Integrated Transcriptomics and Metabolomics Analysis Reveal Anthocyanin Biosynthesis for Petal Color Formation in Catharanthus roseus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Plant Materials
2.2. Measurement of Total Anthocyanin Content
2.3. Extraction, Identification, and Quantitative Analysis of Metabolites
2.4. RNA Extraction and Sequencing
2.5. Analysis and Annotation of RNA-Seq Data
2.6. Structural Gene and Transcription Factor Analysis
2.7. Quantitative Real Time PCR and Expression Validation
3. Results
3.1. Total Anthocyanin Content in Petals with Different Colours
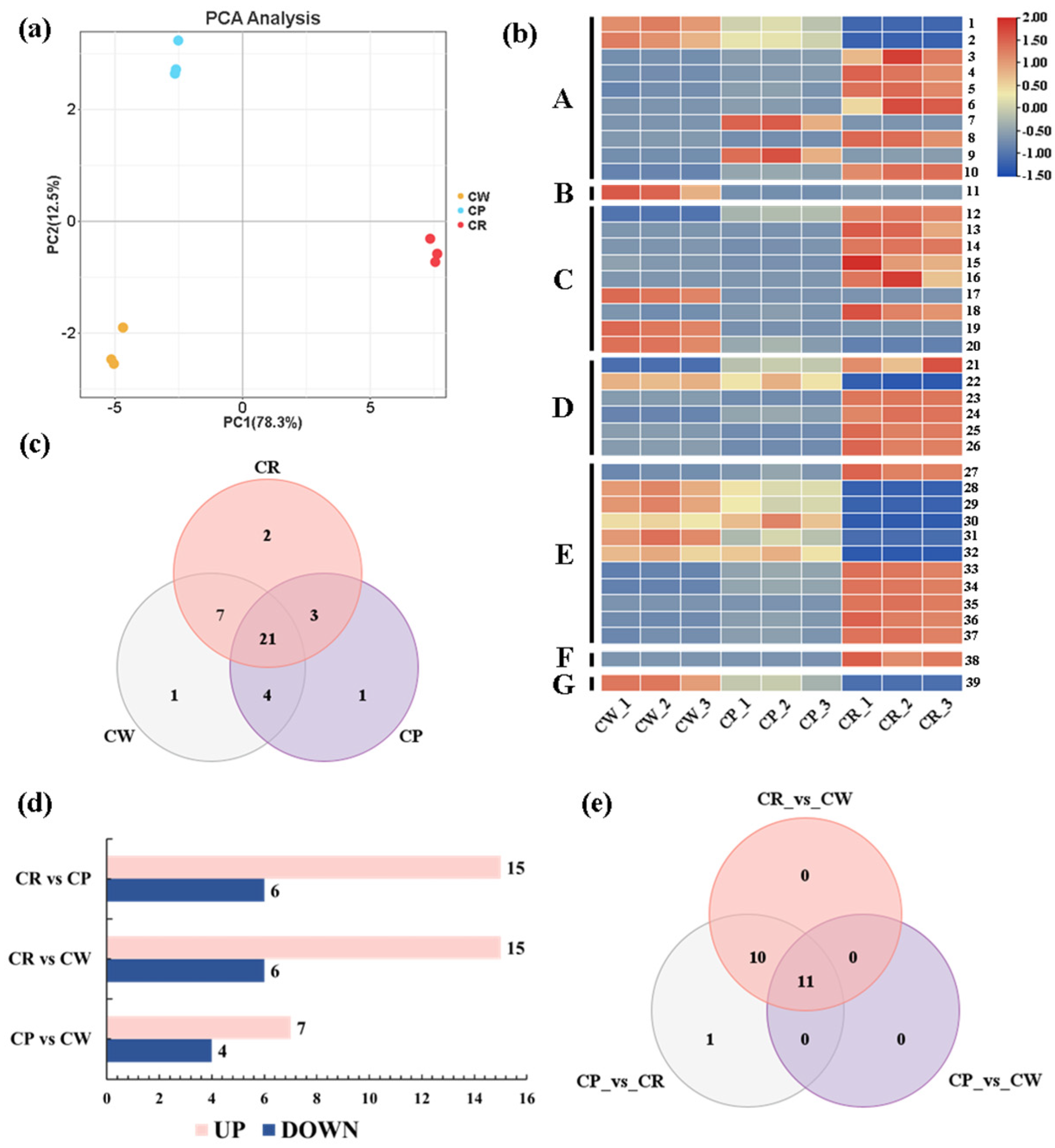
3.2. Metabolite Difference Analysis
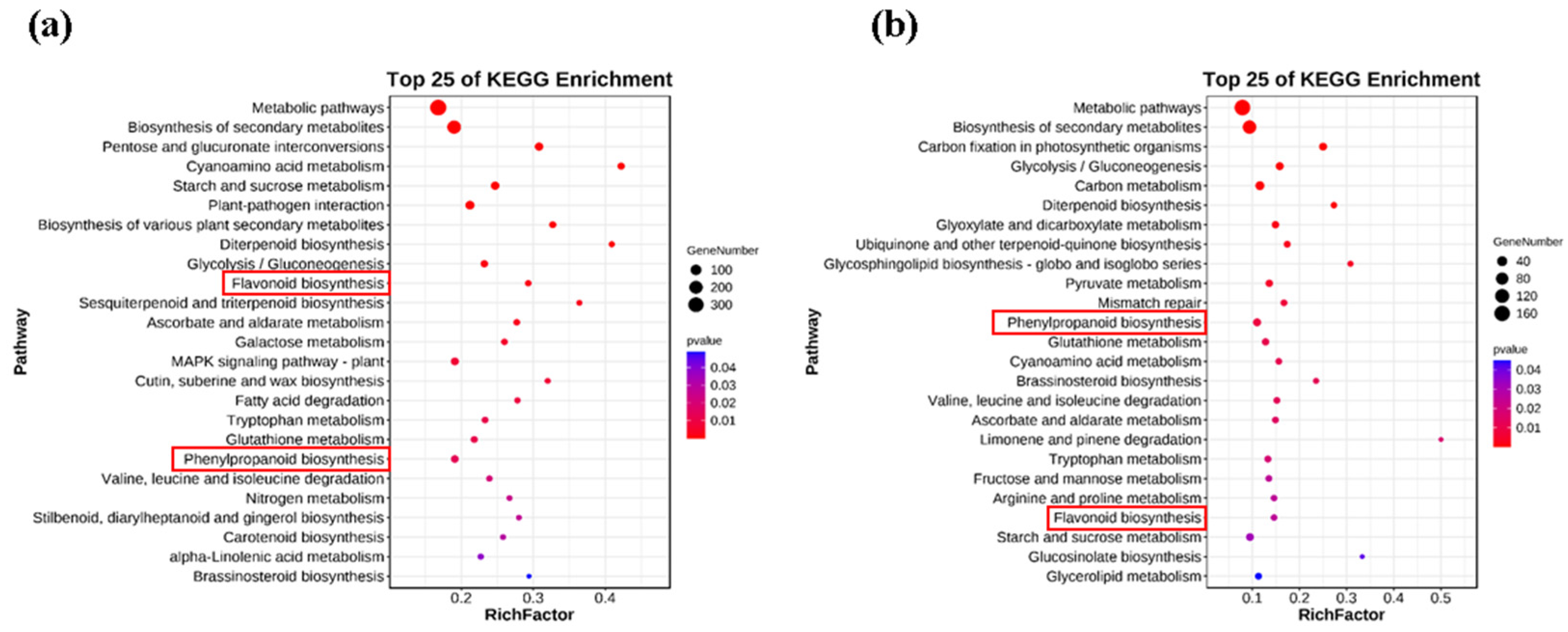
3.3. Overview of Full Transcriptome Sequencing Result
3.4. Differentially Expressed Genes
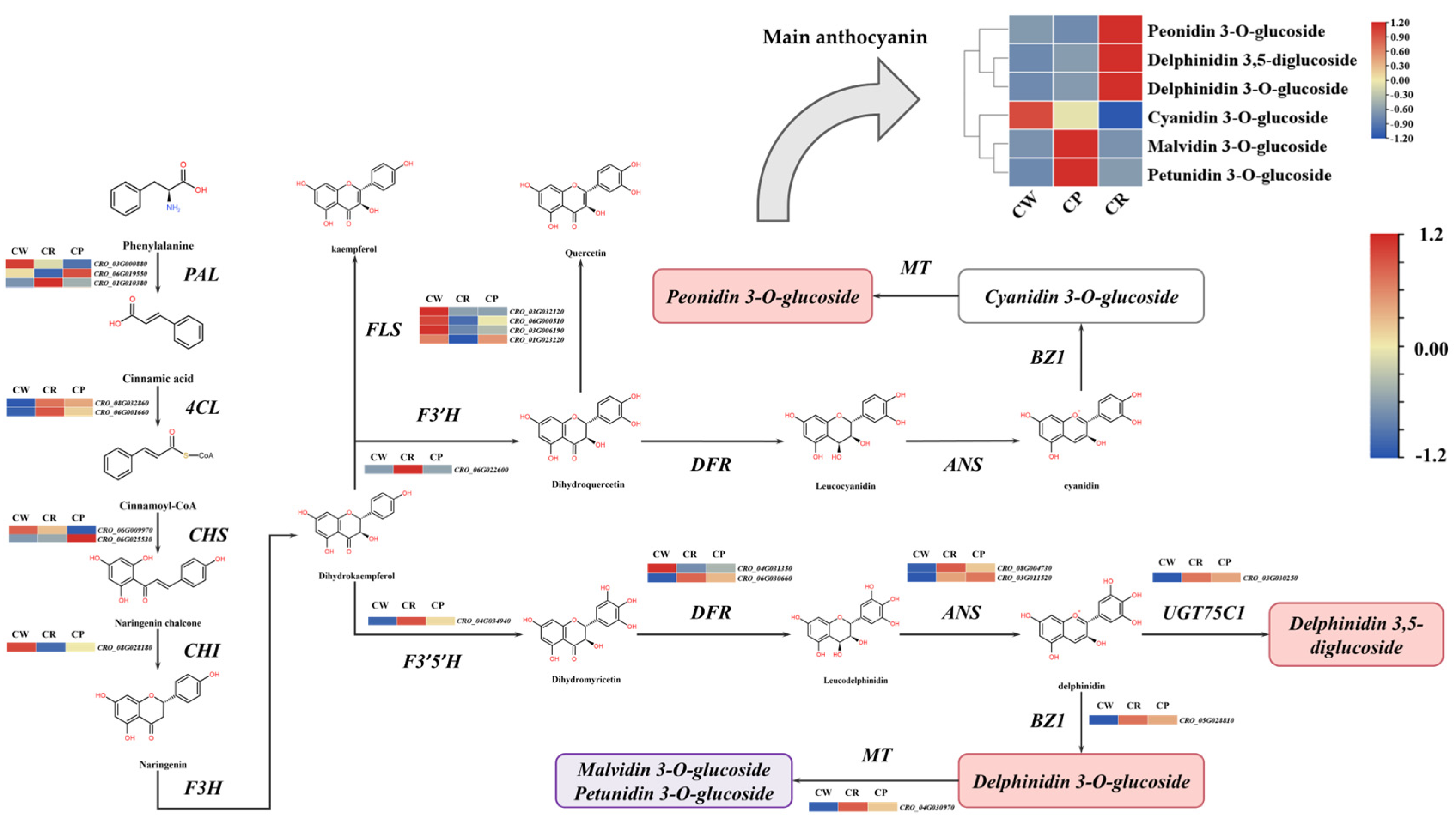
3.5. The Candidate Genes Involved in Anthocyanin Biosynthesis Pathway
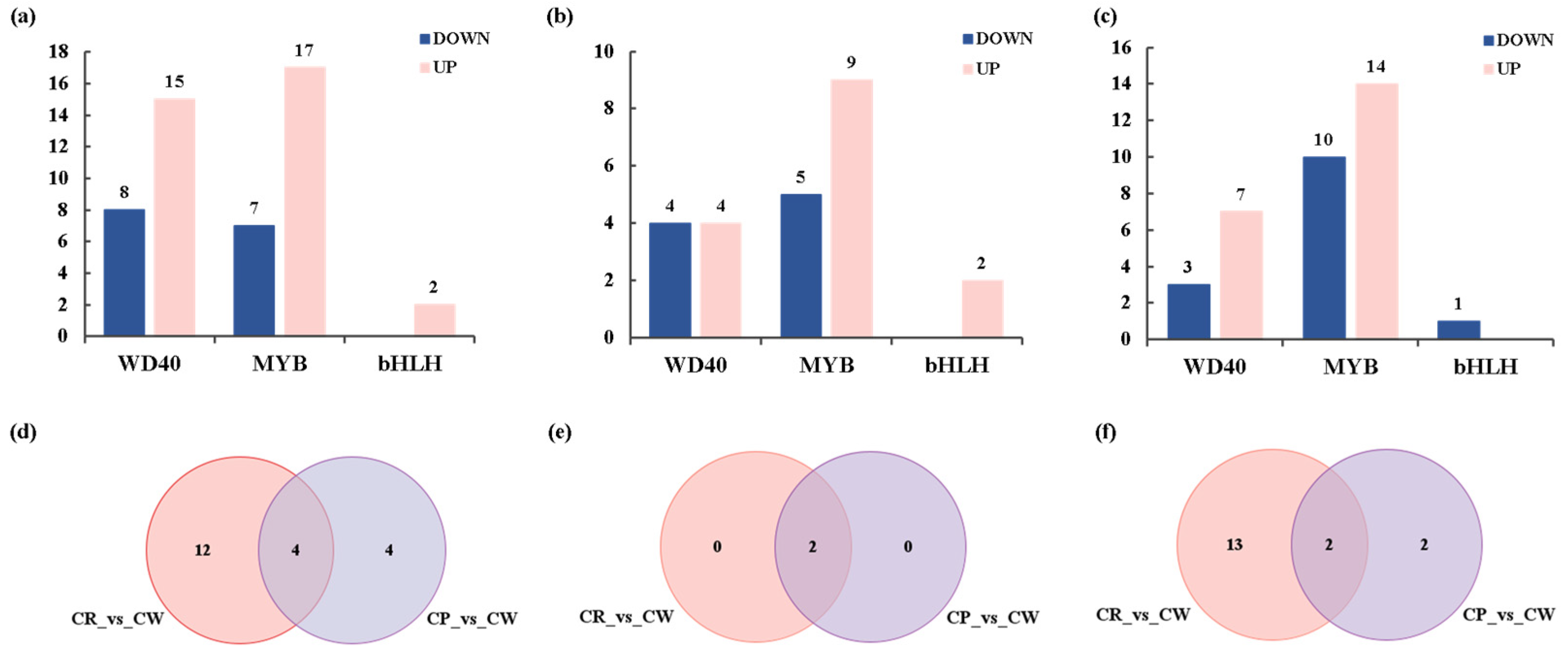
3.6. Transcription Factors Related to Anthocyanin Biosynthesis
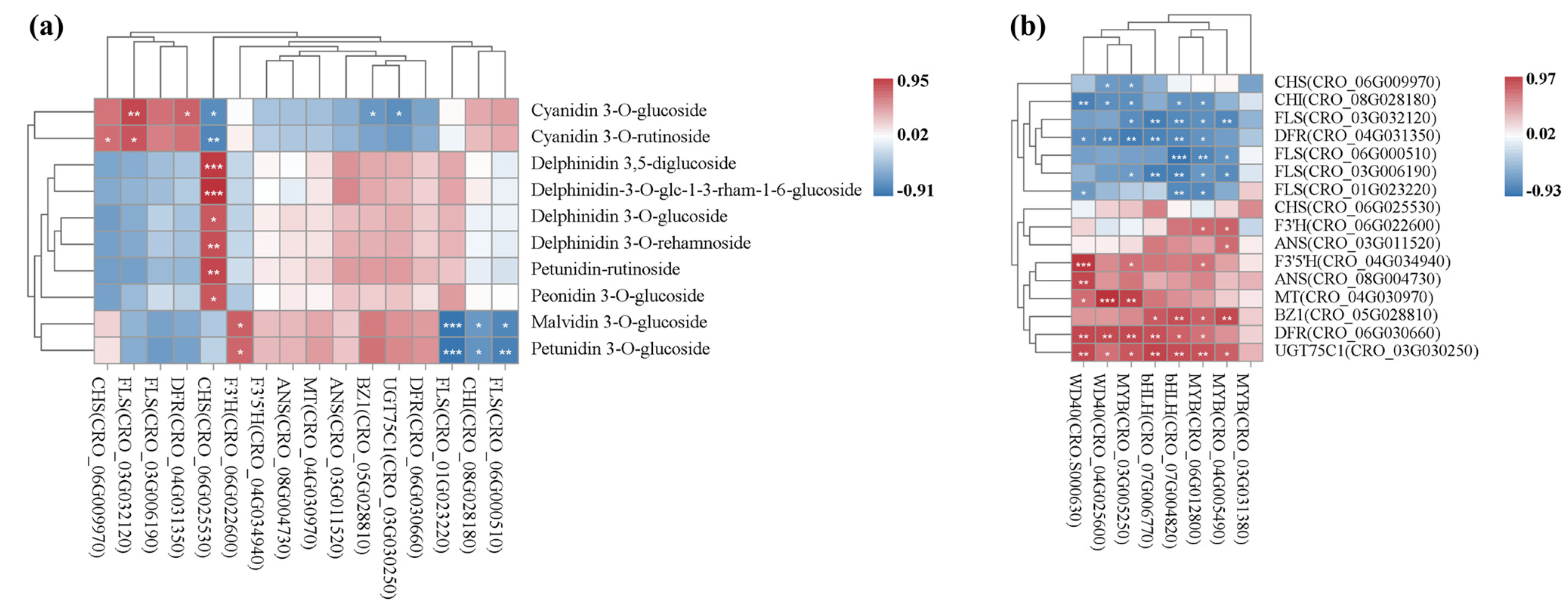
3.7. Integrated Transcriptome and Metabolome Analysis
3.8. Verification of the Results in RNA-Seq by qRT-PCR
4. Discussion
4.1. Effects of Anthocyanin Content and Types on the Catharanthus roseus Petals
4.2. Key Structural Genes Responsible for Anthocyanin Synthesis in Catharanthus roseus Petals
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, C.; Wood, J.C.; Vu, A.H.; Hamilton, J.P.; Rodriguez Lopez, C.E.; Payne, R.M.E.; Serna Guerrero, D.A.; Gase, K.; Yamamoto, K.; Vaillancourt, B.; et al. Single-Cell Multi-Omics in the Medicinal Plant Catharanthus roseus. Nat. Chem. Biol. 2023, 19, 1031–1041. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, B.; Singh, R. Catharanthus roseus (L.) G. Don: A Review of Its Ethnobotany, Phytochemistry, Ethnopharmacology and Toxicities. J. Ethnopharmacol. 2022, 284, 114647. [Google Scholar] [CrossRef]
- Wang, L.S.; Shiraishi, A.; Hashimoto, F.; Aoki, N.; Shimizu, K.; Sakata, Y. Analysis of Petal Anthocyanins to Investigate Flower Coloration of Zhongyuan (Chinese) and Daikon Island (Japanese) Tree Peony Cultivars. J. Plant Res. 2001, 114, 33–43. [Google Scholar] [CrossRef]
- Polturak, G.; Heinig, U.; Grossman, N.; Battat, M.; Leshkowitz, D.; Malitsky, S.; Rogachev, I.; Aharoni, A. Transcriptome and Metabolic Profiling Provides Insights into Betalain Biosynthesis and Evolution in Mirabilis Jalapa. Mol. Plant 2018, 11, 189–204. [Google Scholar] [CrossRef]
- Chiou, C.-Y.; Pan, H.-A.; Chuang, Y.-N.; Yeh, K.-W. Differential Expression of Carotenoid-Related Genes Determines Diversified Carotenoid Coloration in Floral Tissues of Oncidium Cultivars. Planta 2010, 232, 937–948. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Holton, T.A.; Brugliera, F.; Tanaka, Y. Cloning and Expression of Flavonol Synthase from Petunia hybrida. Plant J. 1993, 4, 1003–1010. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The Flavonoid Biosynthetic Pathway in Arabidopsis: Structural and Genetic Diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current Understanding of the Pathways of Flavonoid Biosynthesis in Model and Crop Plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef]
- Kitts, D.; Tomiuk, S. Studies on Mitigating Lipid Oxidation Reactions in a Value-Added Dairy Product Using a Standardized Cranberry Extract. Agriculture 2013, 3, 236–252. [Google Scholar] [CrossRef]
- Peng, X.Q.; Ai, Y.J.; Pu, Y.T.; Wang, X.J.; Li, Y.H.; Wang, Z.; Zhuang, W.B.; Yu, B.J.; Zhu, Z.Q. Transcriptome and Metabolome Analyses Reveal Molecular Mechanisms of Anthocyanin-Related Leaf Color Variation in Poplar (Populus deltoides) Cultivars. Front. Plant Sci. 2023, 14, 1103468. [Google Scholar] [CrossRef]
- Li, S.; Deng, B.; Tian, S.; Guo, M.; Liu, H.; Zhao, X. Metabolic and Transcriptomic Analyses Reveal Different Metabolite Biosynthesis Profiles between Leaf Buds and Mature Leaves in Ziziphus jujuba Mill. Food Chem. 2021, 347, 129005. [Google Scholar] [CrossRef]
- Jiao, F.; Zhao, L.; Wu, X.; Song, Z.; Li, Y. Metabolome and Transcriptome Analyses of the Molecular Mechanisms of Flower Color Mutation in Tobacco. BMC Genom. 2020, 21, 611. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Q.; Lang, L.; Jiang, C.; Wang, X.; Sun, H. Integrated Metabolome and Transcriptome Analysis of the Anthocyanin Biosynthetic Pathway in Relation to Color Mutation in Miniature Roses. BMC Plant Biol. 2021, 21, 257. [Google Scholar] [CrossRef]
- Yu, M.; Man, Y.; Lei, R.; Lu, X.; Wang, Y. Metabolomics Study of Flavonoids and Anthocyanin-Related Gene Analysis in Kiwifruit (Actinidia chinensis) and Kiwiberry (Actinidia arguta). Plant Mol. Biol. Rep. 2020, 38, 353–369. [Google Scholar] [CrossRef]
- Jiang, L.; Yue, M.; Liu, Y.; Ye, Y.; Zhang, Y.; Lin, Y.; Wang, X.; Chen, Q.; Tang, H. Alterations of Phenylpropanoid Biosynthesis Lead to the Natural Formation of Pinkish-Skinned and White-Fleshed Strawberry (Fragaria × Ananassa). Int. J. Mol. Sci. 2022, 23, 7375. [Google Scholar] [CrossRef]
- Grotewold, E. The Genetics and Biochemistry of Floral Pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F.; Chandler, S. Recent Progress of Flower Colour Modification by Biotechnology. Int. J. Mol. Sci. 2009, 10, 5350–5369. [Google Scholar] [CrossRef]
- Dasgupta, K.; Thilmony, R.; Stover, E.; Oliveira, M.L.; Thomson, J. Novel R2R3-MYB Transcription Factors from Prunus Americana Regulate Differential Patterns of Anthocyanin Accumulation in Tobacco and Citrus. GM Crops Food 2017, 8, 85–105. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB Transcription Factors That Colour Our Fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB–BHLH–WD40 Protein Complex and the Evolution of Cellular Diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.-P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of Tomato Fruit with Health-Promoting Anthocyanins by Expression of Select Transcription Factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Feng, C.; Ding, D.; Feng, C.; Kang, M. The Identification of an R2R3-MYB Transcription Factor Involved in Regulating Anthocyanin Biosynthesis in Primulina Swinglei Flowers. Gene 2020, 752, 144788. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, H.; Ma, Z.; Huang, J. DELLA Proteins Promote Anthocyanin Biosynthesis via Sequestering MYBL2 and JAZ Suppressors of the MYB/BHLH/WD40 Complex in Arabidopsis thaliana. Mol. Plant 2016, 9, 711–721. [Google Scholar] [CrossRef]
- Donoso, A.; Rivas, C.; Zamorano, A.; Peña, Á.; Handford, M.; Aros, D. Understanding Alstroemeria Pallida Flower Colour: Links between Phenotype, Anthocyanins and Gene Expression. Plants 2020, 10, 55. [Google Scholar] [CrossRef]
- Fu, M.; Yang, X.; Zheng, J.; Wang, L.; Yang, X.; Tu, Y.; Ye, J.; Zhang, W.; Liao, Y.; Cheng, S.; et al. Unraveling the Regulatory Mechanism of Color Diversity in Camellia Japonica Petals by Integrative Transcriptome and Metabolome Analysis. Front. Plant Sci. 2021, 12, 685136. [Google Scholar] [CrossRef]
- Rabiner, L.R. A Tutorial on Hidden Markov Models and Selected Applications in Speech Recognition. Proc. IEEE 1989, 77, 257–286. [Google Scholar] [CrossRef]
- Pollier, J.; Vanden Bossche, R.; Rischer, H.; Goossens, A. Selection and Validation of Reference Genes for Transcript Normalization in Gene Expression Studies in Catharanthus roseus. Plant Physiol. Biochem. 2014, 83, 20–25. [Google Scholar] [CrossRef]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and Anthocyanins: Colored Pigments as Food, Pharmaceutical Ingredients, and the Potential Health Benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Yao, Y.; An, L.; Li, X.; Bai, Y.; Cui, Y.; Wu, K. Accumulation and Regulation of Anthocyanins in White and Purple Tibetan Hulless Barley (Hordeum vulgare L. Var. Nudum Hook. f.) Revealed by Combined de Novo Transcriptomics and Metabolomics. BMC Plant Biol. 2022, 22, 391. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Liu, L.; Wei, Z.; Liu, J.; Li, M.; Yan, Z.; Gao, D. Anthocyanin Accumulation and Molecular Analysis of Correlated Genes by Metabolomics and Transcriptomics in Sister Line Apple Cultivars. Life 2022, 12, 1246. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ji, G.; Xu, Z.; Feng, B.; Zhou, Q.; Fan, X.; Wang, T. Metabolomics and Transcriptomics Provide Insights into Anthocyanin Biosynthesis in the Developing Grains of Purple Wheat (Triticum aestivum L.). J. Agric. Food Chem. 2021, 69, 11171–11184. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The War of the Whorls: Genetic Interactions Controlling Flower Development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Ausin, I.; Alonso-Blanco, C.; Martinez-Zapater, J.-M. Environmental Regulation of Flowering. Int. J. Dev. Biol. 2005, 49, 689–705. [Google Scholar] [CrossRef]
- Iwashina, T. Contribution to Flower Colors of Flavonoids Including Anthocyanins: A Review. Nat. Prod. Commun. 2015, 10, 529–544. [Google Scholar] [CrossRef]
- Qian, R.; Ye, Y.; Hu, Q.; Ma, X.; Zhang, X.; Zheng, J. Metabolomic and Transcriptomic Analyses Reveal New Insights into the Role of Metabolites and Genes in Modulating Flower Colour of Clematis Tientaiensis. Horticulturae 2023, 9, 14. [Google Scholar] [CrossRef]
- Zhong, H.; Liu, Z.; Zhang, F.; Zhou, X.; Sun, X.; Li, Y.; Liu, W.; Xiao, H.; Wang, N.; Lu, H.; et al. Metabolomic and Transcriptomic Analyses Reveal the Effects of Self- and Hetero-Grafting on Anthocyanin Biosynthesis in Grapevine. Hortic. Res. 2022, 9, uhac103. [Google Scholar] [CrossRef]
- Honda, C.; Kotoda, N.; Wada, M.; Kondo, S.; Kobayashi, S.; Soejima, J.; Zhang, Z.; Tsuda, T.; Moriguchi, T. Anthocyanin Biosynthetic Genes Are Coordinately Expressed during Red Coloration in Apple Skin. Plant Physiol. Biochem. 2002, 40, 955–962. [Google Scholar] [CrossRef]
- Ban, Y.; Kondo, S.; Ubi, B.E.; Honda, C.; Bessho, H.; Moriguchi, T. UDP-Sugar Biosynthetic Pathway: Contribution to Cyanidin 3-Galactoside Biosynthesis in Apple Skin. Planta 2009, 230, 871–881. [Google Scholar] [CrossRef]
- Aza-González, C.; Herrera-Isidrón, L.; Núñez-Palenius, H.G.; De La Vega, O.M.; Ochoa-Alejo, N. Anthocyanin Accumulation and Expression Analysis of Biosynthesis-Related Genes during Chili Pepper Fruit Development. Biol. Plant 2013, 57, 49–55. [Google Scholar] [CrossRef]
- Rahim, M.A.; Busatto, N.; Trainotti, L. Regulation of Anthocyanin Biosynthesis in Peach Fruits. Planta 2014, 240, 913–929. [Google Scholar] [CrossRef]
- Zhuang, H.; Lou, Q.; Liu, H.; Han, H.; Wang, Q.; Tang, Z.; Ma, Y.; Wang, H. Differential Regulation of Anthocyanins in Green and Purple Turnips Revealed by Combined De Novo Transcriptome and Metabolome Analysis. Int. J. Mol. Sci. 2019, 20, 4387. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.; Shi, J.; Zhang, S.; Pingcuo, G.; Wang, S.; Zhao, F.; Cui, Y.; Zeng, X. Transcriptomic and Metabolomic Profiling Provide Novel Insights into Fruit Development and Flesh Coloration in Prunus Mira Koehne, a Special Wild Peach Species. BMC Plant Biol. 2019, 19, 463. [Google Scholar] [CrossRef]
- Luo, P.; Ning, G.; Wang, Z.; Shen, Y.; Jin, H.; Li, P.; Huang, S.; Zhao, J.; Bao, M. Disequilibrium of Flavonol Synthase and Dihydroflavonol-4-Reductase Expression Associated Tightly to White vs. Red Color Flower Formation in Plants. Front. Plant Sci. 2016, 6, 1257. [Google Scholar] [CrossRef]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of Apple ANR Genes into Tobacco Inhibits Expression of Both CHI and DFR Genes in Flowers, Leading to Loss of Anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Xia, E.-H.; Zhang, H.-B.; Yao, Q.-Y.; Gao, L.-Z. De Novo Transcriptome Sequencing of Camellia Sasanqua and the Analysis of Major Candidate Genes Related to Floral Traits. Plant Physiol. Biochem. 2017, 120, 103–111. [Google Scholar] [CrossRef]
- Li, Z.; Vickrey, T.L.; McNally, M.G.; Sato, S.J.; Clemente, T.E.; Mower, J.P. Assessing Anthocyanin Biosynthesis in Solanaceae as a Model Pathway for Secondary Metabolism. Genes 2019, 10, 559. [Google Scholar] [CrossRef]
- Boase, M.R.; Lewis, D.H.; Davies, K.M.; Marshall, G.B.; Patel, D.; Schwinn, K.E.; Deroles, S.C. Isolation and Antisense Suppression of Flavonoid 3′,5′-Hydroxylase Modifies Flower Pigments and Colour in Cyclamen. BMC Plant Biol. 2010, 10, 107. [Google Scholar] [CrossRef]

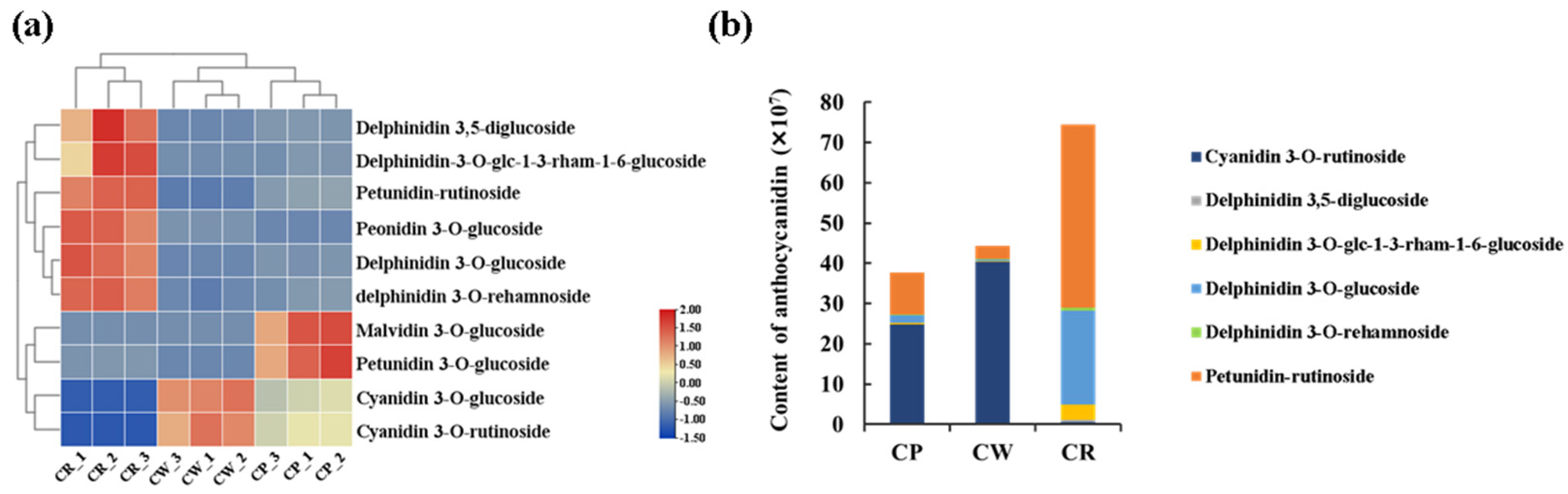

| Name | Class | Reference Ion | m/z | RT [min] | Attribution | |
|---|---|---|---|---|---|---|
| 1 | Cyanidin 3-O-glucoside | Anthocyanins | [M + H] + 1 | 449.11 | 9.78 | CW, CP |
| 2 | Cyanidin 3-O-rutinoside | Anthocyanins | [M + H] + 1 | 595.17 | 9.06 | CW, CP, CR |
| 3 | Delphinidin 3,5-diglucoside | Anthocyanins | [M + H] + 1 | 627.16 | 7.89 | CW, CP, CR |
| 4 | Delphinidin 3-O-glucoside | Anthocyanins | [M + H] + 1 | 465.10 | 9.66 | CW, CP, CR |
| 5 | Delphinidin 3-O-glc-1-3-rham-1-6-glucoside | Anthocyanins | [M + H] + 1 | 773.21 | 8.52 | CW, CP, CR |
| 6 | Delphinidin 3-O-rehamnoside | Anthocyanins | [M + H] + 1 | 449.11 | 8.78 | CW, CP, CR |
| 7 | Malvidin 3-O-glucoside | Anthocyanins | [M + H] + 1 | 493.13 | 8.73 | CP |
| 8 | Peonidin 3-O-glucoside | Anthocyanins | [M + H] + 1 | 463.12 | 8.65 | CW, CR |
| 9 | Petunidin 3-O-glucoside | Anthocyanins | [M + H] + 1 | 479.12 | 8.20 | CP, CR |
| 10 | Petunidin-rutinoside | Anthocyanins | [M + H] + 1 | 625.18 | 10.74 | CW, CP, CR |
| 11 | Licochalcone B | Chalcones | [M + H] + 1 | 287.09 | 9.10 | CW, CR |
| 12 | 3,5,7,3′,4′,5′-Hexahydroxy-6,8-dimethylflavanone | Flavanone | [M − H] − 1 | 347.08 | 9.55 | CP, CR |
| 13 | 4′-Methoxy-7-O-beta-D-glucopyranosyl-8,3′-dihydroxyflavanone | Flavanone | [M + H] + 1 | 465.14 | 8.76 | CW, CR |
| 14 | diosmin | Flavanone | [M + H] + 1 | 609.18 | 8.47 | CW, CP, CR |
| 15 | Eriodictyol | Flavanone | [M + H] + 1 | 289.07 | 9.11 | CW, CP, CR |
| 16 | Hesperetin | Flavanone | [M + H] + 1 | 303.09 | 7.35 | CR |
| 17 | Isosakuranin | Flavanone | [M + H] + 1 | 449.14 | 9.73 | CW |
| 18 | Isoscutellarein 7-(6′″-acetylallosyl-(1->2)-glucoside) | Flavanone | [M − H] − 1 | 651.16 | 10.34 | CW, CR |
| 19 | Sakuranetin | Flavanone | [M + H] + 1 | 287.09 | 9.73 | CW, CP, CR |
| 20 | Eriodictyol 7,3′-dimethyl ether | Flavanone | [M + H] + 1 | 317.10 | 10.65 | CW, CR |
| 21 | 5,7,3′,6′-Tetrahydroxy-8,2′-dimethoxyflavone 6′-glucoside | Flavone | [M − H] − 1 | 507.11 | 10.25 | CP, CR |
| 22 | Luteolin | Flavone | [M − H] − 1 | 285.04 | 12.95 | CW, CP, CR |
| 23 | Pectolinarin | Flavone | [M + H] + 1 | 623.20 | 8.96 | CW, CP, CR |
| 24 | Rhamnetin | Flavone | [M + H] + 1 | 317.07 | 10.74 | CW, CP, CR |
| 25 | Tricin 5-O-glucoside | Flavone | [M + H] + 1 | 493.13 | 10.84 | CW, CR |
| 26 | cirsimarin | Flavone | [M + H] + 1 | 477.14 | 9.19 | CW, CR |
| 27 | Isorhamnetin | Flavonol | [M + H] + 1 | 317.07 | 9.83 | CW, CP, CR |
| 28 | Kaempferol | Flavonol | [M + H] + 1 | 287.06 | 9.06 | CW, CP, CR |
| 29 | Mauritianin | Flavonol | [M + H] + 1 | 741.22 | 9.06 | CW, CP, CR |
| 30 | Kaempferol 3-galactoside | Flavonol | [M + H] + 1 | 449.11 | 9.98 | CW, CP |
| 31 | kaempferol 3-glucoside | Flavonol | [M − H] − 1 | 447.09 | 10.11 | CW, CP, CR |
| 32 | kaempferol 3-rhamnoside | Flavonol | [M + H] + 1 | 433.11 | 9.66 | CW, CP |
| 33 | Quercetin | Flavonol | [M + H] + 1 | 303.05 | 9.36 | CW, CP, CR |
| 34 | Quercetin 3-galactoside | Flavonol | [M + H] + 1 | 465.10 | 9.37 | CW, CP, CR |
| 35 | Quercetin 3-O-glc-1-3-rham-1-6-glucoside | Flavonol | [M − H] − 1 | 463.09 | 9.67 | CR |
| 36 | Quercetin-3-O-glucoside | Flavonol | [M + H] + 1 | 773.21 | 7.81 | CW, CP, CR |
| 37 | Rutin | Flavonol | [M + H] + 1 | 611.16 | 8.78 | CW, CP, CR |
| 38 | Iridin | Isoflavone | [M − H] − 1 | 521.13 | 9.99 | CW, CP, CR |
| 39 | Isokaempferide | Isoflavonol | [M + H] + 1 | 301.07 | 11.06 | CW, CP, CR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Y.; Tang, Y.; Huang, X.; Zeng, L.; Liao, Z. Integrated Transcriptomics and Metabolomics Analysis Reveal Anthocyanin Biosynthesis for Petal Color Formation in Catharanthus roseus. Agronomy 2023, 13, 2290. https://doi.org/10.3390/agronomy13092290
Xiao Y, Tang Y, Huang X, Zeng L, Liao Z. Integrated Transcriptomics and Metabolomics Analysis Reveal Anthocyanin Biosynthesis for Petal Color Formation in Catharanthus roseus. Agronomy. 2023; 13(9):2290. https://doi.org/10.3390/agronomy13092290
Chicago/Turabian StyleXiao, Yuchen, Yueli Tang, Xianhui Huang, Lingjiang Zeng, and Zhihua Liao. 2023. "Integrated Transcriptomics and Metabolomics Analysis Reveal Anthocyanin Biosynthesis for Petal Color Formation in Catharanthus roseus" Agronomy 13, no. 9: 2290. https://doi.org/10.3390/agronomy13092290
APA StyleXiao, Y., Tang, Y., Huang, X., Zeng, L., & Liao, Z. (2023). Integrated Transcriptomics and Metabolomics Analysis Reveal Anthocyanin Biosynthesis for Petal Color Formation in Catharanthus roseus. Agronomy, 13(9), 2290. https://doi.org/10.3390/agronomy13092290







