AC R-CNN: Pixelwise Instance Segmentation Model for Agrocybe cylindracea Cap
Abstract
:1. Introduction
2. Materials and Methods
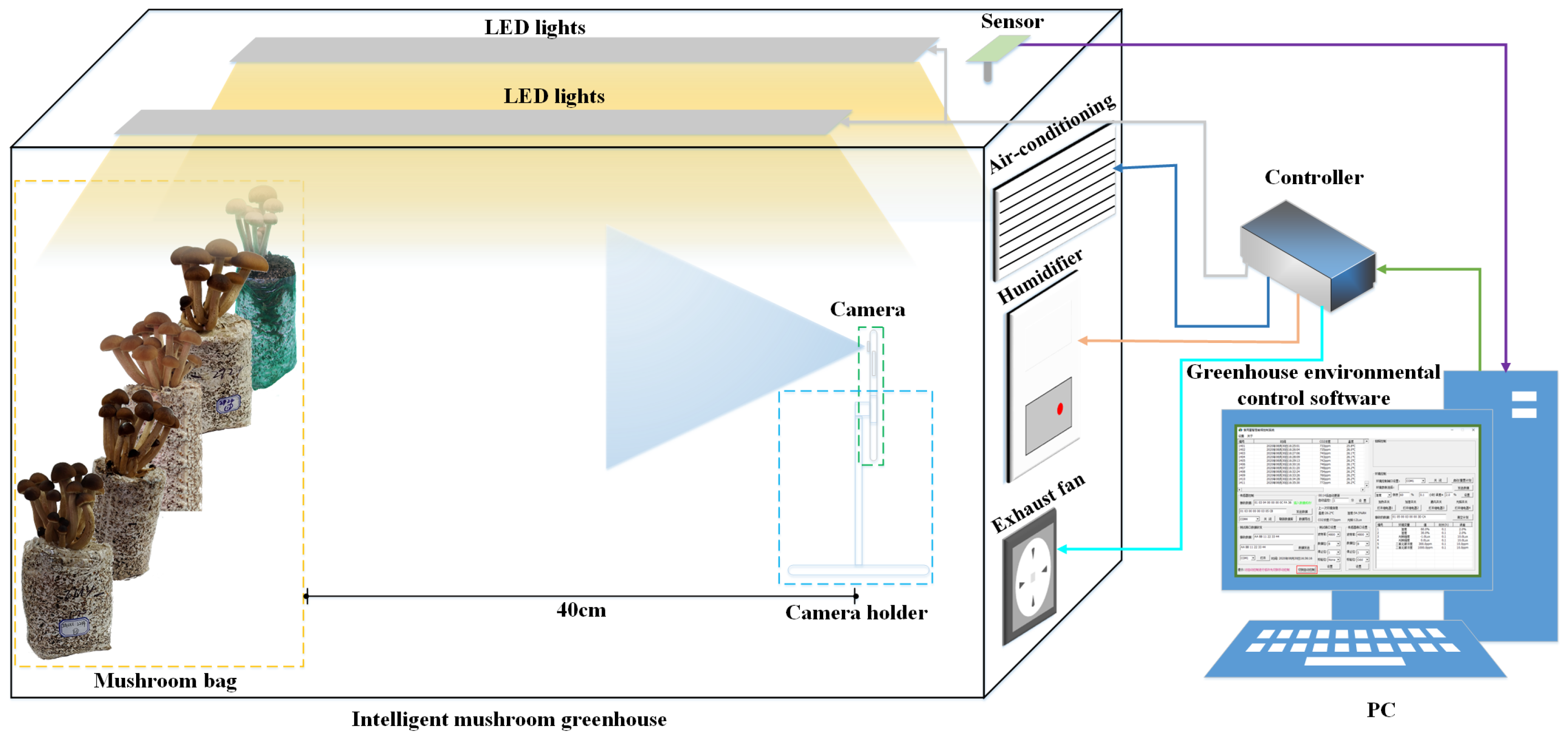
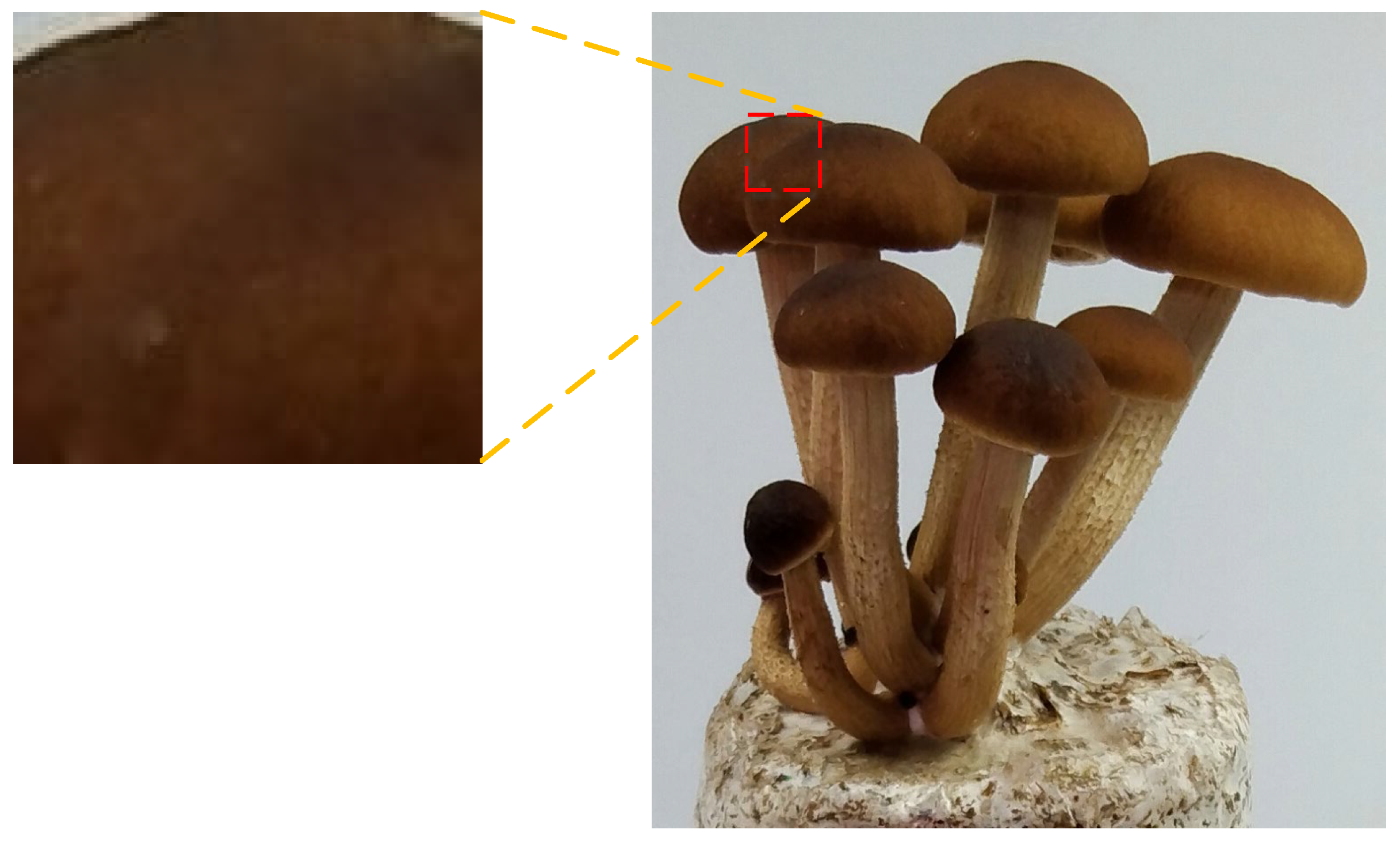
2.1. Data Acquisition
2.2. AC R-CNN Model
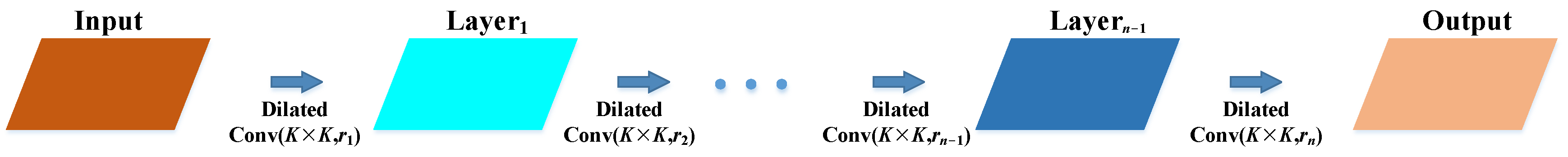
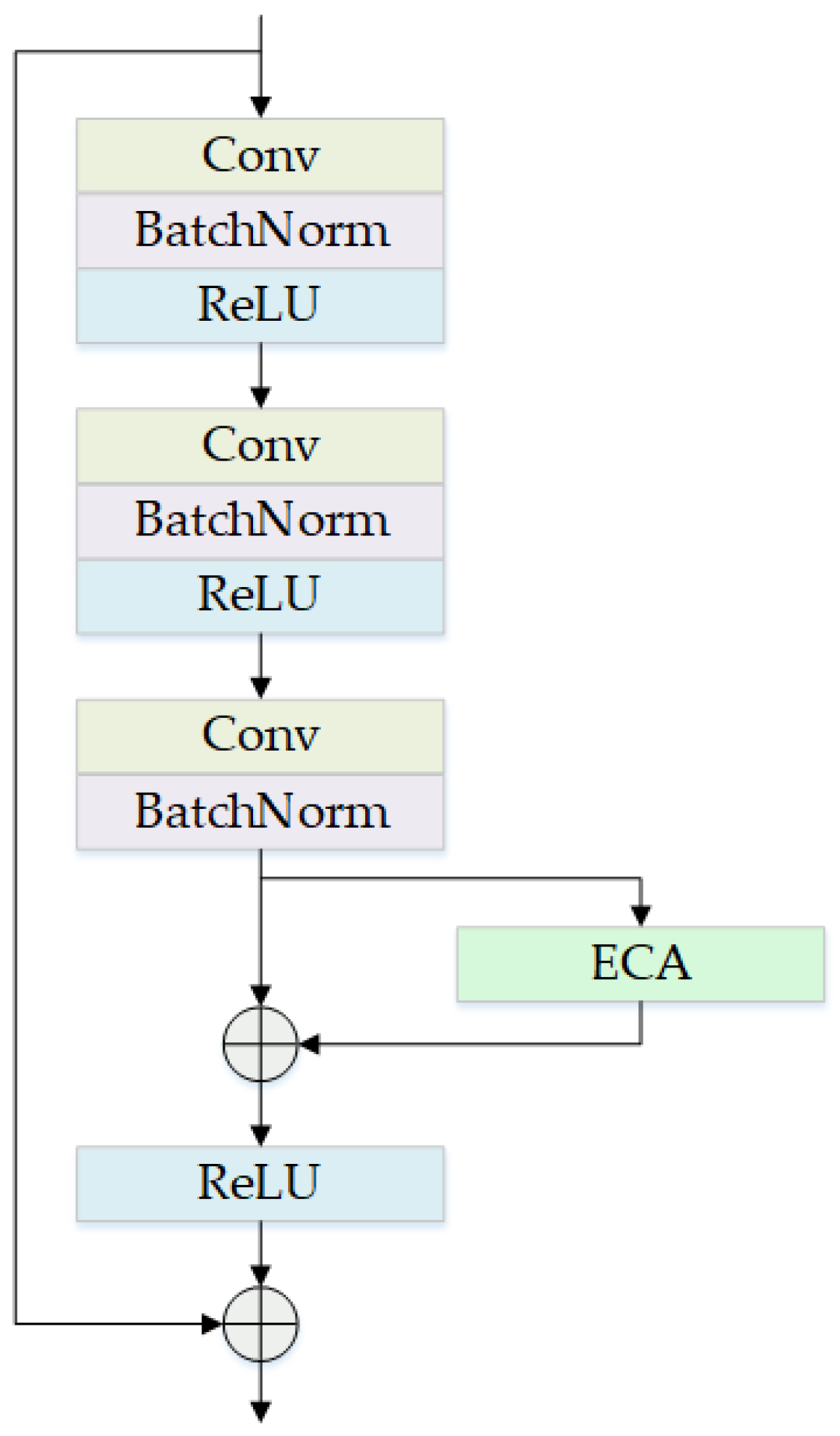
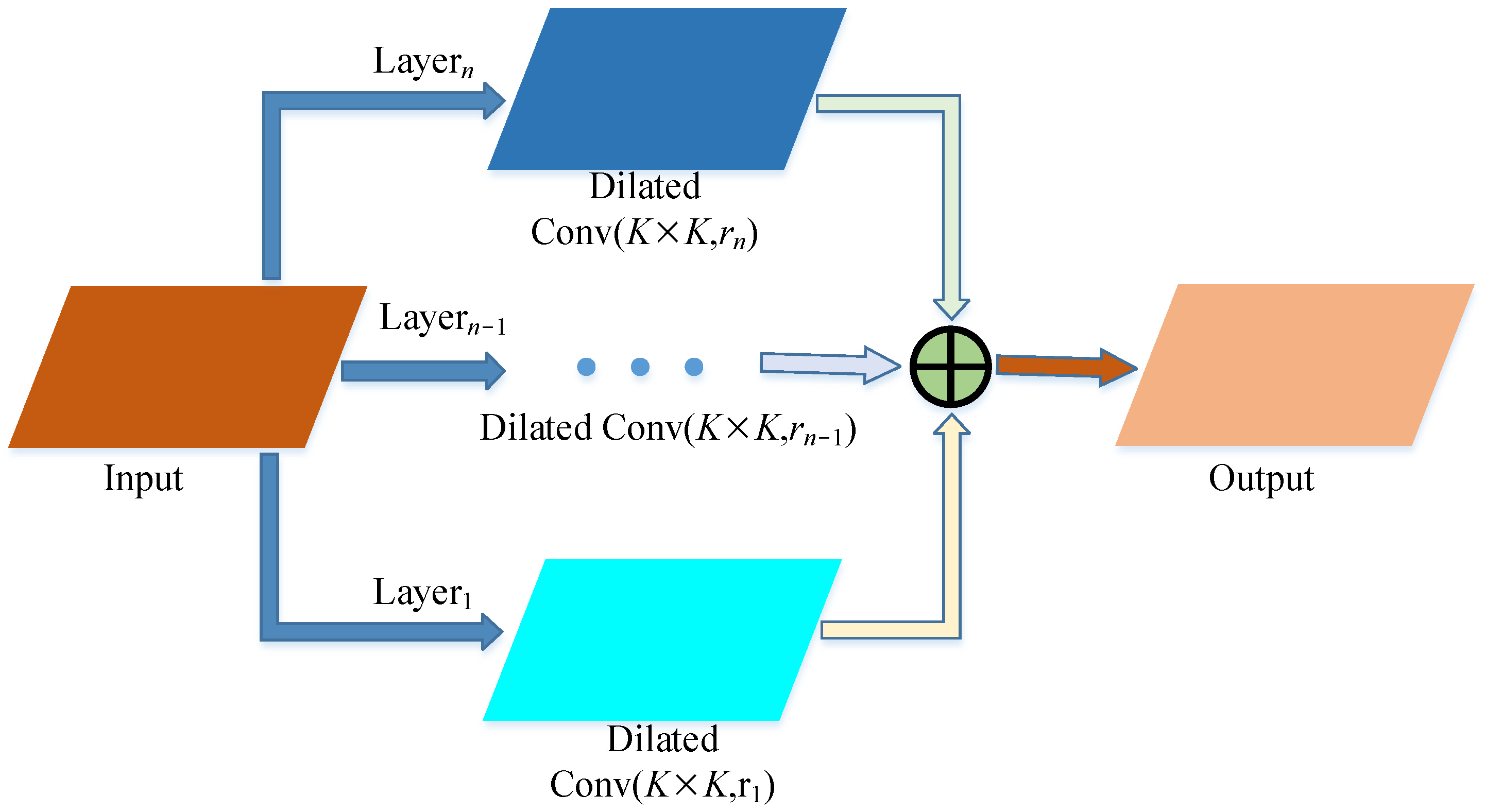
2.2.1. Backbone Improvements
- (1)
- Add expanded convolution module
- (2)
- Add attention module
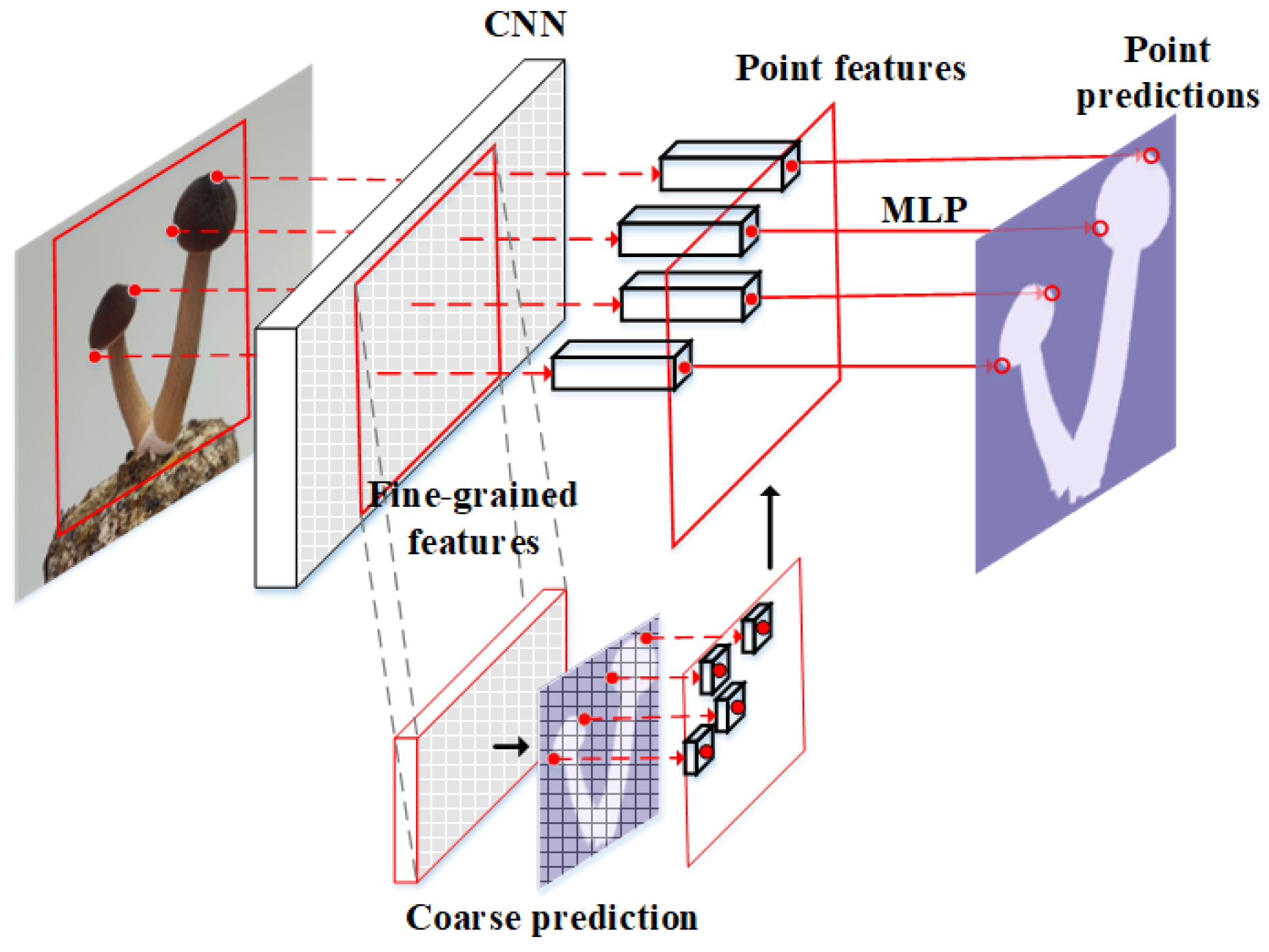
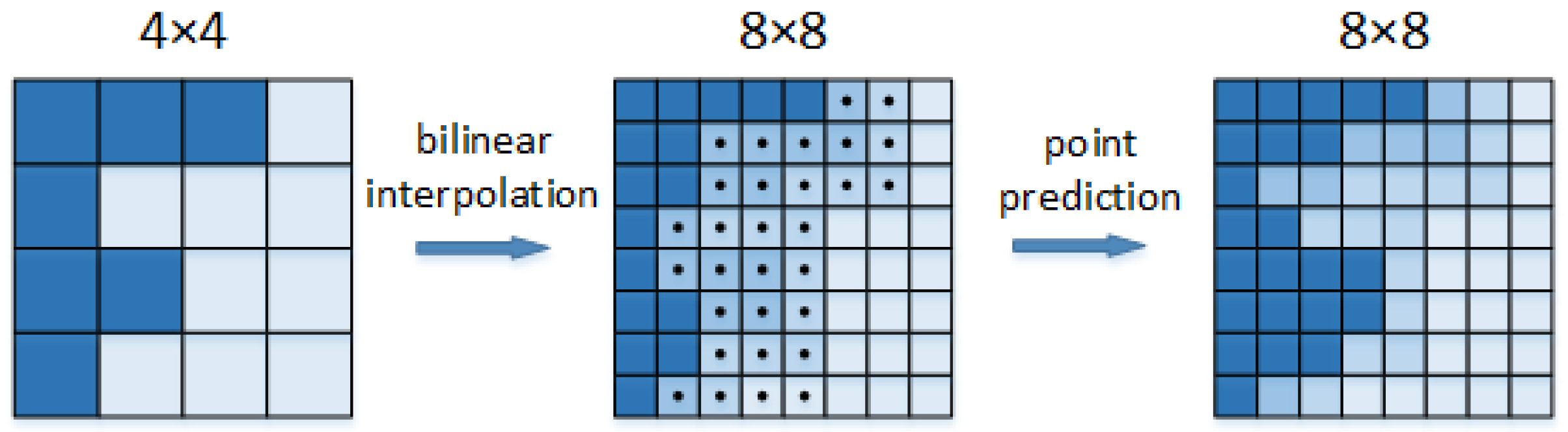
2.2.2. Add PointRend Module
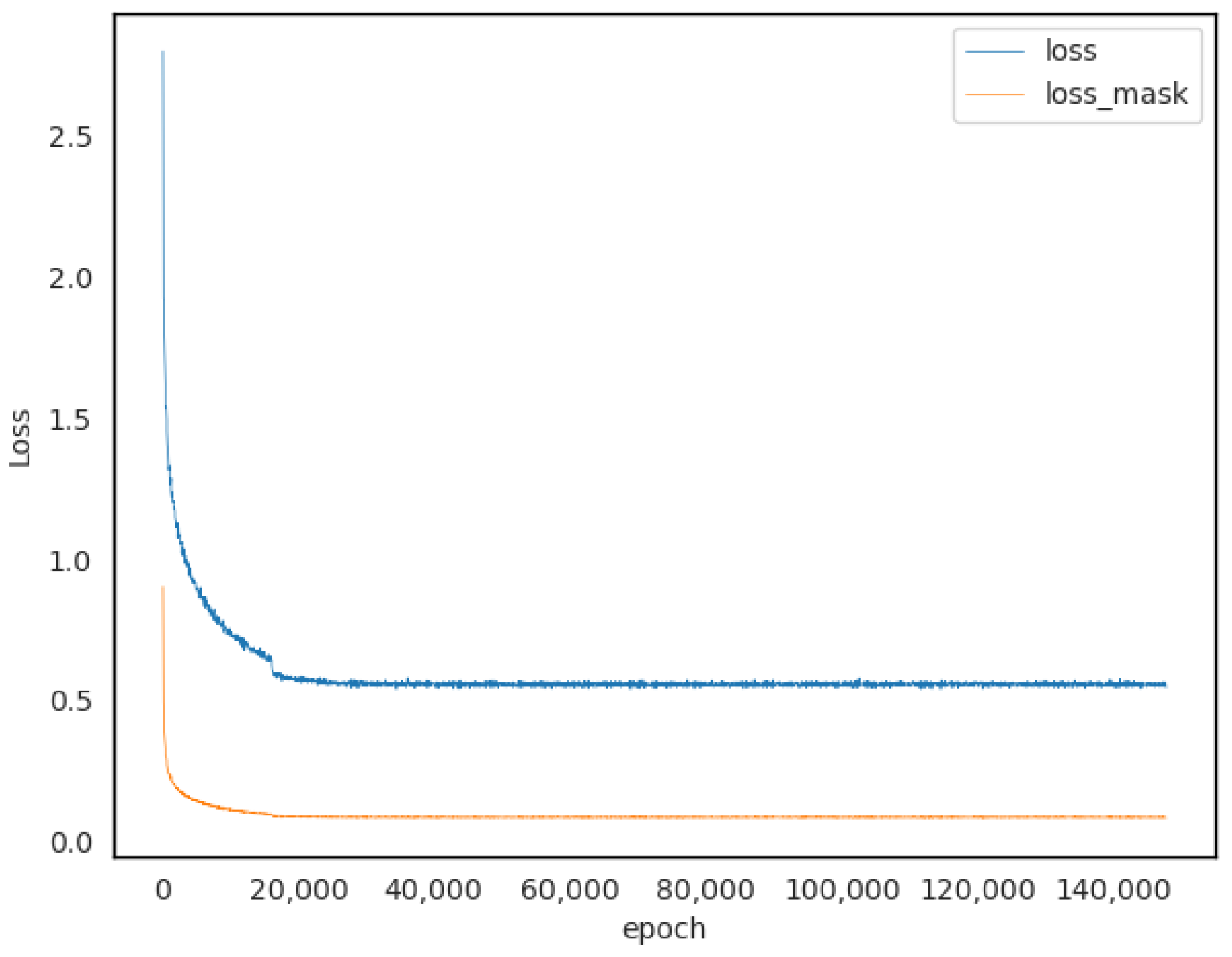
2.3. Model Training and Evaluation
3. Results
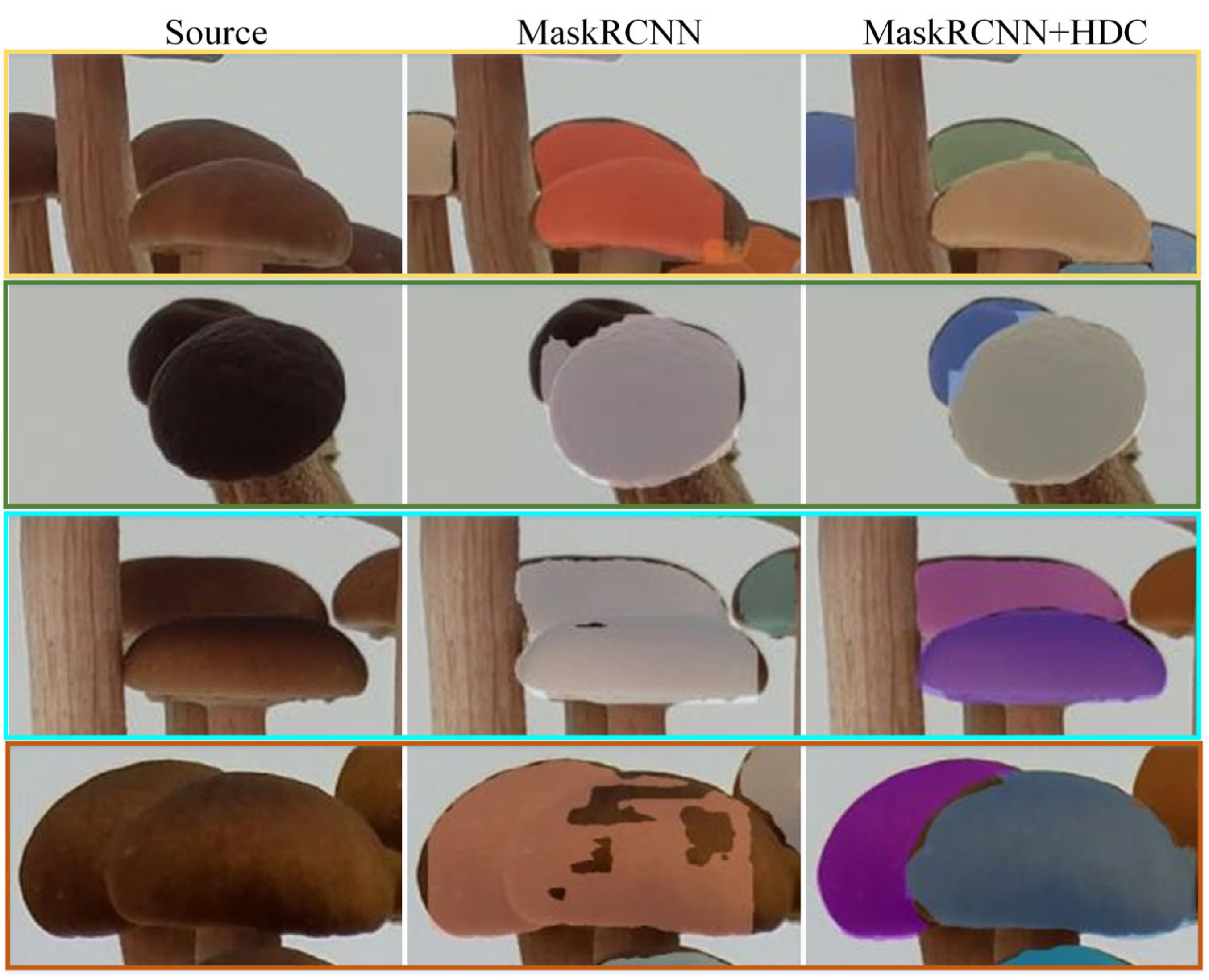
3.1. Segmentation Effect
3.2. Comparison with State of the Art
3.3. Ablation Experiment
3.3.1. HDC Module Effect
3.3.2. Attention Module Effect
3.3.3. PointRend Effect
4. Discussion
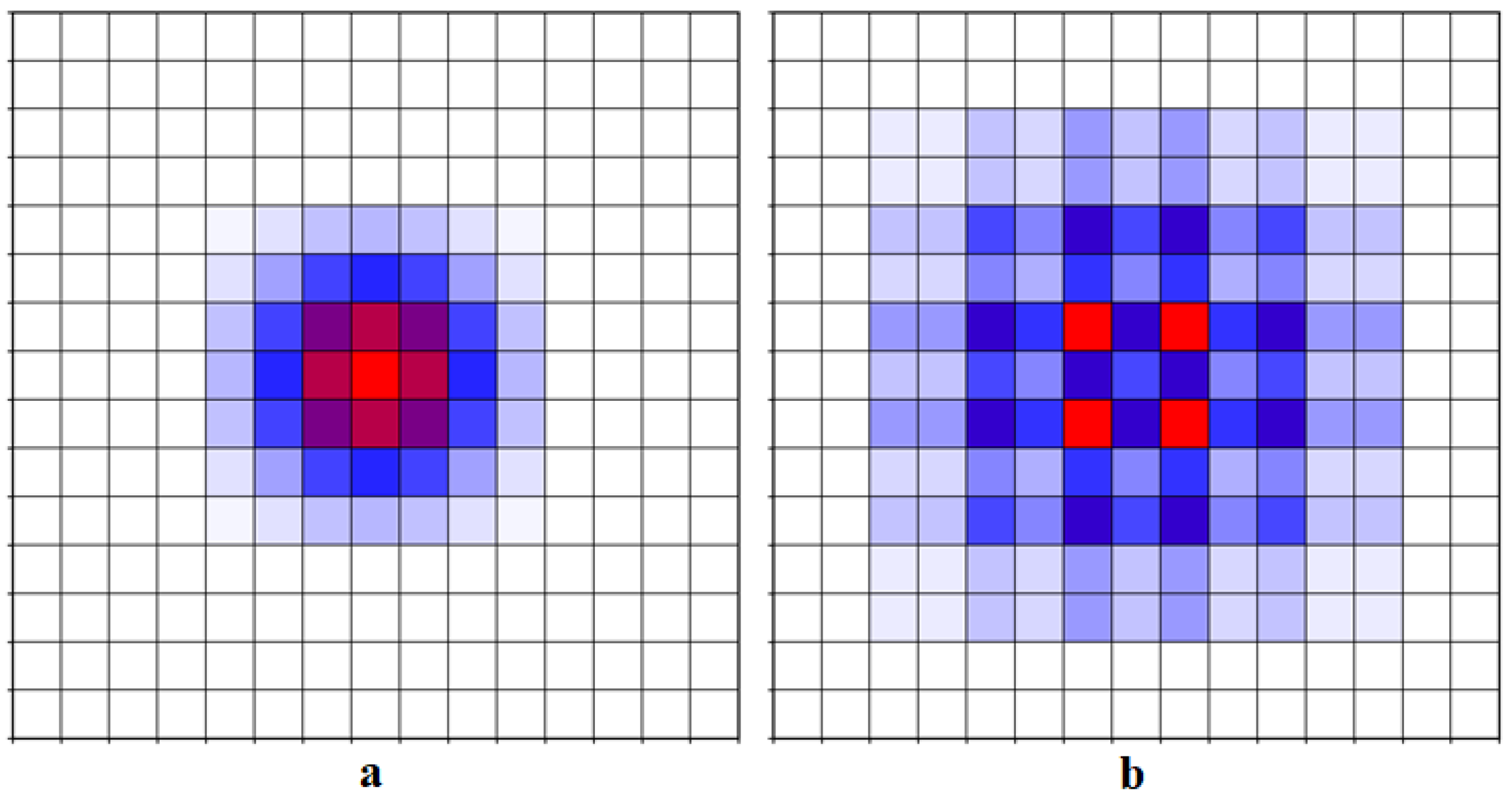
4.1. Discussion of Dilated Convolutional Layer Structure
4.2. Model Deficiencies
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Q.M.; Song, H.Y.; Chen, R.X.; Chen, M.; Zhai, Z.; Zhou, J.; Gao, Y.; Hu, D. Species concept of Cyclocybe chaxingu, an edible mushroom cultivated in China. Mycosystema 2021, 40, 981–991. [Google Scholar] [CrossRef]
- Qian, L.; Abudureheman, B.; Xue, S.; Heng, Z.; Jianlin, Z.; Rui, T. Research Progress of Agrocybe aegerita. Mod. Food 2023, 29, 42–44. [Google Scholar] [CrossRef]
- Shon, Y.H.; Nam, K.S. Antimutagenicity and induction of anticarcinogenic phase II enzymes by basidiomycetes. J. Ethnopharmacol. 2001, 77, 103–109. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, S.; Jin, Y.; Yin, Y.; Yu, G.; Lan, X.; Cui, M.; Liang, Y.; Wong, B.H.C.; Guo, L.; et al. Purification and characterization of an antitumor protein with deoxyribonuclease activity from edible mushroom Agrocybe aegerita. Mol. Nutr. Food Res. 2012, 56, 1729–1738. [Google Scholar] [CrossRef] [PubMed]
- Chien, R.C.; Tsai, S.Y.; Lai, E.Y.; Mau, J.L. Antiproliferative Activities of Hot Water Extracts from Culinary-Medicinal Mushrooms, Ganoderma tsugae and Agrocybe cylindracea (Higher Basidiomycetes) on Cancer Cells. Int. J. Med. Mushrooms 2015, 17, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Yi, W.; Hu, D. Computer vision and machine learning applied in the mushroom industry: A critical review. Comput. Electron. Agric. 2022, 198, 107015. [Google Scholar] [CrossRef]
- Lee, B.R.; Lee, Y.P.; Kim, D.W.; Song, H.Y.; Yoo, K.; Won, M.H.; Kang, T.; Lee, K.J.; Kim, K.H.; Joo, J.H.; et al. Amelioration of streptozotocin-induced diabetes by Agrocybe chaxingu polysaccharide. Mol. Cells 2010, 29, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Nath, M.; Barh, A.; Sharma, A.; Verma, P.; Bairwa, R.K.; Kamal, S.; Sharma, V.P.; Annepu, S.K.; Sharma, K.; Bhatt, D.; et al. Identification of Eight High Yielding Strains via Morpho-Molecular Characterization of Thirty-Three Wild Strains of Calocybe indica. Foods 2023, 12, 2119. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, T.; Huang, X.; Yin, J.; Nie, S. Heteroglycans from the fruiting bodies of Agrocybe cylindracea: Fractionation, physicochemical properties and structural characterization. Food Hydrocolloid 2021, 114, 106568. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, X.; Mu, X.; Hu, M.; Wang, J.; Huang, X.; Nie, S. Protective effects of flavonoids isolated from Agrocybe aegirita on dextran sodium sulfate-induced colitis. Efood 2021, 2, 288–295. [Google Scholar] [CrossRef]
- Zhu, H. Breeding of Excellent Strains of Cyclocybe cylindracea and Their High-yielding Cultivation. Master Type. Ph.D. Thesis, Jiangxi Agricultural University, Nanchang, China, 2022. [Google Scholar]
- Fang, Y.; Yang, S.; Wang, X.; Li, Y.; Fang, C.; Shan, Y.; Feng, B.; Liu, W. Instances as Queries. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, BC, Canada, 11–17 October 2021; pp. 6910–6919. [Google Scholar]
- Huang, Z.; Huang, L.; Gong, Y.; Huang, C.; Wang, X. Mask Scoring R-CNN. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 6409–6418. [Google Scholar]
- Bolya, D.; Zhou, C.; Xiao, F.; Lee, Y.J. YOLACT: Real-Time Instance Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9157–9166. [Google Scholar] [CrossRef]
- Chen, H.; Sun, K.; Tian, Z.; Shen, C.; Huang, Y.; Yan, Y. BlendMask: Top-Down Meets Bottom-Up for Instance Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2020; pp. 8573–8581. [Google Scholar]
- Chen, K.; Pang, J.; Wang, J.; Xiong, Y.; Li, X.; Sun, S.; Feng, W.; Liu, Z.; Shi, J.; Ouyang, W.; et al. Hybrid Task Cascade for Instance Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 4974–4983. [Google Scholar]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Girshick, R. Fast R-CNN. In Proceedings of the IEEE Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Liu, X.; Zhao, D.; Jia, W.; Ji, W.; Ruan, C.; Sun, Y. Cucumber Fruits Detection in Greenhouses Based on Instance Segmentation. IEEE Access 2019, 7, 139635–139642. [Google Scholar] [CrossRef]
- Pérez-Borrero, I.; Marín-Santos, D.; Gegúndez-Arias, M.E.; Cortés-Ancos, E. A fast and accurate deep learning method for strawberry instance segmentation. Comput. Electron. Agric. 2020, 178, 105736. [Google Scholar] [CrossRef]
- Zu, L.; Zhao, Y.; Liu, J.; Su, F.; Zhang, Y.; Liu, P. Detection and Segmentation of Mature Green Tomatoes Based on Mask R-CNN with Automatic Image Acquisition Approach. Sensors 2021, 21, 7842. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; He, D. Fusion of Mask RCNN and attention mechanism for instance segmentation of apples under complex background. Comput. Electron. Agric. 2022, 196, 106864. [Google Scholar] [CrossRef]
- Xu, P.; Fang, N.; Liu, N.; Lin, F.; Yang, S.; Ning, J. Visual recognition of cherry tomatoes in plant factory based on improved deep instance segmentation. Comput. Electron. Agric. 2022, 197, 106991. [Google Scholar] [CrossRef]
- Cong, P.; Li, S.; Zhou, J.; Lv, K.; Feng, H. Research on Instance Segmentation Algorithm of Greenhouse Sweet Pepper Detection Based on Improved Mask RCNN. Agronomy 2023, 13, 196. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, C.; Sun, X.; Fu, L. A Refined Apple Binocular Positioning Method with Segmentation-Based Deep Learning for Robotic Picking. Agronomy 2023, 13, 1469. [Google Scholar] [CrossRef]
- Wang, C.; Yang, G.; Huang, Y.; Liu, Y.; Zhang, Y. A transformer-based mask R-CNN for tomato detection and segmentation. J. Intell. Fuzzy Syst. 2023, 44, 8585–8595. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Xu, D.; Zhang, J.; Wen, J. An Improved Mask RCNN Model for Segmentation of ‘Kyoho’ (Vitis labruscana) Grape Bunch and Detection of Its Maturity Level. Agriculture 2023, 13, 914. [Google Scholar] [CrossRef]
- López-Barrios, J.D.; Escobedo Cabello, J.A.; Gómez-Espinosa, A.; Montoya-Cavero, L. Green Sweet Pepper Fruit and Peduncle Detection Using Mask R-CNN in Greenhouses. Appl. Sci. 2023, 13, 6296. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Jia, M.; Li, J.; Hu, T.; Luo, J. Instance Segmentation and Number Counting of Grape Berry Images Based on Deep Learning. Appl. Sci. 2023, 13, 6751. [Google Scholar] [CrossRef]
- Yeh, J.; Lin, K.; Lin, C.; Kang, J. Intelligent Mango Fruit Grade Classification Using AlexNet-SPP With Mask R-CNN-Based Segmentation Algorithm. IEEE Trans. Agrifood Electron. 2023, 1, 41–49. [Google Scholar] [CrossRef]
- Mu, X.; He, L.; Heinemann, P.; Schupp, J.; Karkee, M. Mask R-CNN based apple flower detection and king flower identification for precision pollination. Smart Agric. Technol. 2023, 4, 100151. [Google Scholar] [CrossRef]
- Shen, R.; Zhen, T.; Li, Z. Segmentation of Unsound Wheat Kernels Based on Improved Mask RCNN. Sensors 2023, 23, 3379. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Wang, P.; Chen, P.; Yuan, Y.; Liu, D.; Huang, Z.; Hou, X.; Cottrell, G. Understanding Convolution for Semantic Segmentation. In Proceedings of the WACV 2018: IEEE Winter Conference on Applications of Computer Vision, Lake Tahoe, CA, USA, 12–15 March 2018; pp. 1451–1460. [Google Scholar]
- Sanghyun, W.; Park, J.; Joon-Young, L.; So, K.I. CBAM: Convolutional Block Attention Module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Hou, Q.; Zhou, D.; Feng, J. Coordinate Attention for Efficient Mobile Network Design. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 13713–13722. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient Channel Attention for Deep Convolutional Neural Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 11534–11542. [Google Scholar]





| Dataset | Number of Caps in Dataset | ||
|---|---|---|---|
| Occluded Caps | Unoccluded Caps | Total | |
| Training set | 2165 | 1202 | 3367 |
| Test set | 327 | 191 | 518 |
| Method | AP50 | AP75 | F1 | Run Time (s) |
|---|---|---|---|---|
| Mask R-CNN | 0.762 | 0.585 | 0.749 | 1.578 |
| Mask Scoring R-CNN | 0.763 | 0.605 | 0.751 | 1.483 |
| YOLACT | 0.748 | 0.382 | 0.757 | 1.518 |
| InstaBoost | 0.710 | 0.517 | 0.684 | 1.743 |
| QueryInst | 0.735 | 0.585 | 0.855 | 1.732 |
| BlendMask | 0.742 | 0.452 | 0.762 | 1.485 |
| AC R-CNN (Ours) | 0.883 | 0.781 | 0.886 | 1.505 |
| PointRend | HDC | ECA | AP50 | AP75 |
|---|---|---|---|---|
| – | – | – | 0.762 | 0.585 |
| √ | – | – | 0.818 | 0.649 |
| √ | √ | – | 0.852 | 0.727 |
| √ | √ | √ | 0.883 | 0.781 |
| Attention Module | AP50 | AP75 | F1 |
|---|---|---|---|
| CBAM | 0.746 | 0.527 | 0.782 |
| SE | 0.778 | 0.631 | 0.805 |
| CA | 0.769 | 0.627 | 0.787 |
| ECA | 0.793 | 0.703 | 0.811 |
| Expansion Coefficients | Method 1-AP50 | Method 2-AP50 |
|---|---|---|
| 1-2-2 | 0.811 | 0.803 |
| 1-2-5 | 0.771 | 0.783 |
| 1-2-2-1 | 0.809 | 0.805 |
| 1-2-2-1-2-2 | 0.806 | 0.796 |
| 1-2-5-1-2-5 | 0.782 | 0.792 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, H.; Yang, S.; Cheng, W.; Wei, Q.; Wang, Y.; Xu, Y. AC R-CNN: Pixelwise Instance Segmentation Model for Agrocybe cylindracea Cap. Agronomy 2024, 14, 77. https://doi.org/10.3390/agronomy14010077
Yin H, Yang S, Cheng W, Wei Q, Wang Y, Xu Y. AC R-CNN: Pixelwise Instance Segmentation Model for Agrocybe cylindracea Cap. Agronomy. 2024; 14(1):77. https://doi.org/10.3390/agronomy14010077
Chicago/Turabian StyleYin, Hua, Shenglan Yang, Wenhao Cheng, Quan Wei, Yinglong Wang, and Yilu Xu. 2024. "AC R-CNN: Pixelwise Instance Segmentation Model for Agrocybe cylindracea Cap" Agronomy 14, no. 1: 77. https://doi.org/10.3390/agronomy14010077
APA StyleYin, H., Yang, S., Cheng, W., Wei, Q., Wang, Y., & Xu, Y. (2024). AC R-CNN: Pixelwise Instance Segmentation Model for Agrocybe cylindracea Cap. Agronomy, 14(1), 77. https://doi.org/10.3390/agronomy14010077






