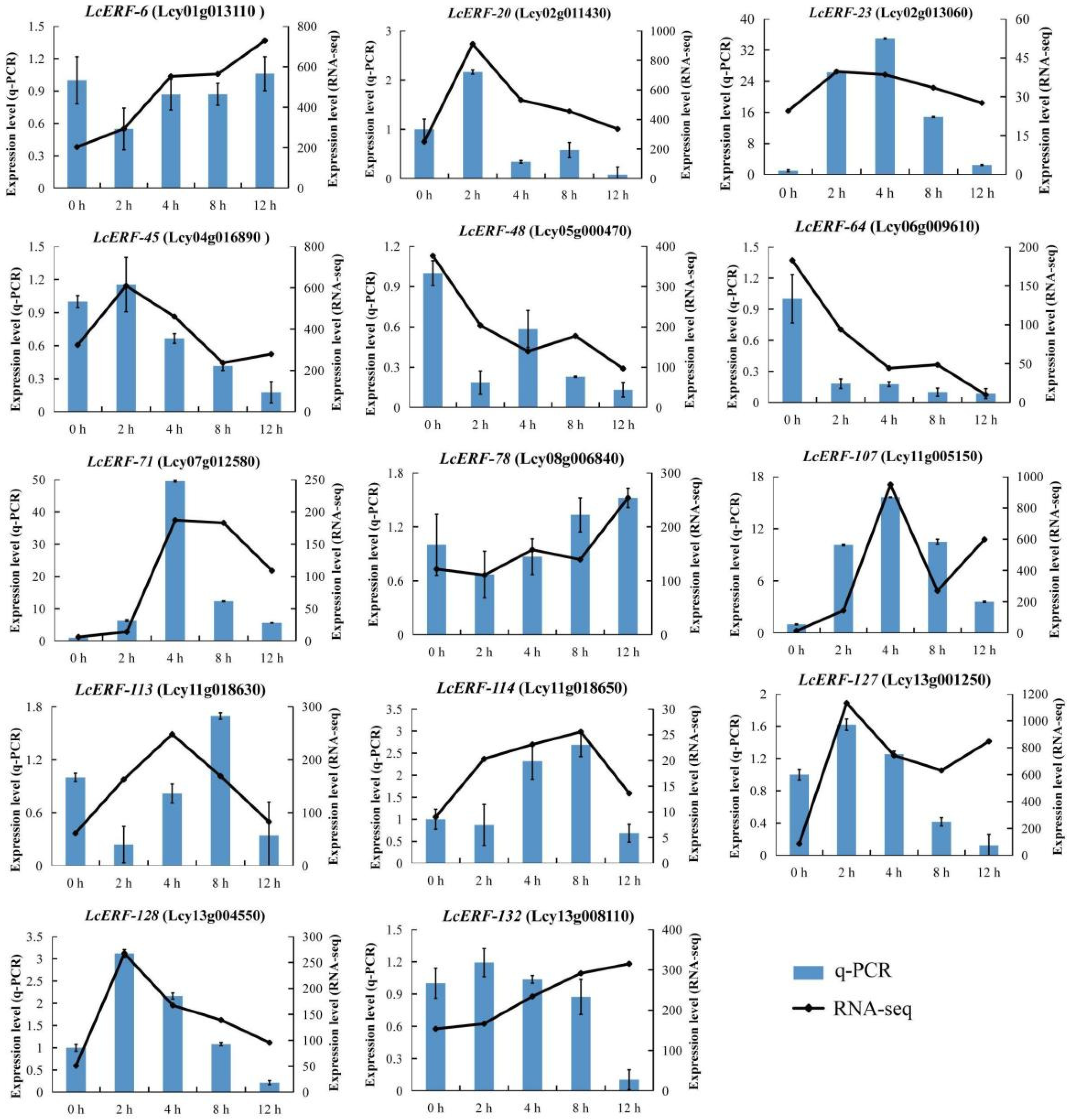
3.6. Expression Analyses of LcAP2/ERF During the Low-Temperature-Induced Stress Condition
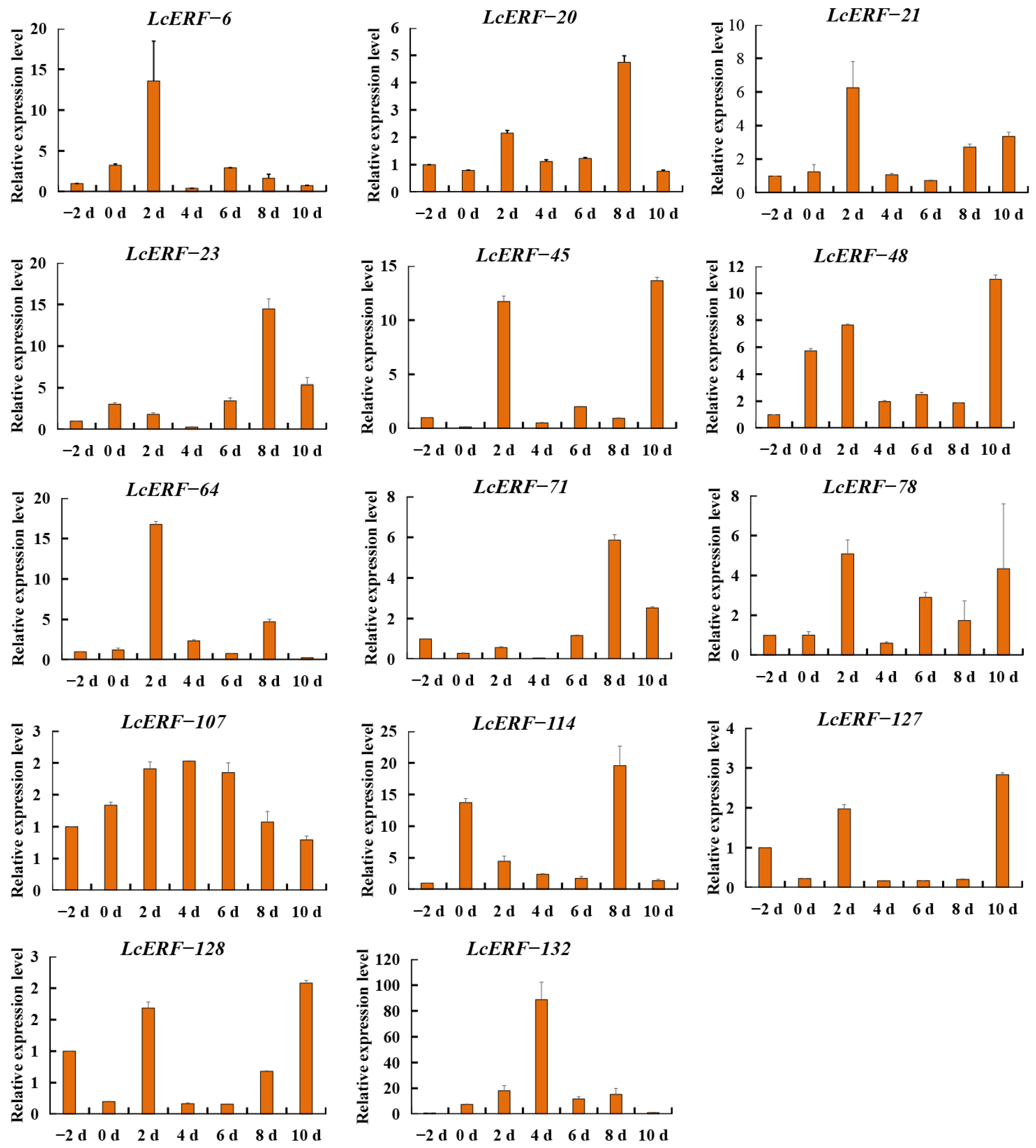
3.7. Analysis of LcAP2/ERF Expression During Fruit Development in L. cylindrica
3.8. The Subcellular Localization of LcAP2/ERF Transcription Factors
4. Discussion
4.1. Characterization of Luffa LcAP2/ERF Genes
4.2. Analysis of L. cylindrica AP2/ERF Gene Promoters and Duplication Events
4.3. Expression Profiles of LcAP2/ERF Genes in L. cylindrica and the Changes in Expression Induced by Low-Temperature Stress
4.4. LcAP2/ERF Genes Associated with L. cylindrica Fruit Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Rao, X.; Zhang, R.; Gu, S.; Shen, Q.; Wu, H.; Lv, S.; Xie, L.; Li, X.; Wang, X. Genome-Wide Identification, Evolution, and Expression Analyses of AP2/ERF Family Transcription Factors in Erianthus fulvus. IJMS 2023, 24, 7102. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liao, J.; Ling, Q.; Xi, Y.; Qian, Y. Genome-wide identification and expression profiling analysis of maize AP2/ERF superfamily genes reveal essential roles in abiotic stress tolerance. BMC Genom. 2022, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chen, Y.; Wang, Y.; Shen, Y.; Yang, J.; Jia, B.; Sun, X.; Sun, M. A comprehensive investigation of the regulatory roles of OsERF096, an AP2/ERF transcription factor, in rice cold stress response. Plant Cell Rep. 2023, 42, 2011–2022. [Google Scholar] [CrossRef]
- Mantiri, F.R.; Kurdyukov, S.; Lohar, D.P.; Sharopova, N.; Saeed, N.A.; Wang, X.D.; Vandenbosch, K.A.; Rose, R.J. The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol. 2008, 146, 1622–1636. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999, 27, 470–478. [Google Scholar] [CrossRef]
- Je, B.I.; Piao, H.L.; Park, S.J.; Park, S.H.; Kim, C.M.; Xuan, Y.H.; Park, S.H.; Huang, J.; Do Choi, Y.; An, G.; et al. RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 2010, 22, 1777–1791. [Google Scholar] [CrossRef]
- Ma, L.; Shi, Q.; Ma, Q.; Wang, X.; Chen, X.; Han, P.; Luo, Y.; Hu, H.; Fei, X.; Wei, A. Genome-wide analysis of AP2/ERF transcription factors that regulate fruit development of Chinese prickly ash. BMC Plant Biol. 2024, 24, 565. [Google Scholar] [CrossRef]
- Qi, X.; Liu, L.; Liu, C.; Song, L.; Dong, Y.; Chen, L.; Li, M. Sweet cherry AP2/ERF transcription factor, PavRAV2, negatively modulates fruit size by directly repressing PavKLUH expression. Physiol. Plant 2023, 175, e14065. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; An, L.; Li, F.; Ahmad, W.; Aslam, M.; Ul Haq, M.Z.; Yan, Y.; Ahmad, R.M. Wide-Range Portrayal of AP2/ERF Transcription Factor Family in Maize (Zea mays L.) Development and Stress Responses. Genes 2023, 14, 194. [Google Scholar] [CrossRef]
- Dong, L.; Cheng, Y.; Wu, J.; Cheng, Q.; Li, W.; Fan, S.; Jiang, L.; Xu, Z.; Kong, F.; Zhang, D.; et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 2015, 66, 2635–2647. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, T.; Yin, K.Q.; Chen, Z.; Gu, H.; Qu, L.J.; Qin, G. Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol. 2012, 195, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Qin, J.; Tong, S.; Wang, W.; Jiang, Y. One AP2/ERF Transcription Factor Positively Regulates Pi Uptake and Drought Tolerance in Poplar. IJMS 2022, 23, 5241. [Google Scholar] [CrossRef]
- Chen, K.; Tang, W.; Zhou, Y.; Chen, J.; Xu, Z.; Ma, R.; Dong, Y.; Ma, Y.; Chen, M. AP2/ERF transcription factor GmDREB1 confers drought tolerance in transgenic soybean by interacting with GmERFs. Plant Physiol. Biochem. 2022, 170, 287–295. [Google Scholar] [CrossRef]
- Kong, L.; Song, Q.; Wei, H.; Wang, Y.; Lin, M.; Sun, K.; Zhang, Y.; Yang, J.; Li, C.; Luo, K. The AP2/ERF transcription factor PtoERF15 confers drought tolerance via JA-mediated signaling in Populus. New Phytol. 2023, 240, 1848–1867. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shi, S.Z.; Jiang, Y.; Zhong, F.; Liu, G.; Yu, C.; Lian, B.; Chen, Y. Genome-wide investigation of the AP2/ERF superfamily and their expression under salt stress in Chinese willow (Salix matsudana). PeerJ 2021, 9, e11076. [Google Scholar] [CrossRef]
- Magar, M.M.; Liu, H.; Yan, G. Genome-Wide Analysis of AP2/ERF Superfamily Genes in Contrasting Wheat Genotypes Reveals Heat Stress-Related Candidate Genes. Front. Plant Sci. 2022, 13, 853086. [Google Scholar] [CrossRef]
- Zhao, M.; Li, Y.; Zhang, X.; You, X.; Yu, H.; Guo, R.; Zhao, X. Genome-Wide Identification of AP2/ERF Superfamily Genes in Juglans mandshurica and Expression Analysis under Cold Stress. IJMS 2022, 23, 15225. [Google Scholar] [CrossRef]
- Du, C.; Hu, K.; Xian, S.; Liu, C.; Fan, J.; Tu, J.; Fu, T. Dynamic transcriptome analysis reveals AP2/ERF transcription factors responsible for cold stress in rapeseed (Brassica napus L.). Mol. Genet. Genomics 2016, 291, 1053–1067. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular Events of Rice AP2/ERF Transcription Factors. IJMS 2022, 23, 12013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, L.; Pang, S.; Zheng, Q.; Quan, S.; Liu, Y.; Xu, T.; Liu, Y.; Qi, M. Function Analysis of the ERF and DREB Subfamilies in Tomato Fruit Development and Ripening. Front. Plant Sci. 2022, 13, 849048. [Google Scholar] [CrossRef]
- Hu, L.; Liu, S. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet. Mol. Biol. 2011, 34, 624–634. [Google Scholar] [CrossRef]
- Wan, R.; Song, J.; Lv, Z.; Qi, X.; Han, X.; Guo, Q.; Wang, S.; Shi, J.; Jian, Z.; Hu, Q.; et al. Genome-Wide Identification and Comprehensive Analysis of the AP2/ERF Gene Family in Pomegranate Fruit Development and Postharvest Preservation. Genes 2022, 13, 895. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X. Genome-wide identification of AP2/ERF superfamily genes and their expression during fruit ripening of Chinese jujube. Sci. Rep. 2018, 8, 15612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, C.; Deng, M.; Li, S.; Chen, Y.; Gu, X.; Tang, G.; Lin, Y.; Wang, Y.; He, W.; et al. Genome-Wide Analysis of the ERF Family and Identification of Potential Genes Involved in Fruit Ripening in Octoploid Strawberry. IJMS 2022, 23, 10550. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Meng, D.; Li, S.; Feng, X.; Di, Q.; Zhou, M.; Yu, X.; He, C.; Yan, Y.; Wang, J.; Sun, M.; et al. CsBPC2 is essential for cucumber survival under cold stress. BMC Plant Biol. 2023, 23, 566. [Google Scholar] [CrossRef]
- Cheng, X.; Qin, M.; Chen, R.; Jia, Y.; Zhu, Q.; Chen, G.; Wang, A.; Ling, B.; Rong, W. Citrullus colocynthis (L.) schrad.: A promising pharmaceutical resource for multiple diseases. Molecules 2023, 28, 6221. [Google Scholar] [CrossRef]
- Tang, L.; He, Y.; Liu, B.; Xu, Y.; Zhao, G. Genome-wide identification and characterization analysis of WUSCHEL-related homeobox family in melon (Cucumis melo L.). IJMS 2023, 24, 12326. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Kong, F.; Tang, T.; Luo, Y.; Gao, H.; Xu, J.; Xing, G.; Li, L. Physiological and transcriptomic analyses revealed that humic acids improve low-temperature stress tolerance in zucchini (Cucurbita pepo L.) Seedlings. Plants 2023, 12, 548. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Peng, L.; Cao, C.; Bai, C.; Wang, Y.; Li, Z.; Zhu, H.; Wen, Q.; He, S. Identification of WRKY family members and characterization of the low-temperature-stress-responsive WRKY genes in Luffa (Luffa cylindrica L.). Plants 2024, 13, 676. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.; Xing, G.; Liu, J.; Duan, A.; Xu, Z.; Li, M.; Zhuang, J.; Xiong, A. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF Transcription Factor Responses and Tolerance to Various Abiotic Stresses in Plants: A Comprehensive Review. IJMS 2024, 25, 893. [Google Scholar] [CrossRef]
- Miura, K.; Tada, Y. Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 2014, 5, 4. [Google Scholar] [CrossRef]
- Xie, Z.; Yang, C.; Liu, S.; Li, M.; Gu, L.; Peng, X.; Zhang, Z. Identification of AP2/ERF transcription factors in Tetrastigma hemsleyanum revealed the specific roles of ERF46 under cold stress. Front. Plant Sci. 2022, 13, 936602. [Google Scholar] [CrossRef]
- Yin, F.; Zeng, Y.; Ji, J.; Wang, P.; Zhang, Y.; Li, W. The Halophyte Halostachys caspica AP2/ERF Transcription Factor HcTOE3 Positively Regulates Freezing Tolerance in Arabidopsis. Front. Plant Sci. 2021, 12, 638788. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Wong, D.C.J.; Wang, Y.; Zhu, Z.; Xu, G.; Wang, Q.; Li, S.; Liang, Z.; Xin, H. The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. 2019, 99, 988–1002. [Google Scholar] [CrossRef]
- Khan, M.; Hu, J.; Dahro, B.; Ming, R.; Zhang, Y.; Wang, Y.; Alhag, A.; Li, C.; Liu, J.H. ERF108 from Poncirus trifoliata (L.) Raf. functions in cold tolerance by modulating raffinose synthesis through transcriptional regulation of PtrRafS. Plant J. 2021, 108, 705–724. [Google Scholar] [CrossRef]
- Yuste-Lisbona, F.J.; Fernández-Lozano, A.; Pineda, B.; Bretones, S.; Ortíz-Atienza, A.; García-Sogo, B.; Müller, N.A.; Angosto, T.; Capel, J.; Moreno, V.; et al. ENO regulates tomato fruit size through the floral meristem development network. Proc. Natl. Acad. Sci. USA 2020, 117, 8187–8195. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hamid, H.; Chin, K.; Moeder, W.; Yoshioka, K. High throughput chemical screening supports the involvement of Ca2+ in cyclic nucleotide-gated ion channel-mediated programmed cell death in Arabidopsis. Plant Signal Behav. 2011, 6, 1817–1819. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Lee, S.M.; Kim, M.H.; Park, S.K.; Jung, K.H. Genome-Wide Analysis of Cyclic Nucleotide-Gated Channel Genes Related to Pollen Development in Rice. Plants 2022, 11, 3145. [Google Scholar] [CrossRef]
- Hao, L.; Qiao, X. Genome-wide identification and analysis of the CNGC gene family in maize. PeerJ 2018, 6, e5816. [Google Scholar] [CrossRef]
- Saand, M.A.; Xu, Y.P.; Munyampundu, J.P.; Li, W.; Zhang, X.R.; Cai, X.Z. Phylogeny and evolution of plant cyclic nucleotide-gated ion channel (CNGC) gene family and functional analyses of tomato CNGCs. DNA Res. 2015, 22, 471–483. [Google Scholar] [CrossRef]
- Zhang, N.; Lin, H.; Zeng, Q.; Fu, D.; Gao, X.; Wu, J.; Feng, X.; Wang, Q.; Ling, Q.; Wu, Z. Genome-wide identification and expression analysis of the cyclic nucleotide-gated ion channel (CNGC) gene family in Saccharum spontaneum. BMC Genom. 2023, 24, 281. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Du, L.; Shen, L.; He, J.; Xia, X.; Zhang, L.; Yang, X. Genome-Wide Exploration and Expression Analysis of the CNGC Gene Family in Eggplant (Solanum melongena L.) under Cold Stress, with Functional Characterization of SmCNGC1a. IJMS 2023, 24, 13049. [Google Scholar] [CrossRef]
- Baloch, A.A.; Raza, A.M.; Rana, S.S.A.; Ullah, S.; Khan, S.; Zaib Un, N.; Zahid, H.; Malghani, G.K.; Kakar, K.U. BrCNGC gene family in field mustard: Genome-wide identification, characterization, comparative synteny, evolution and expression profiling. Sci. Rep. 2021, 11, 24203. [Google Scholar] [CrossRef]
- Wang, L.; Li, M.; Liu, Z.; Dai, L.; Zhang, M.; Wang, L.; Zhao, J.; Liu, M. Genome-wide identification of CNGC genes in Chinese jujube (Ziziphus jujuba Mill.) and ZjCNGC2 mediated signalling cascades in response to cold stress. BMC Genom. 2020, 21, 191. [Google Scholar] [CrossRef]
- Chen, L.; Wang, W.; He, H.; Yang, P.; Sun, X.; Zhang, Z. Genome-Wide Identification, Characterization and Experimental Expression Analysis of CNGC Gene Family in Gossypium. IJMS 2023, 24, 4617. [Google Scholar] [CrossRef]
- Mao, X.; Wang, C.; Lv, Q.; Tian, Y.; Wang, D.; Chen, B.; Mao, J.; Li, W.; Chu, M.; Zuo, C. Cyclic nucleotide gated channel genes (CNGCs) in Rosaceae: Genome-wide annotation, evolution and the roles on Valsa canker resistance. Plant Cell Rep. 2021, 40, 2369–2382. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Xu, Y.; Wang, J.; Liu, W.; Zhou, Q.; Luo, S.; Huang, W.; He, X.; Li, Q.; Peng, Q.; et al. The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nat. Commun. 2019, 10, 5158. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant 2017, 10, 1293–1306. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; González, V.M.; Hénaff, E.; Câmara, F.; Cozzuto, L.; Lowy, E. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef]
- Montero-Pau, J.; Blanca, J.; Bombarely, A.; Ziarsolo, P.; Esteras, C.; Martí-Gómez, C.; Ferriol, M.; Gómez, P.; Jamilena, M.; Mueller, L. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 2018, 16, 1161–1171. [Google Scholar] [CrossRef]
- Wu, H.; Zhao, G.; Gong, H.; Li, J.; Luo, C.; He, X.; Luo, S.; Zheng, X.; Liu, X.; Guo, J.; et al. A high-quality sponge gourd (Luffa cylindrica) genome. Hortic. Res. 2020, 7, 128. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, B.; Li, Y.; Huang, L.; Zhang, Q.; Zhu, H.; Wen, Q. RNA sequencing analysis of low temperature and low light intensity-responsive transcriptomes of zucchini (Cucurbita pepo L.). Sci. Hortic. 2020, 265, 109263. [Google Scholar] [CrossRef]
- Langdon, W.B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 2015, 8, 1. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, M.C.; Konaté, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or normalized counts a comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Ye, X.; Zhang, Q.; Li, Y.; Chen, M.; Wang, B.; Bai, C.; Li, Z.; Wen, Q.; et al. Genome-wide identification and expression analysis of the WRKY gene family in response to low-temperature and drought stresses in Cucurbita pepo L. Sci. Hortic. 2024, 330, 113048. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Castellanos, M.D.P.; Wickramasinghe, C.D.; Betrán, E. The roles of gene duplications in the dynamics of evolutionary conflicts. Proc. Biol. Sci. 2024, 291, 20240555. [Google Scholar] [CrossRef] [PubMed]
- Dorshorst, B.; Harun-Or-Rashid, M.; Bagherpoor, A.J.; Rubin, C.J.; Ashwell, C.; Gourichon, D.; Tixier-Boichard, M.; Hallböök, F.; Andersson, L. A genomic duplication is associated with ectopic eomesodermin expression in the embryonic chicken comb and two duplex-comb phenotypes. PLoS Genet. 2015, 11, e1004947. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Zhang, Y.; Guo, J. Identification and Characterization of AP2/ERF Transcription Factors in Yellow Horn. IJMS 2022, 23, 14991. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Li, A.; Guo, J.; Wang, H.; Qi, H.; Li, M.; Yang, P.; Song, S. An AP2/ERF transcription factor confers chilling tolerance in rice. Sci. Adv. 2024, 10, eado4788. [Google Scholar] [CrossRef]
- Zhu, J.; Wei, X.; Yin, C.; Zhou, H.; Yan, J.; He, W.; Yan, J.; Li, H. ZmEREB57 regulates OPDA synthesis and enhances salt stress tolerance through two distinct signalling pathways in Zea mays. Plant Cell Environ. 2023, 46, 2867–2883. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Li, D.; Liu, X.; Shu, L.; Zhang, H.; Zhang, S.; Song, Y.; Zhang, Z. Global analysis of the AP2/ERF gene family in rose (Rosa chinensis) genome unveils the role of RcERF099 in Botrytis resistance. BMC Plant Biol. 2020, 20, 533. [Google Scholar] [CrossRef]
- Guo, J.; Xu, W.; Hu, Y.; Huang, J.; Zhao, Y.; Zhang, L.; Huang, C.; Ma, H. Phylotranscriptomics in Cucurbitaceae reveal multiple whole-genome duplications and key morphological and molecular innovations. Mol. Plant 2020, 13, 1117–1133. [Google Scholar] [CrossRef]
- Barrera-Redondo, J.; Lira-Saade, R.; Eguiarte, L.E. Gourds and tendrils of cucurbitaceae: How their shape diversity, molecular and morphological novelties evolved via whole-genome duplications. Mol. Plant 2020, 13, 1108–1110. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Cai, B.; Peng, R.H.; Zhu, B.; Jin, X.F.; Xue, Y.; Gao, F.; Fu, X.Y.; Tian, Y.S.; Zhao, W.; et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Shigyo, M.; Ito, M. Analysis of gymnosperm two-AP2-domain-containing genes. Dev. Genes. Evol. 2004, 214, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Ritonga, F.N.; Ngatia, J.N.; Wang, Y.; Khoso, M.A.; Farooq, U.; Chen, S. AP2/ERF, an important cold stress-related transcription factor family in plants: A review. Physiol. Mol. Biol. Plants 2021, 27, 1953–1968. [Google Scholar] [CrossRef]
- Matías-Hernández, L.; Aguilar-Jaramillo, A.E.; Marín-González, E.; Suárez-López, P.; Pelaz, S. RAV genes: Regulation of floral induction and beyond. Ann. Bot. 2014, 114, 1459–1470. [Google Scholar] [CrossRef]
- An, J.P.; Xu, R.R.; Liu, X.; Su, L.; Yang, K.; Wang, X.F.; Wang, G.L.; You, C.X. Abscisic acid insensitive 4 interacts with ICE1 and JAZ proteins to regulate ABA signaling-mediated cold tolerance in apple. J. Exp. Bot. 2022, 73, 980–997. [Google Scholar] [CrossRef]
- Wu, L.; Chen, X.; Ren, H.; Zhang, Z.; Zhang, H.; Wang, J.; Wang, X.C.; Huang, R. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta 2007, 226, 815–825. [Google Scholar] [CrossRef]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; Raghavendra, A.S. Mechanism of Stomatal Closure in Plants Exposed to Drought and Cold Stress. Adv. Exp. Med. Biol. 2018, 1081, 215–232. [Google Scholar] [CrossRef]
- Cui, Y.; Zhai, Y.; He, J.; Song, M.; Flaishman, M.A.; Ma, H. AP2/ERF genes associated with superfast fig (Ficus carica L.) fruit ripening. Front. Plant Sci. 2022, 13, 1040796. [Google Scholar] [CrossRef]
- Zhang, Y.; Ming, R.; Khan, M.; Wang, Y.; Dahro, B.; Xiao, W.; Li, C.; Liu, J.H. ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione S-transferase U17 gene. Plant Biotechnol. J. 2022, 20, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Pei, Y.; Xu, X.; Du, X.; Xue, Q.; Gao, Z.; Shu, P.; Wu, Y.; Liu, Z.; Jian, Y.; et al. Ethylene-MPK8-ERF.C1-PR module confers resistance against Botrytis cinerea in tomato fruit without compromising ripening. New Phytol. 2024, 242, 592–609. [Google Scholar] [CrossRef] [PubMed]
- Tournier, B.; Sanchez-Ballesta, M.T.; Jones, B.; Pesquet, E.; Regad, F.; Latché, A.; Pech, J.C.; Bouzayen, M. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett. 2003, 550, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gao, M.; Zhao, Y.; Chen, Y.; Wu, L.; Yin, H.; Xiong, S.; Wang, S.; Wang, J.; Yang, Y.; et al. LcERF19, an AP2/ERF transcription factor from Litsea cubeba, positively regulates geranial and neral biosynthesis. Hortic. Res. 2022, 9, uhac093. [Google Scholar] [CrossRef]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Körbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef]
- Caarls, L.; Van der Does, D.; Hickman, R.; Jansen, W.; Verk, M.C.; Proietti, S.; Lorenzo, O.; Solano, R.; Pieterse, C.M.; Van Wees, S.C. Assessing the Role of ETHYLENE RESPONSE FACTOR Transcriptional Repressors in Salicylic Acid-Mediated Suppression of Jasmonic Acid-Responsive Genes. Plant Cell Physiol. 2017, 58, 266–278. [Google Scholar] [CrossRef]
- Lv, B.; Wei, K.; Hu, K.; Tian, T.; Zhang, F.; Yu, Z.; Zhang, D.; Su, Y.; Sang, Y.; Zhang, X.; et al. MPK14-mediated auxin signaling controls lateral root development via ERF13-regulated very-long-chain fatty acid biosynthesis. Mol. Plant 2021, 14, 285–297. [Google Scholar] [CrossRef]
- Gambhir, P.; Singh, V.; Parida, A.; Raghuvanshi, U.; Kumar, R.; Sharma, A.K. Ethylene response factor ERF.D7 activates auxin response factor 2 paralogs to regulate tomato fruit ripening. Plant Physiol. 2022, 190, 2775–2796. [Google Scholar] [CrossRef]
- Zhou, S.M.; Wang, F.; Yan, S.Y.; Zhu, Z.M.; Gao, X.F.; Zhao, X.L. Phylogenomics and plastome evolution of Indigofera (Fabaceae). Front. Plant Sci. 2023, 14, 1186598. [Google Scholar] [CrossRef]
- Guo, Z.; Cai, L.; Liu, C.; Chen, Z.; Guan, S.; Ma, W.; Pan, G. Low-temperature stress affects reactive oxygen species, osmotic adjustment substances, and antioxidants in rice (Oryza sativa L.) at the reproductive stage. Sci. Rep. 2022, 12, 6224. [Google Scholar] [CrossRef]
- Fei, J.; Wang, Y.S.; Cheng, H.; Su, Y.B.; Zhong, Y.J.; Zheng, L. The Kandelia obovata transcription factor KoWRKY40 enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 2022, 22, 274. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Tang, Y.; Tam, N.F.; Gan, K.; Wu, J.; Wu, W.; Fu, Y.; Li, M.; Hu, Z.; Li, F.; et al. Microcosm study on cold adaptation and recovery of an exotic mangrove plant, Laguncularia racemosa in China. Mar. Environ. Res. 2022, 176, 105611. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Chen, Y.; Liu, Z.; Liu, Z.; Shu, P.; Wang, R.; Hao, Y.; Su, D.; Pirrello, J.; Liu, Y.; et al. SlERF.F12 modulates the transition to ripening in tomato fruit by recruiting the co-repressor TOPLESS and histone deacetylases to repress key ripening genes. Plant Cell 2022, 34, 1250–1272. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.R.; Allan, A.C.; Chen, K.S.; Ferguson, I.B. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol. 2010, 153, 1280–1292. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, L.; Zhang, M.; Wu, T.; Song, T.; Yao, Y.; Zhang, J.; Tian, J. MdWER interacts with MdERF109 and MdJAZ2 to mediate methyl jasmonate- and light-induced anthocyanin biosynthesis in apple fruit. Plant J. 2024, 118, 1327–1342. [Google Scholar] [CrossRef]
- Pei, Y.; Xue, Q.; Shu, P.; Xu, W.; Du, X.; Wu, M.; Liu, K.; Pirrello, J.; Bouzayen, M.; Hong, Y.; et al. Bifunctional transcription factors SlERF.H5 and H7 activate cell wall and repress gibberellin biosynthesis genes in tomato via a conserved motif. Dev. Cell 2024, 59, 1345–1359.e6. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Zhong, H.; Cao, C.; Wang, Y.; Zhang, Q.; Wen, Q.; Zhu, H.; Li, Z. Identification of AP2/ERF Transcription Factors and Characterization of AP2/ERF Genes Related to Low-Temperature Stress Response and Fruit Development in Luffa. Agronomy 2024, 14, 2509. https://doi.org/10.3390/agronomy14112509
Liu J, Zhong H, Cao C, Wang Y, Zhang Q, Wen Q, Zhu H, Li Z. Identification of AP2/ERF Transcription Factors and Characterization of AP2/ERF Genes Related to Low-Temperature Stress Response and Fruit Development in Luffa. Agronomy. 2024; 14(11):2509. https://doi.org/10.3390/agronomy14112509
Chicago/Turabian StyleLiu, Jianting, Haifeng Zhong, Chengjuan Cao, Yuqian Wang, Qianrong Zhang, Qingfang Wen, Haisheng Zhu, and Zuliang Li. 2024. "Identification of AP2/ERF Transcription Factors and Characterization of AP2/ERF Genes Related to Low-Temperature Stress Response and Fruit Development in Luffa" Agronomy 14, no. 11: 2509. https://doi.org/10.3390/agronomy14112509
APA StyleLiu, J., Zhong, H., Cao, C., Wang, Y., Zhang, Q., Wen, Q., Zhu, H., & Li, Z. (2024). Identification of AP2/ERF Transcription Factors and Characterization of AP2/ERF Genes Related to Low-Temperature Stress Response and Fruit Development in Luffa. Agronomy, 14(11), 2509. https://doi.org/10.3390/agronomy14112509




