Effects of Anthracnose on the Structure and Diversity of Endophytic Microbial Communities in Postharvest Avocado Fruits
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Treatments
2.2. Methods
2.2.1. DNA Extraction and Polymerase Chain Reaction (PCR) Amplification
2.2.2. Library Construction and Sequencing
2.2.3. Processing of Sequencing Data
2.2.4. ASVs Noise Reduction and SPECIES Annotation
2.2.5. Analysis of Alpha Diversity and Beta Diversity
3. Results
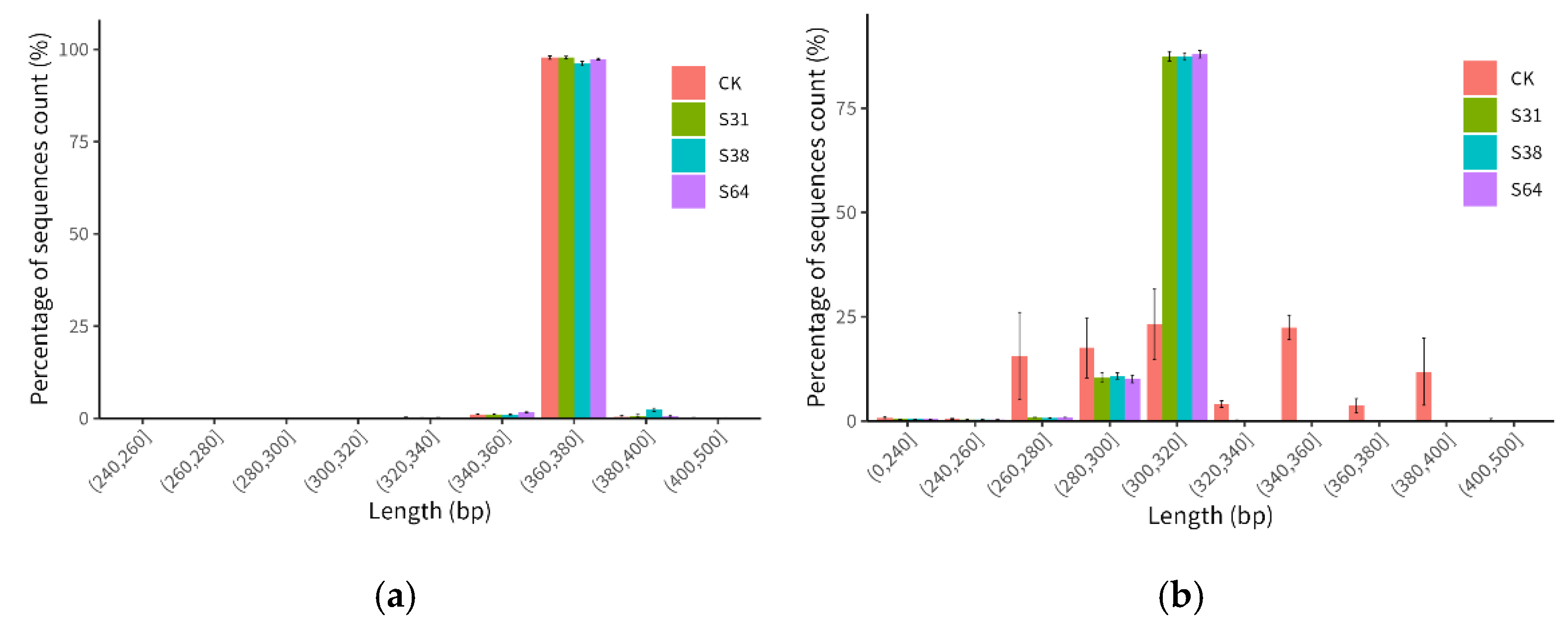
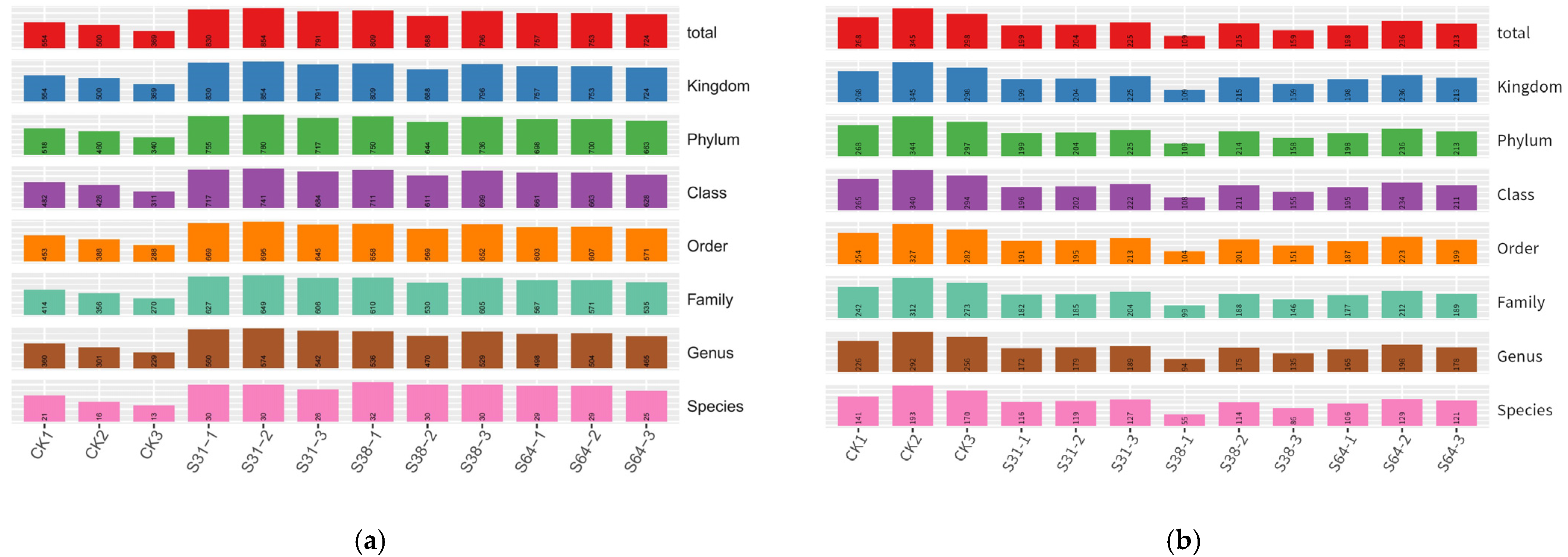
3.1. Analysis of Sequencing Data
3.2. Diversity Analysis of Endophytes
3.2.1. Analysis of Alpha Diversity
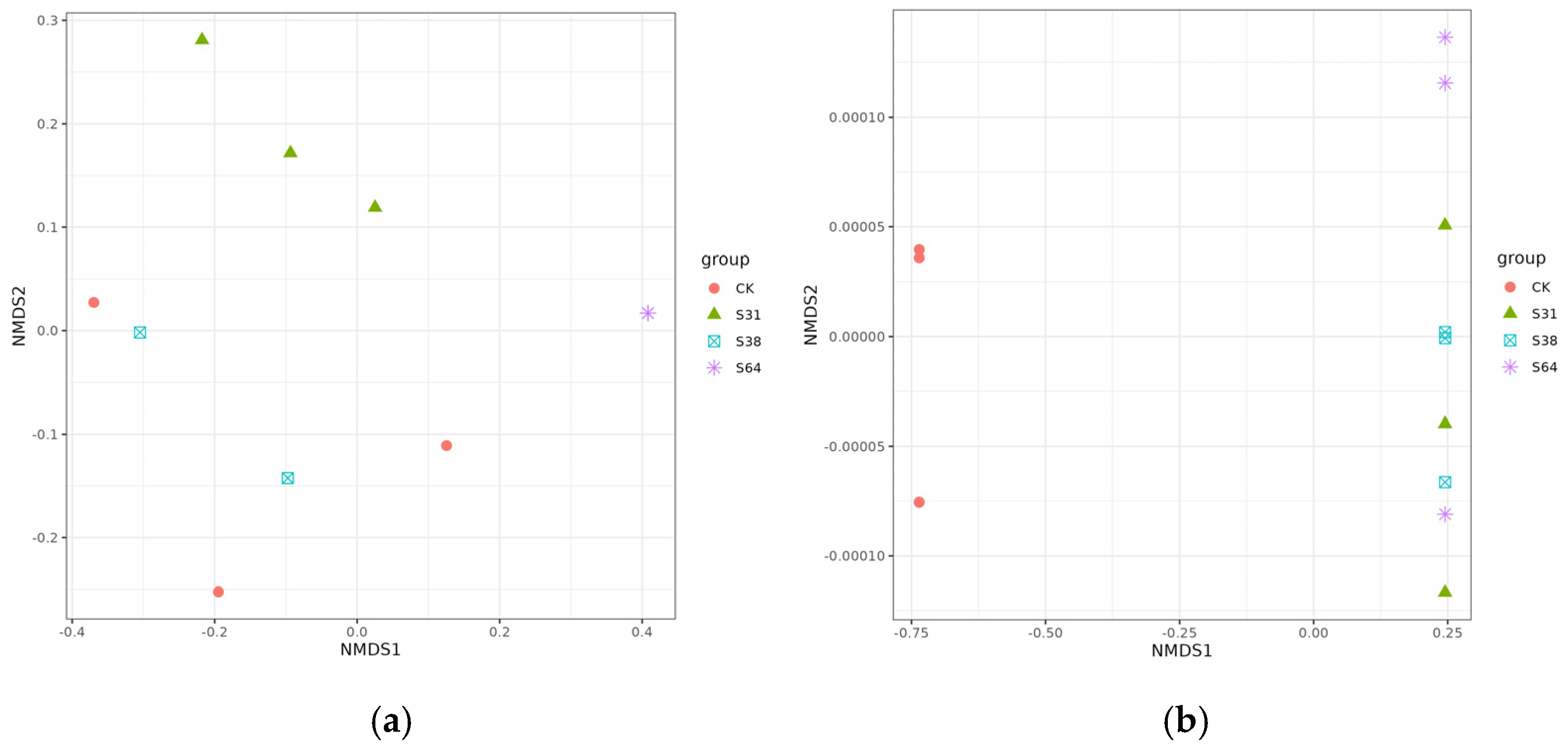
3.2.2. Analysis of Beta Diversity
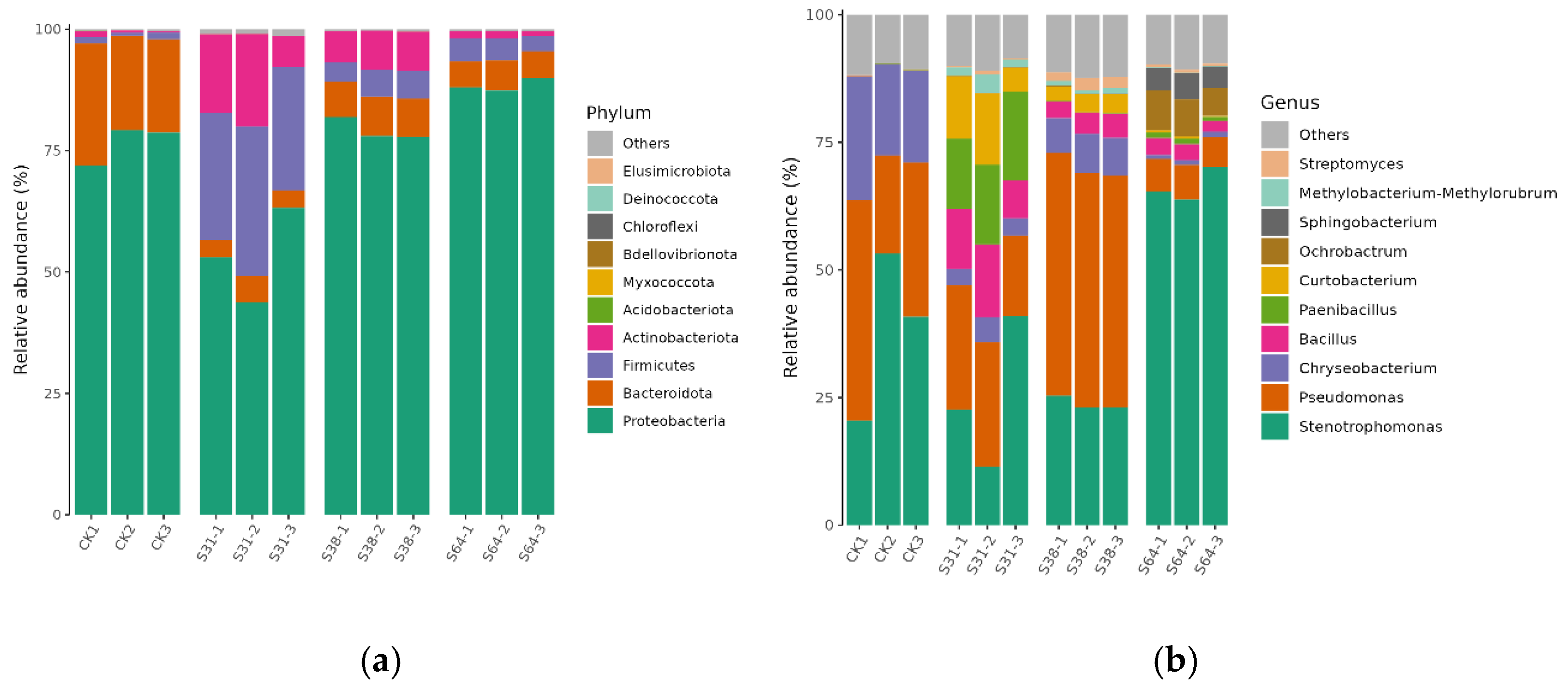
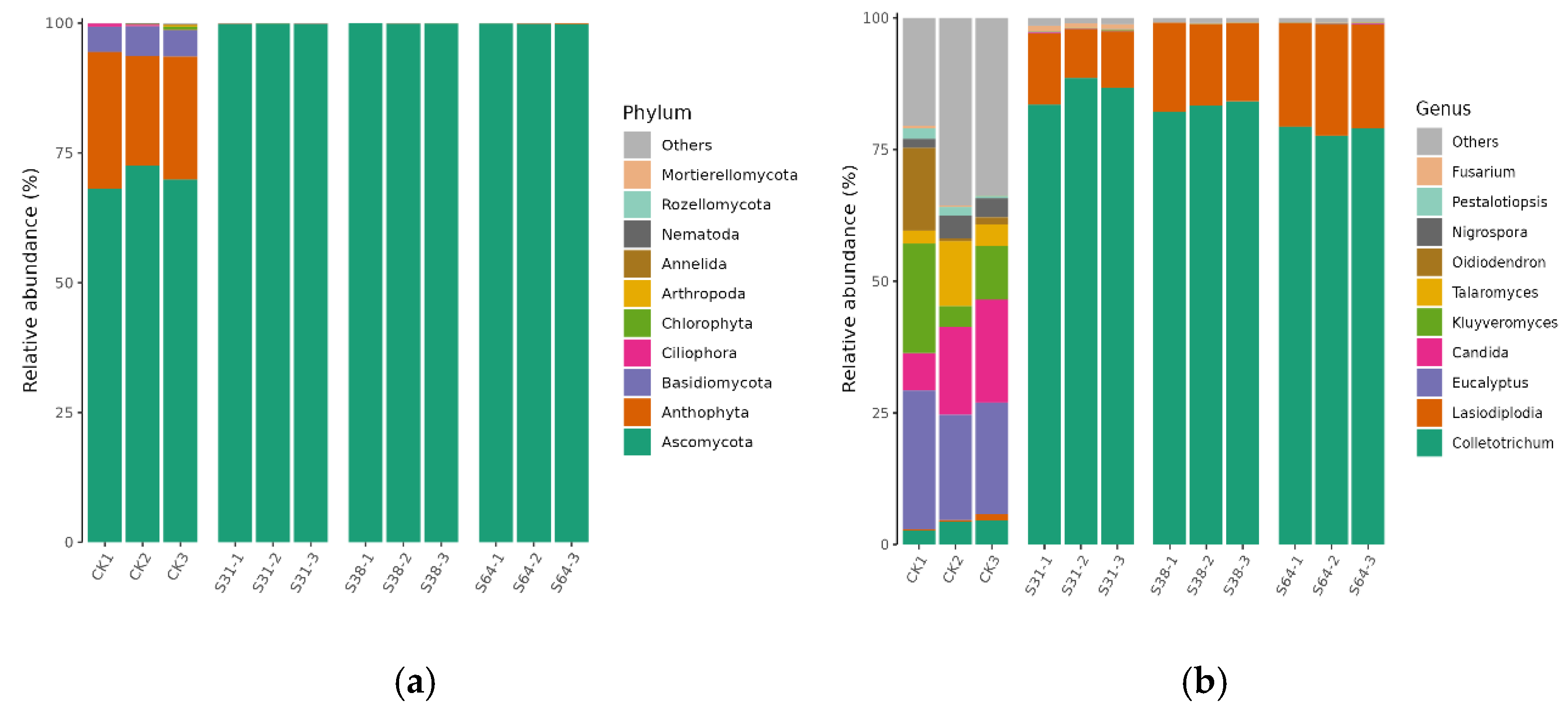
3.3. Composition of Endophyte Community Structure
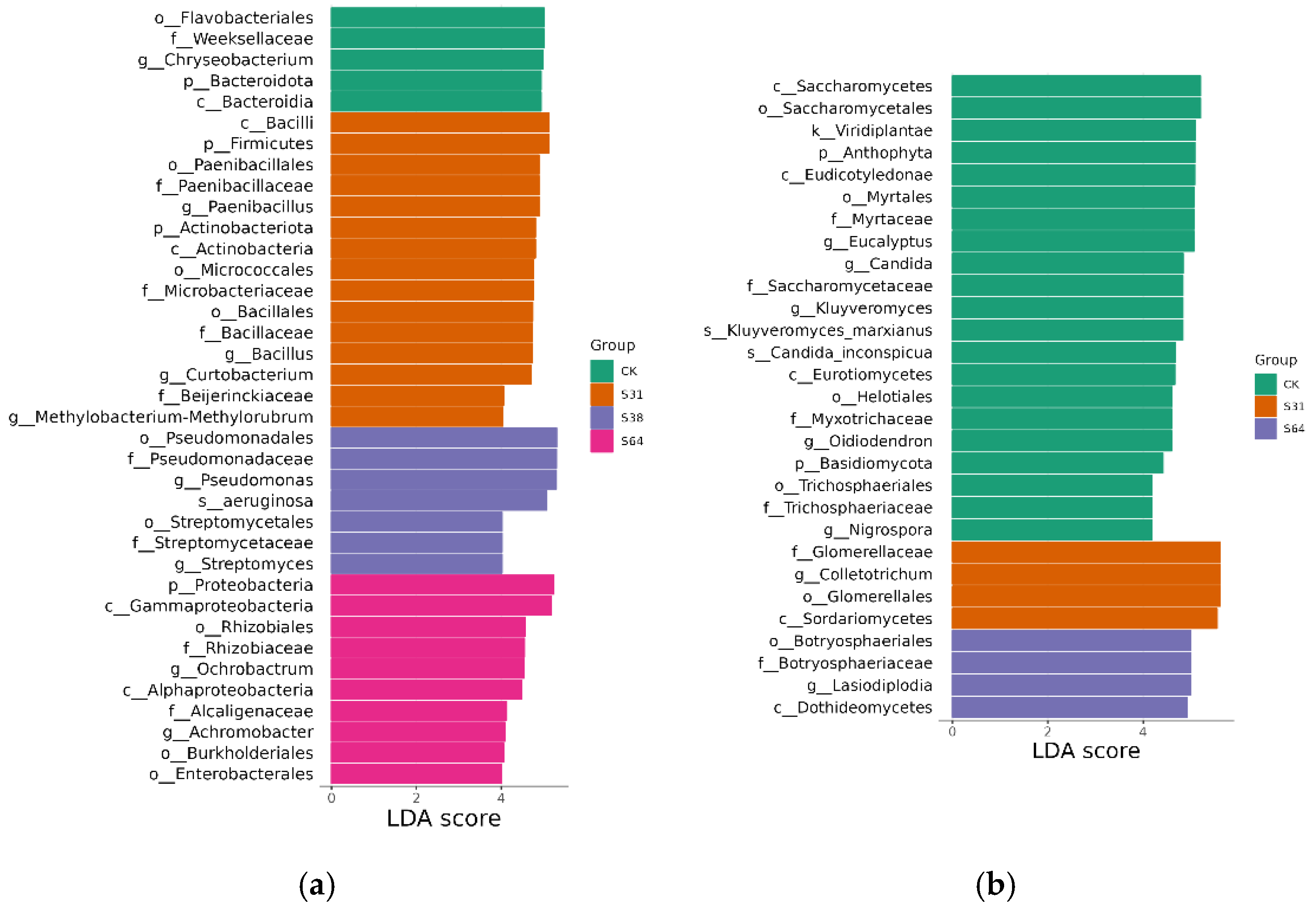
3.4. Analysis of Differences Between Groups of Endophytes
4. Discussion
4.1. Endophyte Diversity in Healthy and Infected Avocados
4.2. Structural Composition and Differences of Endophytic Bacterial Communities in Avocados
4.3. Structural Composition and Differences of Endophytic Fungal Communities in Avocados
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, Q.Y.; Zhang, Y.; Wang, Y.; Wang, D.; Lee, R.P.; Gao, K.; Byrns, R.; Heber, D. California hass avocado: Profiling of carotenoids, tocopherol, fatty acid, and fat content during maturation and from different growing areas. J. Agric. Food Chem. 2009, 57, 10408–10413. [Google Scholar] [CrossRef] [PubMed]
- Marlett, J.A.; Cheung, T.-F. Database and quick methods of assessing typical dietary fiber intakes using data for 228 commonly consumed foods. J. Am. Diet. Assoc. 1997, 97, 1139–1151. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Maymon, M.; Freeman, S. Epidemiology, pathology and identification of Colletotrichum including a novel species associated with avocado (Persea americana) anthracnose in lsrael. Sci. Rep. 2017, 7, 15839. [Google Scholar] [CrossRef] [PubMed]
- Ippolito, A.; Nigro, F. Impact of preharvest application of biological control agents on postharvest diseases of fresh fruits and vegetables. Crop Prot. 2000, 19, 715–723. [Google Scholar] [CrossRef]
- Dessalegn, Y.; Ayalew, A.; Woldetsadik, K. Integrating plant defense inducing chemical, inorganic salt and hot water treatments for the management of postharvest mango anthracnose. Postharvest Biol. Technol. 2013, 85, 83–88. [Google Scholar] [CrossRef]
- EFSA. Conclusion on the peer review of the pesticide risk assessment of the active substance prochloraz. EFSA J. 2011, 9, 2323. [Google Scholar] [CrossRef]
- Shimshoni, J.A.; Bommuraj, V.; Chen, Y.; Sperling, R.; Barel, S.; Feygenberg, O.; Maurer, D.; Alkan, N. Postharvest fungicide for avocado fruits: Antifungal efficacy and peel to pulp distribution kinetics. Foods 2020, 9, 124. [Google Scholar] [CrossRef]
- Wisniewski, M.; Droby, S.; Norelli, J.; Liu, J.; Schena, L. Alternative management technologies for postharvest disease control: The journey from simplicity to complexity. Postharvest Biol. Technol. 2016, 122, 3–10. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Dürr, C.; Schwanck, A.A.; Robin, M.-H.; Sarthou, J.-P.; Cellier, V.; Messéan, A.; Aubertot, J.-N. Integrated management of damping-off diseases. A review. Agron. Sustain. Dev. 2017, 37, 10. [Google Scholar] [CrossRef]
- Bill, M.; Gokul, J.K.; Viljoen, F.; Korsten, L. Fungal microbiome shifts on avocado fruit associated with a combination of postharvest chemical and physical interventions. J. Appl. Microbiol. 2022, 133, 1905–1918. [Google Scholar] [CrossRef]
- Peñuelas, J.; Terradas, J. The foliar microbiome. Trends Plant Sci. 2014, 19, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Massart, S.; Martinez-Medina, M.; Jijakli, M.H. Biological control in the microbiome era: Challenges and opportunities. Biol. Control 2015, 89, 98–108. [Google Scholar] [CrossRef]
- Ibrahim, M.; Kaushik, N.; Sowemimo, A.; Chhipa, H.; Koekemoer, T.; Van De Venter, M.; Odukoya, O.A. Antifungal and antiproliferative activities of endophytic fungi isolated from the leaves of Markhamia tomentosa. Pharm. Biol. 2017, 55, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, F.; Zhang, M.; Ou, T.; Wang, R.; Strobel, G.; Xiang, Z.; Zhou, Z.; Xie, J. Diversity of cultivable endophytic bacteria in mulberry and their potential for antimicrobial and plant growth-promoting activities. Microbiol. Res. 2019, 229, 126328. [Google Scholar] [CrossRef] [PubMed]
- Petrini, O. Fungal endophytes of tree leaves. In Microbial Ecology of Leaves; Springer: New York, NY, USA, 1991; pp. 179–197. [Google Scholar] [CrossRef]
- Da Silva, S.F.; Olivares, F.L.; Canellas, L.P. The biostimulant manufactured using diazotrophic endophytic bacteria and humates is effective to increase sugarcane yield. Chem. Biol. Technol. Agric. 2017, 4, 24. [Google Scholar] [CrossRef]
- Waqas, M.; Khan, A.L.; Kamran, M.; Hamayun, M.; Lee, I.J. Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules 2012, 17, 10754–10773. [Google Scholar] [CrossRef]
- Armada, E.; Roldán, A.; Azcon, R. Differential activity of autochthonous bacteria in controlling drought stress in native Lavandula and Salvia plants species under drought conditions in natural arid soil. Microb. Ecol. 2014, 67, 410–420. [Google Scholar] [CrossRef]
- Moronta-Barrios, F.; Gionechetti, F.; Pallavicini, A.; Marys, E.; Venturi, V. Bacterial microbiota of rice roots: 16s-based taxonomic profiling of endophytic and rhizospheric diversity, endophytes isolation and simplified endophytic community. Microorganisms 2018, 6, 14. [Google Scholar] [CrossRef]
- Zheng, C.J.; Li, L.; Zou, J.P.; Han, T.; Qin, L.P. Identification of a quinazoline alkaloid produced by Penicillium Vinaceum, an endophytic fungus from Crocus Sativus. Pharm. Biol. 2012, 50, 129–133. [Google Scholar] [CrossRef]
- Sare, A.R.; Jijakli, M.H.; Massart, S. Microbial ecology to support integrative efficacy improvement of biocontrol agents for postharvest diseases management. Postharvest Biol. Technol. 2021, 179, 111572. [Google Scholar] [CrossRef]
- Prusky, D.; Plumbley, R.A.; Kobiler, I. The relationship between antifungal diene levels and fungal inhibition during quiescent infection of unripe avocado fruits by Colletotrichum gloeosporioides. Plant Pathol. 1991, 40, 45–52. [Google Scholar] [CrossRef]
- Bejarano-Bolívar, A.A.; Lamelas, A.; Wobeser, E.A.; Sánchez-Rangel, D.; Méndez-Bravo, A.; Eskalen, A.; Reverchon, F. Shifts in the structure of rhizosphere bacterial communities of avocado after fusarium dieback. Rhizosphere 2021, 18, 100333. [Google Scholar] [CrossRef]
- Shetty, K.G.; Rivadeneira, D.V.; Jayachandran, K.; Walker, D.M. Isolation and molecular characterization of the fungal endophytic microbiome from conventionally and organically grown avocado trees in south florida. Mycol. Prog. 2016, 15, 977–986. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, W.; Zhou, B.; Du, H.; Xi, L.; Zou, M.; Zou, H.; Xin, L.; Gao, Z.; Chen, Y. Variable characteristics of microbial communities on the surface of sweet cherries under different storage conditions. Postharvest Biol. Technol. 2021, 173, 111408. [Google Scholar] [CrossRef]
- Sim, H.L.; Hong, Y.-K.; Yoon, W.B.; Yuk, H.-G. Behavior of Salmonella spp. and natural microbiota on fresh-cut dragon fruits at different storage temperatures. Int. J. Food Microbiol. 2013, 160, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.F.; Wei, Z.X.; Li, X.L.; Luo, Y.J.; Han, G.H. Analysis of diversity and community structure of endophytes in Sclerotinia sclerotiorum-infected and healthy mulberry fruits by high-throughput sequencing. Food Sci. 2021, 42, 61–68. [Google Scholar] [CrossRef]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.J. Study on Latent Infection, Pathogenic Mechanism of Botryodiplodia theobromae and Control for Stem-End Rot of Mango Fruit. Ph.D. Thesis, Hainan University, Haikou, China, 2013. [Google Scholar]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Chen, F. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Kazutaka, K.; Misakwa, K.; Kei-ichi, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Rivas, M.N.; Burton, O.T.; Wise, P.; Zhang, Y.-Q.; Hobson, S.A.; Lloret, M.G.; Chehoud, C.; Kuczynski, J.; DeSantis, T.; Warrington, J. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol. 2013, 131, 201–212, ISBN 972-97465-2-4. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Liu, H.; Li, J.; Carvalhais, L.C.; Percy, C.D.; Prakash Verma, J.; Schenk, P.M.; Singh, B.K. Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens. New Phytol. 2021, 229, 2873–2885. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Riera, N.; Jin, T.; Li, J.; Wang, N. Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 2017, 5, 97. [Google Scholar] [CrossRef]
- Shi, W.; Li, M.; Wei, G.; Tian, R.; Li, C.; Wang, B.; Lin, R.; Shi, C.; Chi, X.; Zhou, B. The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome 2019, 7, 14. [Google Scholar] [CrossRef]
- Li, X.G.; Ding, C.F.; Zhang, T.L.; Wang, X.X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil. Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Xiang, L.G.; Zhou, H.; Wang, H.C.; Li, Z.; Chen, Q.L.; Yu, Z.H. Bacterial community structure and diversity of rhizosphere soil and stem of healthy and bacterial wilt tobacco plants. Acta Microbiol. Sin. 2019, 59, 1984–1999. [Google Scholar] [CrossRef]
- Wang, J.; Shi, C.; Fang, D.; Che, J.; Wu, W.; Lyu, L.; Li, W. The impact of storage temperature on the development of microbial communities on the surface of blueberry fruit. Foods 2023, 12, 1611. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Tosh, P.K. In A clinician’s primer on the role of the microbiome in human health and disease. Mayo Clin. Proc. 2014, 89, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Murray, R.; Trüper, H. Proteobacteria classis nov. a name for the phylogenetic taxon that includes the “purple bacteria and their relatives”. Int. J. Syst. Evol. Microbiol. 1988, 38, 321–325. [Google Scholar] [CrossRef]
- Gupta, R.S. The phylogeny of proteobacteria: Relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol. Rev. 2000, 24, 367–402. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A common factor in human diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Ludwig, W.; Schleifer, K.H.; Whitman, W.B. Revised road map to the phylum Firmicutes. In Bergey’s Manual® of Systematic Bacteriology; Springer, New York, NY, USA, 2009; pp. 1–13. [CrossRef]
- Li, C.-M.; Cheng, T.-H.; Liang, Y.-R.; Huang, C.-L. Enhancement of Actinobacteriota and suppression of root-knot nematode infection in cucumbers through chitin application. Res. Sq. 2023, 1, 1–60. [Google Scholar] [CrossRef]
- Mwanza, E.P.; Hugo, A.; Charimba, G.; Hugo, C.J. Pathogenic potential and control of Chryseobacterium species from clinical, fish, food and environmental sources. Microorganisms 2022, 10, 895. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef]
- Chase, A.B.; Arevalo, P.; Polz, M.F.; Berlemont, R.; Martiny, J.B. Evidence for ecological flexibility in the cosmopolitan genus Curtobacterium. Front. Microbiol. 2016, 7, 1874. [Google Scholar] [CrossRef]
- Loria, R.; Bukhalid, R.A.; Fry, B.A.; King, R.R. Plant pathogenicity in the genus Streptomyces. Plant Dis. 1997, 81, 836–846. [Google Scholar] [CrossRef]
- Höfte, M.; Vos, P. Plant pathogenic Pseudomonas species. In Plant-Associated Bacteria; Springer: Dordrecht, The Netherlands, 2006; pp. 507–533. [Google Scholar] [CrossRef]
- Green, P.N.; Ardley, J.K. Review of the genus Methylobacterium and closely related organisms: A proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 2727–2748. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, M.B.; Berg, G.; Van Der Lelie, D.; Dow, J.M. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Shu, L.J.; Yang, Y.L. Bacillus classification based on matrix-assisted laser desorption ionization time-of-flight mass spectrometry-effects of culture conditions. Sci Rep. 2017, 7, 15546. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.P.; Pembroke, J.T. The genus Ochrobactrum as major opportunistic pathogens. Microorganisms 2020, 8, 1797. [Google Scholar] [CrossRef]
- Carmichael, P.C.; Siyoum, N.; Chidamba, L.; Korsten, L. Exploring the microbial communities associated with botrytis cinerea during berry development in table grape with emphasis on potential biocontrol yeasts. Eur. J. Plant Pathol. 2019, 154, 919–930. [Google Scholar] [CrossRef]
- Shen, Y.; Nie, J.; Li, Z.; Li, H.; Wu, Y.; Dong, Y.; Zhang, J. Differentiated surface fungal communities at point of harvest on apple fruits from rural and peri-urban orchards. Sci. Rep. 2018, 8, 2165. [Google Scholar] [CrossRef]
- Jones, E.G.; Suetrong, S.; Sakayaroj, J.; Bahkali, A.H.; Abdel-Wahab, M.A.; Boekhout, T.; Pang, K.-L. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 2015, 73, 1–72. [Google Scholar] [CrossRef]
- Salvatore, M.M.; Alves, A.; Andolfi, A. Secondary metabolites produced by Neofusicoccum species associated with plants: A review. Agriculture 2021, 11, 149. [Google Scholar] [CrossRef]
- Modrzewska, B.; Kurnatowski, P. Selected pathogenic characteristics of fungi from the genus Candida. Ann. Parasitol. 2013, 59, 57–66. Available online: https://agro.icm.edu.pl/agro/element/bwmeta1.element.agro-00217c2e-7e37-4a13-a56e-004b5b173c33 (accessed on 23 May 2024).
- Hambleton, S.; Egger, K.N.; Currah, R.S. The genus Oidiodendron: Species delimitation and phylogenetic relationships based on nuclear ribosomal DNA analysis. Mycologia 1998, 90, 854–868. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.; Hyde, K.D.; Groenewald, J.Z.; Xu, J.; Crous, P.W. Pestalotiopsis revisited. Stud. Mycol. 2014, 79, 121–186. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, M.M.; Andolfi, A.; Nicoletti, R. The thin line between pathogenicity and endophytism: The case of Lasiodiplodia theobromae. Agriculture 2020, 10, 488. [Google Scholar] [CrossRef]
- Angeli, D.; Sare, A.R.; Jijakli, M.H.; Pertot, I.; Massart, S. Insights gained from metagenomic shotgun sequencing of apple fruit epiphytic microbiota. Postharvest Biol. Technol. 2019, 153, 96–106. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, X.; Li, D.; Gu, X.; Godana, E.A.; Dhanasekaran, S.; Zhao, L.; Zhang, H. Transcriptome analysis of postharvest grapes in response to Talaromyces rugulosus O1 infection. Postharvest Biol. Technol. 2021, 178, 111542. [Google Scholar] [CrossRef]
- Wang, M.; Liu, F.; Crous, P.W.; Cai, L. Phylogenetic reassessment of Nigrospora: Ubiquitous endophytes, plant and human pathogens. Persoonia-Mol. Phylogeny Evol. Fungi 2017, 39, 118–142. [Google Scholar] [CrossRef]
- Ma, L.-J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef]
- Nicoletti, R. Endophytic fungi of citrus plants. Agriculture 2019, 9, 247. [Google Scholar] [CrossRef]
- Nicoletti, R.; Di Vaio, C.; Cirillo, C. Endophytic fungi of olive tree. Microorganisms 2020, 8, 1321. [Google Scholar] [CrossRef]
- James, M.; Michael, W.; Jolanda, R.; Bernard, S. Invasive everywhere? Phylogeographic analysis of the globally distributed tree pathogen Lasiodiplodia theobromae. Forests 2017, 8, 145. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M. Functional overlap of the arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; De Bruijn, I.; Dekkers, E.; Van Der Voort, M.; Schneider, J.H.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Ritpitakphong, U.; Falquet, L.; Vimoltust, A.; Berger, A.; Métraux, J.P.; L’Haridon, F. The microbiome of the leaf surface of arabidopsis protects against a fungal pathogen. New Phytol. 2016, 210, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Blaustein, R.A.; Lorca, G.L.; Meyer, J.L.; Gonzalez, C.F.; Teplitski, M. Defining the core citrus leaf-and root-associated microbiota: Factors associated with community structure and implications for managing huanglongbing (citrus greening) disease. Appl. Environ. Microbiol. 2017, 83, e00210-17. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Ranamukhaarachchi, S.L.; Hannaway, D.B. Efficacy of antagonist strains of Bacillus megaterium, Enterobacter cloacae, Pichia guilliermondii and Candida ethanolica against bacterial wilt disease of tomato. J. Phytol. 2011, 3, 1–10. [Google Scholar]
- Dawoud, M.E.; Kamel, Z.; Hanaa, A.; Farahat, M.G. Growth promotion and biocontrol of leaf spot and leaf speck diseases in tomato by Pseudomonas spp. Egypt. J. Exp. Biol. 2012, 8, 61–70. Available online: https://www.egyseb.net/fulltext/15-1430045299.pdf (accessed on 23 May 2024).
- Elsharkawy, M.M.; Shimizu, M.; Takahashi, H.; Hyakumachi, M. Induction of systemic resistance against Cucumber mosaic virus by Penicillium simplicissimum GP17-2 in arabidopsis and tobacco. Plant Pathol. 2012, 61, 964–976. [Google Scholar] [CrossRef]
- Elsharkawy, M.M.; Shimizu, M.; Takahashi, H.; Ozaki, K.; Hyakumachi, M. Induction of systemic resistance against Cucumber mosaic virus in Arabidopsis thaliana by Trichoderma asperellum SKT-1. Plant Pathol. J. 2013, 29, 193–200. Available online: https://pmc.ncbi.nlm.nih.gov/articles/PMC4174775/ (accessed on 23 May 2024). [CrossRef]





| Sample | Reads | Effective Tags | Merged (%) | GC | Q20 | Q30 | MeanError |
|---|---|---|---|---|---|---|---|
| CK-1 | 146,146 | 133,002 | 91.01 | 52.70 | 98.95 | 96.07 | 0.43 |
| CK-2 | 140,549 | 128,325 | 91.30 | 53.15 | 98.91 | 96.00 | 0.44 |
| CK-3 | 51,018 | 46,707 | 91.55 | 53.27 | 98.96 | 96.16 | 0.43 |
| S31-1 | 148,218 | 135,609 | 91.49 | 54.53 | 98.92 | 96.02 | 0.44 |
| S31-2 | 132,163 | 121,490 | 91.92 | 54.57 | 98.97 | 96.16 | 0.43 |
| S31-3 | 107,988 | 99,417 | 92.06 | 54.27 | 98.95 | 96.11 | 0.43 |
| S38-1 | 148,125 | 135,528 | 91.50 | 54.04 | 98.93 | 96.07 | 0.44 |
| S38-2 | 66,239 | 60,937 | 92.00 | 54.09 | 98.93 | 96.09 | 0.44 |
| S38-3 | 147,885 | 133,274 | 90.12 | 54.11 | 98.93 | 96.06 | 0.44 |
| S64-1 | 148,010 | 134,515 | 90.88 | 54.33 | 98.87 | 95.85 | 0.46 |
| S64-2 | 147,765 | 132,018 | 89.34 | 54.33 | 98.83 | 95.73 | 0.48 |
| S64-3 | 147,694 | 133,083 | 90.11 | 54.32 | 98.88 | 95.91 | 0.46 |
| Sample | Reads | Effective Tags | Merged (%) | GC | Q20 | Q30 | MeanError |
|---|---|---|---|---|---|---|---|
| CK-1 | 140,771 | 128,284 | 91.13 | 53.57 | 98.98 | 96.32 | 0.37 |
| CK-2 | 136,655 | 127,336 | 93.18 | 54.16 | 99.20 | 97.05 | 0.29 |
| CK-3 | 137,931 | 127,800 | 92.66 | 53.51 | 99.14 | 96.84 | 0.31 |
| S31-1 | 69,493 | 65,271 | 93.92 | 52.65 | 99.27 | 97.33 | 0.26 |
| S31-2 | 139,577 | 129,987 | 93.13 | 52.60 | 99.24 | 97.22 | 0.27 |
| S31-3 | 100,345 | 94,603 | 94.28 | 52.59 | 99.31 | 97.43 | 0.25 |
| S38-1 | 137,993 | 128,966 | 93.46 | 52.62 | 99.26 | 97.31 | 0.26 |
| S38-2 | 147,063 | 137,563 | 93.54 | 52.61 | 99.25 | 97.27 | 0.27 |
| S38-3 | 137,068 | 127,698 | 93.16 | 52.60 | 99.29 | 97.38 | 0.26 |
| S64-1 | 142,588 | 133,044 | 93.31 | 52.29 | 99.27 | 97.31 | 0.26 |
| S64-2 | 146,918 | 137,941 | 93.89 | 52.30 | 99.25 | 97.28 | 0.27 |
| S64-3 | 122,538 | 115,421 | 94.19 | 52.27 | 99.29 | 97.38 | 0.26 |
| Sample | Chao1 | ACE | Shannon | Simpson | Coverage |
|---|---|---|---|---|---|
| CK-1 | 653.92 | 654.21 | 2.44 | 0.83 | 99.91 |
| CK-2 | 580.56 | 570.49 | 1.90 | 0.68 | 99.92 |
| CK-3 | 433.21 | 434.87 | 2.22 | 0.77 | 99.80 |
| S31-1 | 900.60 | 878.10 | 3.27 | 0.90 | 99.93 |
| S31-2 | 911.04 | 907.29 | 3.57 | 0.93 | 99.92 |
| S31-3 | 863.67 | 852.60 | 2.97 | 0.82 | 99.88 |
| S38-1 | 844.82 | 845.83 | 2.85 | 0.84 | 99.94 |
| S38-2 | 759.64 | 758.12 | 2.87 | 0.84 | 99.79 |
| S38-3 | 862.42 | 831.99 | 2.97 | 0.85 | 99.93 |
| S64-1 | 848.88 | 839.71 | 2.10 | 0.58 | 99.91 |
| S64-2 | 861.99 | 857.84 | 2.19 | 0.60 | 99.89 |
| S64-3 | 833.72 | 823.87 | 1.91 | 0.52 | 99.90 |
| Sample | Chao1 | ACE | Shannon | Simpson | Coverage |
|---|---|---|---|---|---|
| CK-1 | 313.23 | 320.12 | 3.16 | 0.90 | 99.96 |
| CK-2 | 367.44 | 369.68 | 3.68 | 0.94 | 99.97 |
| CK-3 | 400.67 | 370.38 | 3.56 | 0.93 | 99.96 |
| S31-1 | 274.93 | 277.04 | 0.86 | 0.33 | 99.90 |
| S31-2 | 250.41 | 254.62 | 0.65 | 0.24 | 99.96 |
| S31-3 | 276.69 | 288.81 | 0.72 | 0.27 | 99.93 |
| S38-1 | 174.33 | 191.63 | 0.70 | 0.33 | 99.96 |
| S38-2 | 290.02 | 295.10 | 0.73 | 0.32 | 99.94 |
| S38-3 | 202.36 | 215.82 | 0.69 | 0.31 | 99.96 |
| S64-1 | 265.10 | 265.69 | 0.95 | 0.41 | 99.95 |
| S64-2 | 281.22 | 292.50 | 0.98 | 0.42 | 99.95 |
| S64-3 | 258.29 | 260.00 | 0.95 | 0.41 | 99.95 |
| Phylum | CK | S31 | S38 | S64 |
|---|---|---|---|---|
| Proteobacteria | 76.61 ± 4.09 a | 53.33 ± 9.69 a | 79.27 ± 2.30 a | 88.49 ± 1.38 a |
| Bacteroidota | 21.31 ± 3.35 b | 4.16 ± 1.02 c | 7.74 ± 0.40 b | 5.66 ± 0.48 b |
| Firmicutes | 1.06 ± 0.42 c | 27.4 ± 2.93 b | 5.08 ± 1.01 c | 4.16 ± 0.89 b |
| Actinobacteriota | 0.69 ± 0.46 c | 13.89 ± 6.66 c | 7.53 ± 0.89 bc | 1.30 ± 0.27 c |
| Genus | CK | S31 | S38 | S64 |
|---|---|---|---|---|
| Stenotrophomonas | 38.16 ± 16.50 a | 25.02 ± 14.82 a | 23.82 ± 1.33 b | 66.44 ± 3.35 a |
| Chryseobacterium | 20.02 ± 3.66 b | 3.86 ± 0.97 de | 7.24 ± 0.43 d | 0.94 ± 0.17 e |
| Others | 10.65 ± 1.19 bc | 9.85 ± 1.18 cd | 11.94 ± 0.56 c | 10.05 ±0.64 b |
| Bacillus | 0.09 ±0.02 c | 11.14 ± 3.49 cd | 4.07 ± 0.71 e | 2.89 ± 0.64 ef |
| Paenibacillus | 0.06 ± 0.02 c | 15.56 ± 1.85 bc | 0.10 ± 0.04 g | 0.95 ± 0.18 ef |
| Sphingobacterium | 0.05 ± 0.03 c | 0.03 ± 0.01 e | 0.05 ± 0.03 g | 4.60 ± 0.46 de |
| Curtobacterium | 0.03 ± 0.01 c | 10.34 ± 4.93 cd | 3.37 ± 0.47 e | 0.36 ± 0.11 f |
| Ochrobactrum | 0.03 ± 0.01 c | 0.07 ± 0.02 e | 0.07 ± 0.01 g | 6.83 ± 1.24 c |
| Streptomyces | 0.03 ± 0.02 c | 0.42 ± 0.27 e | 2.12 ± 0.41 f | 0.50 ± 0.07 f |
| Pseudomonas | 30.86 ± 11.92 a | 21.48 ± 4.85 ab | 46.35 ± 1.12 a | 6.31 ± 0.50 cd |
| Methylobacterium-Methylorubrum | 0.02 ± 0.01 c | 2.22 ± 1.19 de | 0.86 ± 0.24 g | 0.11 ± 0.01 f |
| Phylum | CK | S31 | S38 | S64 |
|---|---|---|---|---|
| Ascomycota | 70.18 ± 2.25 a | 99.87 ± 0.03 a | 99.92 ± 0.03 a | 99.86 ± 0.05 a |
| Anthophyta | 23.71 ± 2.65 b | 0.06 ± 0.02 b | 0.04 ± 0.01 b | 0.09 ± 0.09 b |
| Basidiomycota | 5.25 ± 0.48 c | 0.06 ± 0.01 b | 0.04 ± 0.02 b | 0.05 ± 0.01 b |
| Genus | CK | S31 | S38 | S64 |
|---|---|---|---|---|
| Others | 29.91 ± 8.23 a | 1.24 ± 0.20 c | 0.89 ± 0.07 c | 0.80 ± 0.05 c |
| Eucalyptus | 22.49 ± 3.36 ab | 0.06 ± 0.02 c | 0.04 ± 0.01 d | 0.08 ± 0.04 d |
| Candida | 14.46 ± 6.49 bc | 0.11 ± 0.03 c | 0.04 ± 0.03 d | 0.08 ± 0.04 d |
| Kluyveromyces | 11.63 ± 8.50 cd | 0.07 ± 0.03 c | 0.03 ± 0.02 d | 0.06 ± 0.02 d |
| Talaromyces | 6.28 ± 5.26 cd | 0.04 ± 0.00 c | 0.02 ± 0.01 d | 0.04 ± 0.02 d |
| Oidiodendron | 5.89 ± 8.49 cd | 0.03 ± 0.01 c | 0.01 ± 0.01 d | 0.03 ± 0.01 d |
| Colletotrichum | 3.88 ± 1.05 de | 86.28 ± 2.57 a | 83.24 ± 0.98 a | 78.68 ± 0.90 a |
| Nigrospora | 3.21 ± 1.33 de | 0.04 ± 0.01 c | 0.01 ± 0.01 d | 0.03 ± 0.00 d |
| Pestalotiopsis | 1.34 ± 0.79 e | 0.03 ± 0.01 c | 0.01 ± 0.01 d | 0.03 ± 0.01 d |
| Lasiodiplodia | 0.58 ± 0.51 e | 11.17 ± 2.16 b | 15.67 ± 1.07 b | 20.12 ± 0.78 b |
| Fusarium | 0.34 ± 0.18 e | 0.92 ± 0.13 c | 0.04 ± 0.02 d | 0.05 ± 0.02 d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Jiang, Z.; He, P.; Tang, X.; Song, H.; Zhang, T.; Wei, Z.; Dong, T.; Zheng, S.; Tu, X.; et al. Effects of Anthracnose on the Structure and Diversity of Endophytic Microbial Communities in Postharvest Avocado Fruits. Agronomy 2024, 14, 2487. https://doi.org/10.3390/agronomy14112487
Chen X, Jiang Z, He P, Tang X, Song H, Zhang T, Wei Z, Dong T, Zheng S, Tu X, et al. Effects of Anthracnose on the Structure and Diversity of Endophytic Microbial Communities in Postharvest Avocado Fruits. Agronomy. 2024; 14(11):2487. https://doi.org/10.3390/agronomy14112487
Chicago/Turabian StyleChen, Xi, Zhuoen Jiang, Peng He, Xiuhua Tang, Haiyun Song, Tao Zhang, Zhejun Wei, Tao Dong, Shufang Zheng, Xinghao Tu, and et al. 2024. "Effects of Anthracnose on the Structure and Diversity of Endophytic Microbial Communities in Postharvest Avocado Fruits" Agronomy 14, no. 11: 2487. https://doi.org/10.3390/agronomy14112487
APA StyleChen, X., Jiang, Z., He, P., Tang, X., Song, H., Zhang, T., Wei, Z., Dong, T., Zheng, S., Tu, X., Qin, J., Chen, J., & Wang, W. (2024). Effects of Anthracnose on the Structure and Diversity of Endophytic Microbial Communities in Postharvest Avocado Fruits. Agronomy, 14(11), 2487. https://doi.org/10.3390/agronomy14112487





