Mapping QTL for Yield and Its Component Traits Using Wheat (Triticum aestivum L.) RIL Mapping Population from TAM 113 × Gallagher
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Field Trials, and Phenotypic Data Collection
2.2. Statistical Analyses
2.3. Genotyping, Linkage Mapping, and QTL Analysis
3. Results
3.1. Phenotypic Evaluation
3.2. Linkage Analysis and QTL Identification
3.2.1. QTL for Yield
3.2.2. QTL for Yield Components and Kernel Traits
3.2.3. QTL for Agronomic Traits
3.3. Pleiotropic QTL
3.4. Interactions of Epistasis and Epistasis-by-Environment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | analysis of variance |
| BM | biomass |
| BMYLD | biomass grain yield |
| BLUE | best linear unbiased estimate |
| BLUP | best linear unbiased predictors |
| CTAB | cetyltrimethyl ammonium bromide |
| DIAM | kernel diameter |
| GbyE | genotype by environment interaction |
| PH | plant height |
| SPW | pike weight including glume |
| HI | harvest index; |
| ICIM-ADD | inclusive composite interval mapping of additive QTL |
| IWGSC | International Wheat Genome Sequencing Consortium |
| KAREA | kernel area |
| KHI | kernel hardness index |
| KLEN | kernel length |
| KPERI | kernel perimeter |
| KPS | kernels per spike |
| KWID | kernel width |
| LOD | logarithm of odds |
| MAS | marker-assistant selection |
| PH | plant height |
| PVE | phenotypic variation explained |
| QTL | quantitative trait loci |
| REML | restricted maximum likelihood method |
| RIL | recombinant inbred line |
| SHDW | single head dry weight |
| SHGW | single head dry grain weight |
| SKW | single kernel weight |
| SPM | spikes per meter |
| SSW | single stem weight |
| TKW | thousand kernel weight |
| YLD | grain yield |
References
- Food and Agriculture Organization of the United Nations (FAO). 2017. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 20 June 2021).
- United Nations (UN). 2017 NATIONS. Available online: https://www.un.org/en/global-issues/population (accessed on 20 June 2021).
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; McMaster, G.S.; Yu, Q. Spatial patterns of relationship between wheat yield and yield components in China. Int. J. Plant Prod. 2018, 12, 61–71. [Google Scholar] [CrossRef]
- Liu, K.; Sun, X.; Ning, T.; Duan, X.; Wang, Q.; Liu, T.; An, Y.; Guan, X.; Tian, J.; Chen, J. Genetic dissection of wheat panicle traits using linkage analysis and a genome-wide association study. Theor. Appl. Genet. 2018, 131, 1073–1090. [Google Scholar] [CrossRef] [PubMed]
- Balcha, A. Genotype by environment interaction for grain yield and association among stability parameters in bread wheat (Triticum aestivum L.). Am. J. Plant. Sci. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Dagnaw, T.; Mulugeta, B.; Haileselassie, T.; Geleta, M.; Tesfaye, K. Phenotypic variability, heritability and associations of agronomic and quality traits in cultivated ethiopian durum wheat (Triticum turgidum L. ssp. Durum, Desf.). Agronomy 2022, 12, 1714. [Google Scholar] [CrossRef]
- Piepho, H.-P.; Möhring, J. Computing heritability and selection response from unbalanced plant breeding trials. Genetics 2007, 177, 1881–1888. [Google Scholar] [CrossRef]
- Nyquist, W.E.; Baker, R.J. Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 1991, 10, 235–322. [Google Scholar] [CrossRef]
- Yazici, E.; Bilgin, O. Heritability estimates for milling quality associations of bread wheat in the Northwest Turkey. Int. J. Agron. 2019, 2, 17–22. [Google Scholar] [CrossRef]
- Dogan, M.; Wang, Z.; Cerit, M.; Valenzuela-Antelo, J.L.; Dhakal, S.; Chu, C.; Xue, Q.; Ibrahim, A.M.H.; Rudd, J.C.; Bernardo, A.; et al. QTL analysis of yield and end-use quality traits in Texas hard red winter wheat. Agronomy 2023, 13, 689. [Google Scholar] [CrossRef]
- Jafarzadeh, J.; Bonnett, D.; Jannink, J.L.; Akdemir, D.; Dreisigacker, S.; Sorrells, M.E. Breeding value of primary synthetic wheat genotypes for grain yield. PLoS ONE 2016, 11, e0162860. [Google Scholar] [CrossRef]
- Tura, H.; Edwards, J.; Gahlaut, V.; Garcia, M.; Sznajder, B.; Baumann, U.; Shahinnia, F.; Reynolds, M.; Langridge, P.; Balyan, H.S.; et al. QTL analysis and fine mapping of a QTL for yield-related traits in wheat grown in dry and hot environments. Theor. Appl. Genet. 2020, 133, 239–257. [Google Scholar] [CrossRef]
- Xin, F.; Zhu, T.; Wei, S.; Han, Y.; Zhao, Y.; Zhang, D.; Ma, L.; Ding, Q. QTL mapping of kernel traits and validation of a major QTL for kernel length-width ratio using snp and bulked segregant analysis in wheat. Sci. Rep. 2020, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Collaku, A.; Harrison, S. Heritability of waterlogging tolerance in wheat. Crop Sci.-CROP SCI 2005, 45, 722–727. [Google Scholar] [CrossRef]
- Ma, D.; Yan, J.; He, Z.; Wu, L.; Xia, X. Characterization of a cell wall invertase gene TaCwi-A1 on common wheat chromosome 2A and development of functional markers. Mol. Breed. 2012, 29, 43–52. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Hu, Y.-G.; Chen, J. Dwarfing genes Rht4 and Rht-B1b affect plant height and key agronomic traits in common wheat under two water regimes. Field Crops Res. 2017, 204, 242–248. [Google Scholar] [CrossRef]
- Gill, B.K.; Klindworth, D.L.; Rouse, M.N.; Zhang, J.; Zhang, Q.; Sharma, J.S.; Chu, C.; Long, Y.; Chao, S.; Olivera, P.D.; et al. Function and evolution of allelic variations of Sr13 conferring resistance to stem rust in tetraploid wheat (Triticum turgidum L.). Plant J. 2021, 106, 1674–1691. [Google Scholar] [CrossRef]
- Loukoianov, A.; Yan, L.; Blechl, A.; Sanchez, A.; Dubcovsky, J. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol. 2005, 138, 2364–2373. [Google Scholar] [CrossRef]
- Whittal, A.; Kaviani, M.; Graf, R.; Humphreys, G.; Navabi, A. Allelic variation of vernalization and photoperiod response genes in a diverse set of North American high latitude winter wheat genotypes. PLoS ONE 2018, 13, e0203068. [Google Scholar] [CrossRef] [PubMed]
- Royo, C.; Ammar, K.; Alfaro, C.; Dreisigacker, S.; del Moral, L.F.G.; Villegas, D. Effect of Ppd-1 photoperiod sensitivity genes on dry matter production and allocation in durum wheat. Field Crops Res. 2018, 221, 358–367. [Google Scholar] [CrossRef]
- Boden, S.A.; Cavanagh, C.; Cullis, B.R.; Ramm, K.; Greenwood, J.; Jean Finnegan, E.; Trevaskis, B.; Swain, S.M. Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat. Plants 2015, 1, 14016. [Google Scholar] [CrossRef]
- Grogan, S.M.; Brown-Guedira, G.; Haley, S.D.; McMaster, G.S.; Reid, S.D.; Smith, J.; Byrne, P.F. Allelic variation in developmental genes and effects on winter wheat heading date in the U.S. great plains. PLoS ONE 2016, 11, e0152852. [Google Scholar] [CrossRef]
- Jaganathan, D.; Bohra, A.; Thudi, M.; Varshney, R.K. Fine mapping and gene cloning in the post-NGS era: Advances and prospects. Theor. Appl. Genet. 2020, 133, 1791–1810. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Varshney, R.K.; Singh, V.K.; Coyne, C.J.; Domoney, C.; Kejnovský, E.; Warkentin, T. From Mendel’s discovery on pea to today’s plant genetics and breeding. Theor. Appl. Genet. 2016, 129, 2267–2280. [Google Scholar] [CrossRef]
- Gao, F.; Wen, W.; Liu, J.; Rasheed, A.; Yin, G.; Xia, X.; Wu, X.; He, Z. Genome-Wide Linkage Mapping of QTL for Yield Components, Plant Height and Yield-Related Physiological Traits in the Chinese Wheat Cross Zhou 8425B/Chinese Spring. Front. Plant Sci. 2015, 6, 1099. [Google Scholar] [CrossRef]
- Guan, P.; Lu, L.; Jia, L.; Kabir, M.R.; Zhang, J.; Lan, T.; Zhao, Y.; Xin, M.; Hu, Z.; Yao, Y.; et al. Global QTL analysis identifies genomic regions on chromosomes 4a and 4b harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 529. [Google Scholar] [CrossRef]
- Wang, S.-X.; Zhu, Y.-L.; Zhang, D.-X.; Shao, H.; Liu, P.; Hu, J.-B.; Zhang, H.; Zhang, H.-P.; Chang, C.; Lu, J.; et al. Genome-wide association study for grain yield and related traits in elite wheat varieties and advanced lines using SNP markers. PLoS ONE 2017, 12, e0188662. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Liu, X.; Chu, C.; Yang, Y.; Rudd, J.C.; Ibrahim, A.M.H.; Xue, Q.; Devkota, R.N.; Baker, J.A.; Baker, S.A.; et al. Genome-wide QTL mapping of yield and agronomic traits in two widely adapted winter wheat cultivars from multiple mega-environments. PeerJ 2021, 9, e12350. [Google Scholar] [CrossRef]
- Yang, Y.; Dhakal, S.; Chu, C.; Wang, S.; Xue, Q.; Rudd, J.C.; Ibrahim, A.M.H.; Jessup, K.; Baker, J.; Fuentealba, M.P.; et al. Genome wide identification of QTL associated with yield and yield components in two popular wheat cultivars TAM 111 and TAM 112. PLoS ONE 2020, 15, e0237293. [Google Scholar] [CrossRef] [PubMed]
- Rudd, J.C.; Devkota, R.N.; Baker, J.A.; Ibrahim, A.M.; Worrall, D.; Lazar, M.D.; Sutton, R.; Rooney, L.W.; Nelson, L.R.; Bean, B.; et al. Registration of ‘TAM 113’ Wheat. J. Plant Reg. 2013, 7, 63–68. [Google Scholar] [CrossRef]
- Marburger, D.A.; Silva, A.d.O.; Hunger, R.M.; Edwards, J.T.; Van der Laan, L.; Blakey, A.M.; Kan, C.C.; Garland-Campbell, K.A.; Bowden, R.L.; Yan, L.; et al. ‘Gallagher’ and ‘Iba’ hard red winter wheat: Half-sibs inseparable by yield gain, separable by producer preference. J. Plant Regist. 2021, 15, 177–195. [Google Scholar] [CrossRef]
- Wang, Z.; Dhakal, S.; Cerit, M.; Wang, S.; Rauf, Y.; Yu, S.; Maulana, F.; Huang, W.; Anderson, J.D.; Ma, X.F.; et al. QTL mapping of yield components and kernel traits in wheat cultivars TAM 112 and Duster. Front. Plant Sci. 2022, 13, 1057701. [Google Scholar] [CrossRef]
- Whan, A.P.; Smith, A.B.; Cavanagh, C.R.; Ral, J.-P.F.; Shaw, L.M.; Howitt, C.A.; Bischof, L. GrainScan: A low cost, fast method for grain size and colour measurements. Plant Methods 2014, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Shiff, S.; Lensky, I.M.; Bonfil, D.J. Using satellite data to optimize wheat yield and quality under climate change. Remote Sens. 2021, 13, 2049. [Google Scholar] [CrossRef]
- Sall, J.; Lehman, A. JMP Start Statistics: A Guide to Statistical and Data Analysis Using JMP and JMP in Software; Duxbury Press: London, UK, 1996. [Google Scholar]
- Alvarado, G.; Rodríguez, F.M.; Pacheco, A.; Burgueño, J.; Crossa, J.; Vargas, M.; Pérez-Rodríguez, P.; Lopez-Cruz, M.A. META-R: A software to analyze data from multi-environment plant breeding trials. Crop J. 2020, 8, 745–756. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Doyle, J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. [Google Scholar]
- IWGSC, I.W.G.S.C. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345. [Google Scholar] [CrossRef]
- Dhakal, S.; Liu, X.; Girard, A.; Chu, C.; Yang, Y.; Wang, S.; Xue, Q.; Rudd, J.C.; Ibrahim, A.M.H.; Awika, J.M.; et al. Genetic dissection of end-use quality traits in two widely adapted wheat cultivars ‘TAM 111’ and ‘TAM 112’. Crop Sci. 2021, 61, 1944–1959. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Van Ooijen, J. JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wageningen, The Netherlans, 2006; Volume 33. [Google Scholar]
- McIntosh, R.; Yamazaki, Y.; Dubcovsky, J.; Rogers, J.; Morris, C.; Appels, R. Catalogue of gene symbols for wheat. In Proceedings of the 12th International Wheat Genetics Symposium, Okohama, Japan, 7–14 September 2013; pp. 8–13. [Google Scholar]
- Isham, K.; Wang, R.; Zhao, W.; Wheeler, J.; Klassen, N.; Akhunov, E.; Chen, J. QTL mapping for grain yield and three yield components in a population derived from two high-yielding spring wheat cultivars. Theor. Appl. Genet. 2021, 134, 2079–2095. [Google Scholar] [CrossRef]
- Piepho, H.P.; Möhring, J.; Melchinger, A.E.; Büchse, A. BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 2008, 161, 209–228. [Google Scholar] [CrossRef]
- Öztürk, İ. Genotypes by Environment Interaction of Bread Wheat (Triticum aestivum L.) Genotypes on Yield and Quality Parameters under Rainfed Conditions. Int. J. Appl. Agric. Res. 2022, 6, 27–40. [Google Scholar] [CrossRef]
- Huang, W.; Carbone, M.A.; Lyman, R.F.; Anholt, R.R.H.; Mackay, T.F.C. Genotype by environment interaction for gene expression in Drosophila melanogaster. Nat. Commun. 2020, 11, 5451. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, E.D.; Ferreira, V.A.; Grassi, E.M.; Picca, A.M.T.; Paccapelo, H.A. Genetic parameters estimation in quantitative traits of a cross of triticale (x Triticosecale W.). Open Agric. 2018, 3, 25–31. [Google Scholar] [CrossRef]
- Garcia, M.; Eckermann, P.; Haefele, S.; Satija, S.; Sznajder, B.; Timmins, A.; Baumann, U.; Wolters, P.; Mather, D.E.; Fleury, D. Genome-wide association mapping of grain yield in a diverse collection of spring wheat (Triticum aestivum L.) evaluated in southern Australia. PLoS ONE 2019, 14, e0211730. [Google Scholar] [CrossRef] [PubMed]
- Al-Tabbal, J.A.; Al-Fraihat, A.H. Heritability studies of yield and yield associated traits in wheat genotypes. J. Agric. Sci. 2012, 4, 11. [Google Scholar] [CrossRef]
- Kutlu, I.; Balkan, A.; Korkut, K.; Bilgin, O.; Başer, İ. Evaluation of reciprocal cross populations for spike-related traits in early consecutive generations of bread wheat. Genetika 2017, 49, 511–528. [Google Scholar] [CrossRef]
- Ali, M.; Zhang, Y.; Rasheed, A.; Wang, J.; Zhang, L. Genomic prediction for grain yield and yield-related traits in chinese winter wheat. Int. J. Mol. Sci. 2020, 21, 1342. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, N.; Bhandari, K. Assessment of different genotypes of wheat under late sowing condition. Heliyon 2022, 8, e08726. [Google Scholar] [CrossRef] [PubMed]
- Gonfa, A.; Tesfaye, K. Relationship between grain yield and yield components of the ethiopian durum wheat genotypes at various growth stages. Trop. Subtrop. Agroecosyst. 2016, 19, 81–91. [Google Scholar]
- Li, R.; Chen, Z.; Zheng, R.; Chen, Q.; Deng, J.; Li, H.; Huang, J.; Liang, C.; Shi, T. QTL mapping and candidate gene analysis for yield and grain weight/size in Tartary buckwheat. BMC Plant Biol. 2023, 23, 58. [Google Scholar] [CrossRef]
- Semagn, K.; Iqbal, M.; Chen, H.; Perez-Lara, E.; Bemister, D.H.; Xiang, R.; Zou, J.; Asif, M.; Kamran, A.; N’Diaye, A. Physical mapping of QTL associated with agronomic and end-use quality traits in spring wheat under conventional and organic management systems. Theor. Appl. Genet. 2021, 134, 3699–3719. [Google Scholar] [CrossRef]
- Cao, J.; Shang, Y.; Xu, D.; Xu, K.; Cheng, X.; Pan, X.; Liu, X.; Liu, M.; Gao, C.; Yan, S.; et al. Identification and validation of new stable QTLs for grain weight and size by multiple mapping models in common wheat. Front. Genet. 2020, 11, 584859. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhang, H.; Li, S.; Zou, Y.; Li, T.; Liu, J.; Ding, P.; Mu, Y.; Tang, H.; Deng, M. Identification of quantitative trait loci for kernel traits in a wheat cultivar Chuannong16. BMC Genet. 2019, 20, 77. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Liu, J.; Guo, Q.; Zhang, Y.; Dreisigacker, S.; Xia, X.; et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhao, G.; Gao, L.; Kong, X.; Guo, Z.; Wu, B.; Jia, J. functional studies of heading date-related gene TaPRR73, a paralog of ppd1 in common wheat. Front. Plant Sci. 2016, 7, 772. [Google Scholar] [CrossRef]
- Kato, K.; Yokoyama, H. Geographical variation in heading characters among wheat landraces, Triticum aestivum L., and its implication for their adaptability. Theor. Appl. Genet. 1992, 84, 259–265. [Google Scholar] [CrossRef]
- Lu, F.; Chen, M.; Zhao, Y.; Wu, S.; Yasir, M.; Zhang, H.; Hu, X.; Rong, J. Genetic mapping and candidate gene prediction of a qtl related to early heading on wild emmer chromosome 7bs in the genetic background of common wheat. Agronomy 2022, 12, 1089. [Google Scholar] [CrossRef]
- Liu, T.; Wu, L.; Gan, X.; Chen, W.; Liu, B.; Fedak, G.; Cao, W.; Chi, D.; Liu, D.; Zhang, H.; et al. Mapping quantitative trait loci for 1000-grain weight in a double haploid population of common wheat. Int. J. Mol. Sci. 2020, 21, 3960. [Google Scholar] [CrossRef]
- Qin, L.; Hao, C.; Hou, J.; Wang, Y.; Li, T.; Wang, L.; Ma, Z.; Zhang, X. Homologous haplotypes, expression, genetic effects and geographic distribution of the wheat yield gene TaGW2. BMC Plant Biol. 2014, 14, 107. [Google Scholar] [CrossRef]
- Jaiswal, V.; Gahlaut, V.; Mathur, S.; Agarwal, P.; Khandelwal, M.K.; Khurana, J.P.; Tyagi, A.K.; Balyan, H.S.; Gupta, P.K. Identification of novel snp in promoter sequence of TaGW2-6A associated with grain weight and other agronomic traits in wheat (Triticum aestivum L.). PLoS ONE 2015, 10, e0129400. [Google Scholar] [CrossRef]
- Yu, S.; Assanga, S.O.; Awika, J.M.; Ibrahim, A.M.H.; Rudd, J.C.; Xue, Q.; Guttieri, M.J.; Zhang, G.; Baker, J.A.; Jessup, K.E.; et al. Genetic mapping of quantitative trait loci for end-use quality and grain minerals in hard red winter wheat. Agronomy 2021, 11, 2519. [Google Scholar] [CrossRef]
- Cao, S.; Xu, D.; Hanif, M.; Xia, X.; He, Z. Genetic architecture underpinning yield component traits in wheat. Theor. Appl. Genet. 2020, 133, 1811–1823. [Google Scholar] [CrossRef] [PubMed]
- Vitale, P.; Fania, F.; Esposito, S.; Pecorella, I.; Pecchioni, N.; Palombieri, S.; Sestili, F.; Lafiandra, D.; Taranto, F.; De Vita, P. QTL analysis of five morpho-physiological traits in bread wheat using two mapping populations derived from common parents. Genes 2021, 12, 604. [Google Scholar] [CrossRef] [PubMed]
- Basavaraddi, P.A.; Savin, R.; Wingen, L.U.; Bencivenga, S.; Przewieslik-Allen, A.M.; Griffiths, S.; Slafer, G.A. Interactions between two QTLs for time to anthesis on spike development and fertility in wheat. Sci. Rep. 2021, 11, 2451. [Google Scholar] [CrossRef]
- Rudd, J.C.; Devkota, R.N.; Baker, J.A.; Peterson, G.L.; Lazar, M.D.; Bean, B.; Worrall, D.; Baughman, T.; Marshall, D.; Sutton, R.; et al. ‘TAM 112’ wheat, resistant to greenbug and wheat curl mite and adapted to the dryland production system in the southern high plains. J. Plant Regist. 2014, 8, 291–297. [Google Scholar] [CrossRef]
- Lv, H.; Wang, Q.; Liu, X.; Han, F.; Fang, Z.; Yang, L.; Zhuang, M.; Liu, Y.; Li, Z.; Zhang, Y. Whole-genome mapping reveals novel qtl clusters associated with main agronomic traits of cabbage (Brassica oleracea var. capitata L.). Front. Plant Sci. 2016, 7, 989. [Google Scholar] [CrossRef] [PubMed]
- Hai, L.; Guo, H.; Wagner, C.; Xiao, S.; Friedt, W. Genomic regions for yield and yield parameters in Chinese winter wheat (Triticum aestivum L.) genotypes tested under varying environments correspond to QTL in widely different wheat materials. Plant. Sci. 2008, 175, 226–232. [Google Scholar] [CrossRef]
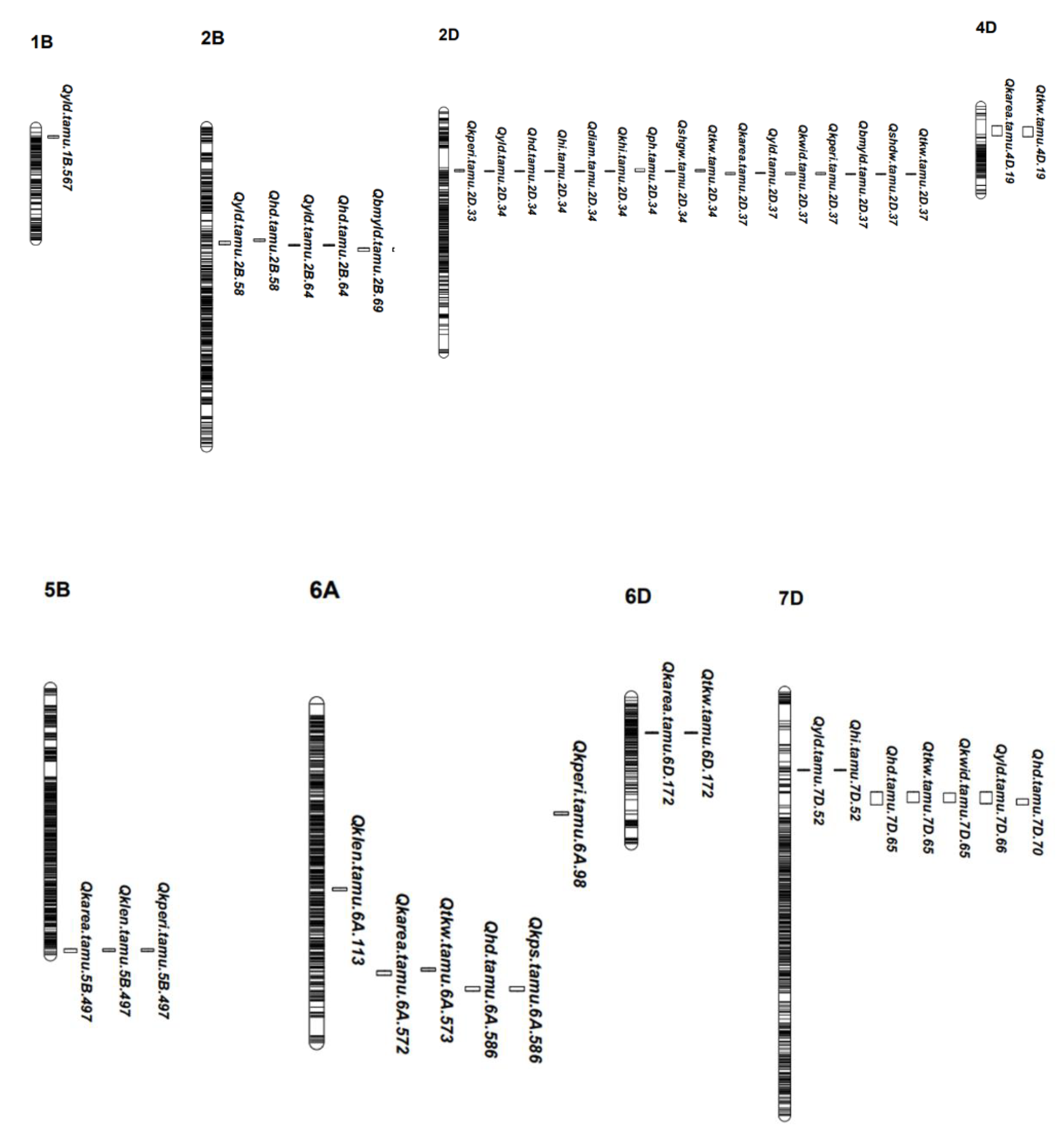
| QTL Region | QTL Name | Chr. a | Peak Mbp b | Trait c | Env d | LOD-Threshold | LOD e | LOD (A) | LOD (AbyE) | PVE f | PVE (A) | PVE (AbyE) g | Add h | Con. i | Plei. j |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Qyld.tamu.1B.567 | 1B | 566.97 | YLD | MET, 20 MCG, 20 CS | 3.5–6.4 | 7.3–19.4 | 9.96 | 9.52 | 5.7–14.6 | 7.37 | 7.32 | (−23.75)–(−8.38) | y | |
| 2 | Qyld.tamu.2B.58 | 2B | 57.25 | YLD | 20 MCG, 19 CS | 3.59–3.86 | 3.68–3.83 | - | - | 3.82–4.34 | - | - | 13.64–16.56 | y | y |
| 2 | Qhd.tamu.2B.58 | 2B | 57.61 | HD | MET, 21 BD,19 MCG | 3.54–6.38 | 4.96–16.11 | 5.11 | 11 | 2.15–1163 | 1.11 | 1.14 | (−1.07)–(−0.48) | y | y |
| 3 | Qhd.tamu.2B.64 | 2B | 63.96 | HD | MET, 19 BSP, 19 MCG | 3.47–6.38 | 3.96–10.27 | 1.57 | 8.7 | 0.78–8.12 | 0.33 | 0.45 | (−0.72)–(−0.26) | y | y |
| 3 | Qyld.tamu.2B.64 | 2B | 63.96 | YLD | MET, 21 BD | 3.54–6.4 | 6.18–7.77 | 3.05 | 4.72 | 4.26–13.11 | 2.2 | 2.07 | 4.58–14.46 | y? | y |
| 4 | Qbmyld.tamu.2B.69 | 2B | 69.34 | BMYLD | 21 BD | 3.54 | 4.24 | - | - | 10.34 | - | - | 37.0428 | y | |
| 4 | Qhi.tamu.2B.69 | 2B | 69.34 | HI | MET, 21 BD | 3.54–4.82 | 7.79–7.92 | 6.71 | 1.22 | 14.12–27.35 | 10.37 | 16.99 | 1.32–3.72 | y? | y |
| 4 | Qkps.tamu.2B.69 | 2B | 69.34 | KPS | 21 BD | 3.54 | 5.59 | - | - | 11.54 | - | - | 1.7685 | y | |
| 4 | Qshgw.tamu.2B.69 | 2B | 69.34 | SHGW | MET | 4.76 | 5.12 | 4.51 | 0.61 | 12.88 | 5.65 | 7.22 | 0.0243 | y | |
| 5 | Qkarea.tamu.2B.121 | 2B | 120.96 | KAREA | MET, 21 BD | 3.54–4.85 | 4.68–4.95 | 2.57 | 2.38 | 5.31–9.39 | 2.5 | 2.81 | −0.0838 | y? | |
| 5 | Qklen.tamu.2B.121 | 2B | 120.97 | KLEN | MET, 21 BD | 3.54–4.86 | 7.01–9.44 | 8.46 | 0.98 | 9.69–14.13 | 7.22 | 2.47 | (−0.08)–(−0.04) | y? | |
| 5 | Qkperi.tamu.2B.121 | 2B | 120.97 | KPERI | MET, 21 BD | 3.54–4.86 | 8.09–9.64 | 7.13 | 2.51 | 13.08–15.74 | 7.95 | −0.13 | (−0.24)–(−0.11) | y? | |
| 6 | Qdiam.tamu.2D.34 | 2D | 34.43 | DIAM | 20 MCG | 3.56 | 23.51 | - | - | 35.05 | - | - | −0.0744 | y | |
| 6 | Qkperi.tamu.2D.34 | 2D | 34.43 | KPERI | MET | 4.84 | 5.7 | 0.52 | 5.18 | 5.07 | 0.55 | 0.09 | −0.0308 | y | |
| 6 | Qhi.tamu.2D.34 | 2D | 34.43 | HI | MET, 19 BSP | 3.47–4.82 | 10.95–15.66 | 2.04 | 13.63 | 3.44–21.80 | 3.05 | 0.38 | (−0.94)–(−0.72) | y? | y |
| 6 | Qshgw.tamu.2D.34 | 2D | 34.43 | SHGW | MET, 19 BSP | 3.51–4.76 | 5.64–7.68 | 4.78 | 2.91 | 6.45–10.54 | 5.83 | 0.63 | (−0.03)–(−0.02) | y? | y |
| 6 | Qtkw.tamu.2D.34 | 2D | 34.43 | TKW | MET, 19 BSP | 3.47–4.83 | 12.17–12.34 | 5.24 | 7.1 | 17.4519.13 | 7.65 | 11.49 | (−1.33)–(−0.48) | y? | y |
| 6 | Qyld.tamu.2D.34 | 2D | 34.43 | YLD | MET, 19 CS | 3.86–6.4 | 17.42–21.12 | 9.22 | 11.9 | 17.57–20.23 | 6.9 | 10.67 | (−31.45)–(−8.11) | y? | y |
| 6 | Qkhi.tamu.2D.34 | 2D | 34.43 | KHI | 20 MCG | 3.56 | 23.12 | - | - | 29.85 | - | - | 5.7068 | y | |
| 6 | Qhd.tamu.2D.34 | 2D | 34.43 | HD | MET, 19 BSP, 19 CS, 19 MCG, 20 CS, 20 MCG, 21 BD | 3.47–6.38 | 21.89–179.84 | 118.67 | 61.18 | 34.82–57.45 | 49.6 | 7.85 | 1.5–4.86 | y | y |
| 6 | Qph.tamu.2D.34 | 2D | 34.43 | PH | 19 BSP, 20 MCG | 3.47–3.48 | 11.35–19.84 | - | - | 15.46–28.78 | - | - | 2.11–3.48 | y | y |
| 7 | Qkarea.tamu.2D.37 | 2D | 36.9 | KAREA | MET, 19 BSP, 20 EMN | 4.85–4.87 | 8.67–17.54 | 12.6 | 4.95 | 14.89–18.15 | 12.71 | 5.45 | (−0.29)–(−0.18) | y | y |
| 7 | Qkperi.tamu.2D.37 | 2D | 36.9 | KPERI | 20 EMN | 3.52 | 4.45 | - | - | 8.81 | - | - | −0.1652 | y | |
| 7 | Qyld.tamu.2D.37 | 2D | 36.9 | YLD | MET, 20 CS, 20 MCG | 3.59–6.4 | 9.13–33.40 | 11.37 | 22.03 | 11.72–26.14 | 8.53 | 17.62 | (−37.64)–(−8.99) | y | y |
| 7 | Qbmyld.tamu.2D.37 | 2D | 37.25 | BMYLD | MET, 19BSP | 4.85 | 4.96–6.35 | 2.48 | 3.87 | 2.35–8.53 | 1.9 | 0.45 | (−20.03)–(−12.42) | y? | y |
| 7 | Qkwid.tamu.2D.37 | 2D | 37.25 | KWID | MET, 20 EMN, 19 BSP | 3.47–4.99 | 5.29–18.58 | 15.95 | 2.63 | 12.68–17.48 | 14.75 | 2.49 | (−0.04)–(−0.02) | y | y |
| 7 | Qshdw.tamu.2D.37 | 2D | 37.25 | SHDW | MET, 21 BD | 3.51–4.9 | 10.32–11.56 | 10.41 | 1.15 | 22.16–23.95 | 12.4 | 11.56 | (−0.08)–(−0.03) | y? | y |
| 7 | Qtkw.tamu.2D.37 | 2D | 37.25 | TKW | MET, 20 MCG, 20 EMN | 3.48–4.83 | 5.62–8.40 | 3.23 | 2.61 | 10.09–12.55 | 4.65 | 5.44 | (−1.01)–(−0.37) | y | y |
| 8 | Qklen.tamu.5B.497 | 5B | 496.62 | KLEN | MET, 19 BSP, 20 EMN | 3.47–4.86 | 3.92–8.34 | 6.34 | 2 | 4.91–7.71 | 5.62 | 1.06 | 0.04–0.05 | y | |
| 9 | Qtkw.tamu.6A.573 | 6A | 573.37 | TKW | 20 MCG, 20 EMN, 21 BD | 3.54 | 4.20–8.36 | - | - | 8.37–12.97 | - | - | (−1.03)–(−0.74) | y | |
| 10 | Qhd.tamu.6A.586 | 6A | 586.24 | HD | MET | 6.38 | 6.94 | 4.99 | 1.94 | 1.88 | 1.08 | 0.8 | 0.4775 | y | |
| 10 | Qkps.tamu.6A.586 | 6A | 586.24 | KPS | MET, 19 BSP | 3.47–4.79 | 6.71–7.14 | 2.59 | 4.56 | 7.82–16.17 | 5.15 | 2.68 | 0.49–0.94 | y? | y |
| 11 | Qkperi.tamu.6A.98 | 6A | 98.34 | KPERI | MET, 19 BSP, 20 EMN | 3.47–4.84 | 5.42–12.40 | 8.43 | 3.98 | 11.06–12.52 | 9.35 | 0.1 | (−0.18)–(−0.12) | y | |
| 12 | Qklen.tamu.6A.113 | 6A | 113.23 | KLEN | MET, 19 BSP, 21 BD | 3.47–4.86 | 5.38–11.21 | 8.22 | 2.99 | 10.99–11.48 | 7.06 | 3.94 | (−0.07)–(−0.04) | y | |
| 13 | Qhi.tamu.7D.52 | 7D | 52.32 | HI | MET, 19 BSP | 3.47–4.82 | 4.89–6.75 | 2.53 | 4.22 | 4.60–9.81 | 3.56 | 1.05 | 0.63–0.77 | y? | y |
| 13 | Qyld.tamu.7D.52 | 7D | 52.32 | YLD | MET, 19 BSP | 3.47–6.4 | 9.68–21.59 | 7.27 | 14.32 | 12.87–16.87 | 5.28 | 7.59 | 7.07–13.16 | y? | y |
| 14 | Qhd.tamu.7D.65 | 7D | 65.22 | HD | 21 BD | 3.54 | 5.28 | - | - | 6.71 | - | - | −0.7312 | y | |
| 14 | Qtkw.tamu.7D.65 | 7D | 65.22 | TKW | MET, 19 BSP | 3.47 | 8.44 | - | - | 12.89 | - | - | 1.1414 | y | |
| 14 | Qkwid.tamu.7D.66 | 7D | 66.02 | KWID | MET, 19 BSP, 19 MCG | 3.47–4.99 | 5.55–9.23 | 5.01 | 4.22 | 6.99–12.64 | 4.03 | 2.96 | 0.015–0.037 | y | y |
| 14 | Qyld.tamu.7D.66 | 7D | 66.02 | YLD | 20 MCG | 3.59 | 5.62 | - | - | 7.83 | - | - | 22.2131 | y | |
| 15 | Qhd.tamu.7D.70 | 7D | 70.03 | HD | MET, 19 BSP, 20 CS | 3.47–6.38 | 3.78–16.11 | 7.55 | 8.57 | 1.81–5.10 | 1.62 | 0.2 | (−0.96)–(−0.58) | y |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerit, M.; Wang, Z.; Dogan, M.; Yu, S.; Valenzuela-Antelo, J.L.; Chu, C.; Wang, S.; Xue, Q.; Ibrahim, A.M.H.; Rudd, J.C.; et al. Mapping QTL for Yield and Its Component Traits Using Wheat (Triticum aestivum L.) RIL Mapping Population from TAM 113 × Gallagher. Agronomy 2023, 13, 2402. https://doi.org/10.3390/agronomy13092402
Cerit M, Wang Z, Dogan M, Yu S, Valenzuela-Antelo JL, Chu C, Wang S, Xue Q, Ibrahim AMH, Rudd JC, et al. Mapping QTL for Yield and Its Component Traits Using Wheat (Triticum aestivum L.) RIL Mapping Population from TAM 113 × Gallagher. Agronomy. 2023; 13(9):2402. https://doi.org/10.3390/agronomy13092402
Chicago/Turabian StyleCerit, Mustafa, Zhen Wang, Mehmet Dogan, Shuhao Yu, Jorge L. Valenzuela-Antelo, Chenggen Chu, Shichen Wang, Qingwu Xue, Amir M. H. Ibrahim, Jackie C. Rudd, and et al. 2023. "Mapping QTL for Yield and Its Component Traits Using Wheat (Triticum aestivum L.) RIL Mapping Population from TAM 113 × Gallagher" Agronomy 13, no. 9: 2402. https://doi.org/10.3390/agronomy13092402
APA StyleCerit, M., Wang, Z., Dogan, M., Yu, S., Valenzuela-Antelo, J. L., Chu, C., Wang, S., Xue, Q., Ibrahim, A. M. H., Rudd, J. C., Metz, R., Johnson, C. D., & Liu, S. (2023). Mapping QTL for Yield and Its Component Traits Using Wheat (Triticum aestivum L.) RIL Mapping Population from TAM 113 × Gallagher. Agronomy, 13(9), 2402. https://doi.org/10.3390/agronomy13092402








