Abstract
Teosinte is the closest wild ancestor of maize and is used as a valuable resource for taxonomical, evolutionary and genetic architectural studies of maize. Teosinte is also a repository of numerous diverse alleles for complex traits, including nutritional value and stress adaptation. Accessions including teosintes, maize inbred lines and coix were investigated for kernel protein and its association with DNA markers. The proposed investigation assumed that wild accessions had different genic/allelic content and consequently expression profile than modern maize because of the domestication syndrome and bottleneck effects. Total protein content in hard stony fruit case teosinte accessions were assessed from kernels with and without seed coats, while protein content from coix and maize lines was evaluated from kernels only. The accessions were also subjected to molecular profiling using 84 SSR markers, and obtained genotypic data were used for population structure and association analysis. The results emphasize that teosintes have higher protein content (18.5% to 26.29%), followed by coix (18.26%), and the least among maize lines (9% to 11%). Among teosintes, without-seed-coat samples had 3–6% higher protein content than with-seed-coat samples. When compared to other teosinte species, Z. mays subsp. mexicana accessions showed higher protein content, ranging from 18.62% to 26.29%. All evaluated accessions were divided into four subpopulations with K = 4, and seven significant (p < 0.01) marker–trait associations were seen with umc1294, umc1171, phi091, umc2182 and bnlg292 markers, which are distributed across chromosomes 4, 5, 7, 8 and 9, respectively. We have observed that the wild relatives carry protein content-enhancing alleles and can be used as productive donor parents in pre-breeding efforts to increase the protein content of maize.
1. Introduction
Maize (Zea mays ssp. mays) is one of the most significant agricultural plants in the world and has been considered a model organism due to its extensive studies on genetics, cytogenetics, and genomics owing to its nucleotide diversity and genetic collinearity within related grasses [1]. Due to its superior agronomic performance and high yield potential, maize has drawn the attention of evolutionists and geneticists who are interested in understanding its origin, domestication and diversification to gain complete knowledge of its evolution, taxonomy and plant architecture [2]. Although the maize genome is diploid, it has undergone segmental allotetraploidization of its two progenitor genomes, which eventually diversified from its sorghum progenitor approximately 11.9 million years ago (mya) [3,4]. Around 10,000 years ago in central Mexico, maize was domesticated from teosinte (Zea mays ssp. parviglumis) in a single domestication event [5] and subsequently distributed in different parts of the world. Since extensive domestication over many years associated with natural as well as human-made selection, maize has undergone numerous morphological, physiological and biochemical alterations in the plant system [6]. Maize kernel composition is one of the most important traits, which has changed drastically and resulted in a loss of genetic diversity during the domestication of its wild relative teosinte [7]. Genetic selection during the domestication of teosinte gradually reduced certain components while increasing other kernel constituents [8].
It is essential to fully utilize and harness genetic resources in particular progenitors and wild relatives since a significant number of genes from wild relatives are valuable for creating genetic variation and population improvement [9]. In addition to teosinte, genus Coix is closest to genus Zea, which is native to South Asia, particularly India, Burma, China and Malaysia. Before maize became a popular staple cereal, coix was widely grown throughout South Asia [10] and has long been utilized as a source of food for humans and animals [11,12]. Coix has also been used as a medicinal herb to treat cancer in China [13] and reduce obesity [14]. Coix (Coix lacryma-jobi L.) is a diploid (2n = 20) and belongs to the Poaceae family, which is most closely related to the genera Zea, Tripsacum and Sorghum [15].
Based on seed coat hardiness, coix is grouped into soft-shelled cultivated species and hard-shelled wild species. Previously, it has been reported that coix contains around 20% protein in seeds, mainly constituting seed storage coix protein belonging to prolamins [16], and apparently, it has been reported as protein-dense species among cereals [17]. Coix has a very recent cultivation history in comparison to sorghum (8000 years ago) and maize (10,000 years ago) as coix was domesticated relatively late (about 7000 years); hence many of its features are still undomesticated [18,19,20].
Historically, introgression of rare alleles from wild relatives of teosinte has resulted in increased genetic gain and significant breeding improvement of maize. Hence, maize germplasm evaluation for different qualitative and quantitative traits has been commonly adapted and has been very effective hitherto. Although phenotypic evaluation is useful in finding desirable genotypes, the need for time and resources has been a main constraint for efficiency; hence, molecular markers’ utilization has been very economical. The use of molecular markers enhances our understanding of germplasm collections. For crop breeding and germplasm management, genome-wide single nucleotide polymorphisms that measure relatedness among distinct inbreds are highly useful [21]. However, SSR markers are also useful to identify the population structure and are considered suitable for association mapping since they are co-dominant, locus-specific, polymorphic, multi-allelic and highly repeatable [22,23]. By utilizing modern breeding tools, we can expedite the breeding process by identifying chromosomal locations and finding significant marker–trait associations through association mapping [24]. Numerous nutritional quality genes have been identified in the past few decades due to extensive genetic analysis and traditional QTL mapping of the nutritional quality of maize kernels [25]. Many studies have identified the genetic determinants of maize kernel quality wherein seven QTLs for protein content [26] and 38 QTLs for kernel quality [18] have been identified. Major genes involved in the synthesis of maize protein are opaque1 (o1) and floury4 (f4), whereas the Mucronate (Mc) gene has been cloned [27,28,29]. Recently, the USDA-ARS group has developed the teosinte-derived near-isogenic lines (NILs) to understand the role of the alleles that determine maize kernel compositional traits using QTL mapping analysis [30]. Even though many QTLs and genes have been identified and cloned, genetic variability studies and chromosomal regions responsible for higher protein content improvement are still scarce and limited. There is limited genetic variability for protein content in cultivated maize, and absolute protein content in cultivated maize is far less than its wild relatives. Thus, higher genetic variability for kernel protein content in teosinte constitutes potential genetic resources to identify likely sites in the genome and to increase the protein content using classical and molecular breeding approaches [8,31,32]. Since maize has been domesticated from teosinte and possesses tremendous genetic variability for several economically important traits [33], hence teosinte accessions must be utilized in marker–trait association studies; however, very few studies have focused on kernel quality so far. Teosintes have twice the protein content than cultivated maize and are assumed to be a huge repository for alleles that control protein content. Several such genes might have been lost during the bottleneck effect of domestication and selective breeding; however, these genes could be rediscovered and reintrogressed [7,34] by understanding their genetic control.
Quantifying kernel protein among different teosinte, coix and maize accessions followed by identifying markers that are linked to genomic regions responsible for genetic diversity may provide valuable information for trait introgression aiming to improve protein content in maize. Therefore, this study hypothesized that uses of SSR markers would allow identifying genomic regions that annotate protein content in wild species and assist in faster allele recovery when incorporated during trait introgression. Our overall goal of this investigation was to domesticate wild alleles from teosinte to enrich protein content in the kernels of cultivated maize. Our specific objectives were to explore the genetic variability for kernel protein content among teosinte, coix and maize accessions as well as identify genomic regions responsible for the variability of kernel protein based on evaluated accessions marker profiles.
2. Materials and Methods
2.1. Experimental Materials
The teosinte accessions used in the investigation were provided by ICAR-National Bureau of Plant Genetic Resources, New Delhi (India). Coix accession was collected locally, and maize accessions belonged to improved inbred lines. A set of 28 accessions was used, which includes two accessions of Zea nicaraguensis species (G1 and G11), five accessions of Zea diploperennis (G2, G9, G10, G15 and G16), 10 accessions of Z. mays subsp. parviglumis (G3, G4, G5, G6, G7, G8, G12, G13, G14 and G28) and seven accessions of Z. mays subsp. Mexicana (G17, G18, G19, G20, G21, G22 and G23). Three maize accessions, namely CAL1444, CML451 and CAL159, were improved inbred lines with parentage of hybrids. Accession G27 of Coix lacryma-jobi was collected from a naturally grown population along the small water tributary of the local wild habitat. These accessions were grown in N.E. Borlaug Crop Research Centre at Pantnagar, where seeds were collected and utilized for further experiments.
2.2. Protein Quantification of Maize and Wild Accessions
Prior to protein analysis, the collected seeds of each teosinte, maize and coix accessions were thoroughly cleaned to remove unfilled and chaffy seeds. The seeds of teosinte and coix accessions were divided into two groups. In the first group, stony seed coats were removed manually and discarded and only kernels were finely ground to a fine powder using a mortar grinder laboratory miller (RM200, Retsch, Haan, Germany) and used for protein estimation. Accessions in the second group comprised teosinte accession seeds that were used along with a stony fruit case for grinding into a fine powder (Figure 1). Since maize kernels are naked, therefore maize accessions were ground into fine powder only once. The fine powder of each accession was weighed 200 mg thrice in three replicates and used further for digestion and protein estimation using the KDI040 Kjeldahl distillation system (Labquest by Borosil® KDI040, Mumbai, India) [35]. The catalyst, which includes a 10:1 combination of K2SO4 and CuSO4, was added into the flask as a part of the digestion mixture, and 5 mL concentrated H2SO4 was then added to the mixture, which helped in the conversion of nitrogen into ammonium sulphate. Then, the flasks were put in a digestion system and cooked for 90 min at 420 °C, plus an additional 20 min for clearing. The heating was then turned off to enable the contents of the flasks to cool before being used for distillation. Nitrogen content in the grains was digested into ammonia, while other organic matter was transformed into CO2 and water. Since ammonia was in the ammonium ion (NH4+) form, which binds to the sulfate ion (SO42−) and stays in the solution, ammonia was not released in an acidic solution. An automated KDI040 Kjeldahl distillation system with 40 percent sodium hydroxide (NaOH) was used for the alkaline distillation. Flasks were carefully removed from the digester and connected to the distillation system for 6 min to release the ammonia from the digested mixture. In the distillation system, a conical flask was attached to the receiving end to trap the ammonia. After NaOH fluid was added to the digesting flask, it turned alkaline, which caused ammonium sulfate to dissolve in ammonia. The released ammonia gas was then trapped into the receiving flask containing 4% excess boric acid with a mixed indicator. Ammonia gas was changed into the ammonium ion by the low pH solution in the receiving flask, and boric acid was changed into the borate ion at the same time. Nitrogen concentration was then calculated by measuring the end-point at which the ammonium borate color changed from green to red, which was performed by titrating the ammonium borate created with standard 0.1 N H2SO4. To calculate nitrogen content, the titer values were recorded. To account for any residual nitrogen that could be in the reagents used to conduct the analysis, a blank sample was often run concurrently along with samples. Once the nitrogen content was established, the proper conversion factor was used to convert it to protein content. Nitrogen content in analyzed samples was estimated by the below-given formula [35], and the crude protein content of the samples was determined by multiplying the nitrogen value by a factor (F) of 6.25.
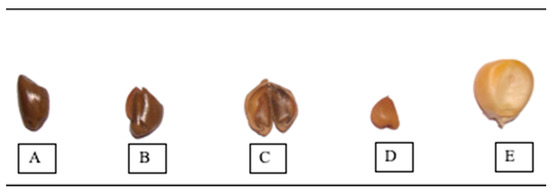
Figure 1.
Illustration of evolution of teosinte seed. (A) Teosinte seed with stony fruit case/seed coat, (B) seed inside stony fruit case, (C) opened seed coat, (D) teosinte kernel and (E) maize kernel.
2.3. Genotyping of Maize and Wild Accessions
The set of 28 accessions used for protein analysis was also analyzed with SSR markers for population structure and marker–trait association studies. The leaf samples of accessions were collected from 30-days-old seedlings and preserved in a deep freezer (−20 °C). Preserved samples were further used for DNA isolation and genomic DNA isolation following the Cetyltrimethyl Ammonium Bromide (CTAB) method with some modifications [36]. A set of 84 SSR random polymorphic markers was selected out of 197 based on their distribution across 10 maize chromosomes from the maize genome database (Table 1). PCR reactions were carried out on Himedia Prima-96™ Thermal Cycler (HiMedia Laboratories, Mumbai, India) and Applied Biosystems Veriti™ 96-Well Thermal Cycler (Applied Biosystems, Inc., Foster City, CA, USA). PCR amplification in a 13.8 μL reaction mixture included 3 μL (200 ng genomic DNA, 0.25 μL Taq DNA polymerase (3 U/μL), 0.35 μL dNTPs mix (2.5 mM each), 1.5 μL Taq DNA polymerase buffer with 15 mM MgCl2 (10×), 0.75 μL forward primer, 0.75 μL reverse primer (10 mM/μL) and 7.2 μL deionized water. The reaction mixture was prepared in thin wall flat-capped PCR tubes of 0.2 mL capacity. The PCR machine was programmed for denaturation at 94 °C for 5 min at the initial stage, followed by 35 cycles at 94 °C for 40 s, primer annealing at 53 to 73 °C for 40 s (differs with primer annealing temperature), elongation at 72 °C for 1 min, and a final extension step at 72 °C for 10 min. After the completion of targeted PCR cycles, the reaction mixtures were immediately transferred to a freezer. Each reaction mixture was added with 2 μL of 6× loading dye and resolved by loading in 3.0 percent agarose gel on a horizontal electrophoresis assembly. A marker of 100 bp size (100 bp ladder) was also run along the PCR samples in each gel. A constant voltage of 50 V was regulated for three hours for the resolution of the amplified profile. The amplification profile on the gel was documented using UVITEC Cambridge Gel Documentation System (Uvitec, Cambridge, UK). The genotypic profile of accessions was developed by scoring gel pictures with a score of “1” denoting amplified areas and “0” denoting unamplifiable regions. For molecular data analysis based on the amplification, DNA bands were scored, and a “0” and “1” matrix was created [37].

Table 1.
Polymorphic markers utilized for genotyping of maize and wild accessions.
2.4. Statistical Analysis
2.4.1. Protein Analysis
The protein content of maize and wild accessions was evaluated using the randomized complete block design (RCBD) and validated by one-way ANOVA using SPSS® Statistics v26 software [38]. Duncan’s new multiple-range test was applied to show significant differences between genotypes.
2.4.2. Population Structure Analysis
The population structure among accessions was identified by the model-based clustering method. The STRUCTURE version 2.3.4 software was used to find sub-populations among accessions utilized for the study. The program creates sub-populations based on genotypic data and assigns each accession to different sub-populations, which indicates the presence of population structure, and identifies migrants and admixtures among accessions. The ‘K’ (unknown) sub-populations in the proposed model each had a unique set of allele frequencies at each locus and were assumed to be in Hardy–Weinberg and linkage equilibrium within sub-populations. Accessions considered for study were grouped into different sub-populations or jointly to two or more sub-populations. The population structure analysis was carried out using the 84 SSR marker data based on the Bayesian model using STRUCTURE 2.3.4 software to calculate the number of sub-populations (K) [39]. An admixture model, 10,000 burn-in iterations and 100,000 Monte Carlo Markov Chain (MCMC) repeats were used in this investigation. Three replications for each genetic cluster (K) ranging from K = 2 to K = 10 were used to test each genotype’s membership [40]. STRUCTURE HARVESTER was used to calculate the ideal number of sub-populations using the delta k technique provided by Earl et al. [41].
2.4.3. Marker–Trait Association Analysis
The TASSEL (Trait Analysis by aSSociation, Evolution, and Linkage) version 5.2.82 was used to perform association analysis [42]. Two statistical models were utilized in TASSEL: the general linear model (GLM) and the mixed linear model (MLM). The difference between these two models lies in association analysis as GLM is based on only Q values obtained from population structure determination, but association analysis based on MLM takes account of relative kinship effects (K) along with population structure; hence, MLM = Q + K. These models were used to determine the genetic associations between phenotypic (protein content) and genotypic (SSR markers) data. Based on their R2 and p-value of significant marker–traits, associations were identified with chromosomal location (bins) association with different phenotypic traits.
3. Results
3.1. Inter- and Intra-Species Protein Content Variation
3.1.1. Inter-Species Variation
The protein content of the evaluated accessions was analyzed using a one-way analysis of variance (ANOVA) with the null hypothesis of no discernible variation between genotypes for protein content. ANOVA results indicate a substantial variation among evaluated accessions for the protein content (Table 2). Teosinte accessions were analyzed with and without seed coats, which indicates significant intra-species variation for protein content between both groups, and the results indicate a significant variation between accession samples with and without seed coats. However, protein content in coix accessions was evaluated in without-seed-coat samples only. The seed coat forms a major portion of the seed weight and contributes 40–50% to the kernel weight, largely because of its thickness (Figure 1). We observed that the protein levels were reduced by 3–6% in teosinte samples with seed coats in comparison to samples without any seed coats; however, these samples were seen to have higher kernel weight (Table 2 and Figure 2). The protein content of teosinte kernels without seed coats was two–three times higher than that of the maize inbred lines CAL1444, CML451 and CAL159.

Table 2.
Mean protein content of maize and wild accessions. a,b,c…Values in columns followed by different letter are significantly different at p < 0.01, Duncan’s multiple range test.
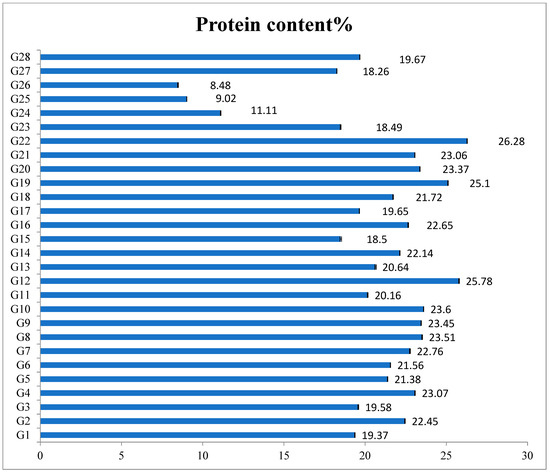
Figure 2.
Protein content of maize and wild accessions (1–28 as mentioned in Table 2).
3.1.2. Intra-Species Variation
Among teosinte accessions, G22 (Z. mays subsp. Mexicana) recorded the highest protein content of 26.29%, and the least of 18.50% was observed in G15 (Z. diploperennis) as shown in Figure 2. Among all evaluated accessions, eighteen, four and three accessions of teosinte showed >20%, >19% and >18% of total protein content, respectively, while maize lines of CAL1444, CML451 and CAL159 showed 9–11% protein content and single coix accession showed 18.26% of protein content. Interestingly, protein content in coix was higher than in the maize lines. Among teosintes, Z. mays subsp. mexicana accessions had higher protein content and were seen to vary from 18.62% to 26.29% in comparison to other teosinte species. Protein content variation in Z. mays subsp. parviglumis also varied from 19.58% to 25.78%, while in Z. diploperennis it varied from 18.50% to 23.60% and Z. nicaraguensis accessions showed protein content from 19.37% (EC938022) to 20.16% (EC944138).
3.2. Population Structure of Maize and Wild Accessions
Analysis of population structure was based on maximum likelihood and DK (Delta K) using marker data of 84 polymorphic SSR markers obtained at K = 4 with the highest DK value of 73.63. Population structure analysis discerned that all accessions were divided into four sub-populations (K = 4) because the mean log-likelihood (LnP(K)) value was 2.48, which was the highest and most stable at K = 4; the standard deviation of log-likelihood (LnP(K)) value was −2772.13, log likelihood of each value of K was 150.86 and mode of the log likelihood of each value of K was 182.50 (Table 3; Figure 3 and Figure 4), wherein each subpopulation has distinct values for protein content and showed clear genetic differences represented in different banding patterns for SSR markers. Sub-population I (Green) had ten accessions (G1, G2, G9, G10, G11, G15, G16, G23, G27 and G28). The sub-population I comprised two accessions of Z. nicaraguensis, five accessions of Z. diploperennis, and one accession of Z. mays subsp. mexicana and one accession each of Coix lacryma-jobi and Z. mays subsp. parviglumis. Sub-population II (Red) grouped four accessions of Z. mays subsp. mexicana and one accession of Z. mays subsp. parviglumis (G13, G19, G20, G21, and G22). Sub-population III comprised eight accessions namely (G3, G4, G5, G6, G7, G8, G17 and G18) with five accessions of Z. mays subsp. parviglumis and three accessions of Z. mays subsp. mexicana. The last sub-population IV (yellow) consisted of five accessions (G12, G14, G24, G25 and G26), of which two accessions were of Z. mays subsp. parviglumis and the remaining three accessions were of maize (Figure 4).

Table 3.
Evanno method statistics to determine the optimal number of subpopulations (K) for a structured study of maize and wild accessions using Structure software. The asterisk indicates the selected population structure. NA means that the values could not be estimated.
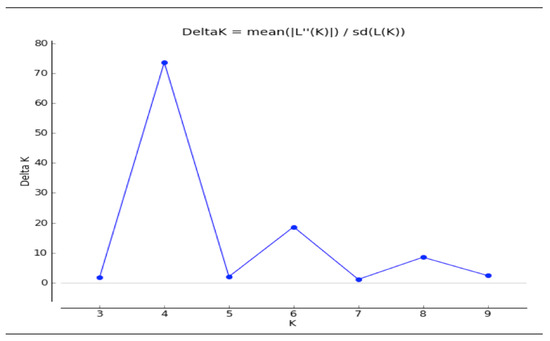
Figure 3.
Indication of ideal number of subpopulations depicted by delta K analysis of population structure.
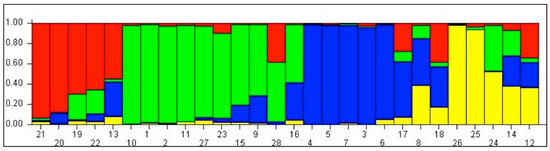
Figure 4.
Structure map for maize and wild accessions (1–28 as mentioned in Table 2) dispersion pattern within K = 4 subpopulations.
3.3. Population Marker Protein Content Association Using Maize and Wild Accessions
Association analysis was performed using TASSEL software, and associations were assessed based on generalized linear model (GLM) and mixed linear model (MLM). MLM analysis was performed based on combining population structure (Q) and genetic marker-based kinship matrix (K), and statistical strength was increased compared to “Q” alone as in the case of GLM. Seven marker–trait associations located on different chromosomal bins having p < 0.01 were identified based on GLM (Table 4), but using MLM, we did not find any significant associations. The Manhattan plot and QQ plot were created based on GLM for protein content of 28 accessions (Figure 5 and Figure 6). Quantile–Quantile (QQ) plot compared two probability distributions graphically by placing their respective quantiles against one another. The QQ plot based on GLM was obtained and depicts four sub-populations among accessions (Figure 6). Markers that showed a significant association with protein content are presented in Table 1, and a total of seven marker–trait associations showed significant association (p < 0.01) and were distributed on chromosome 4, 5, 7, 8 and 9. Correlation (R2) between associated markers and protein content varied from 0.22 (umc1171b) to 0.76 (umc1294c), with most trait-marker associations very weak.

Table 4.
Marker–trait associations having p < 0.01 for protein content using maize and wild accessions using TASSEL software based on GLM.
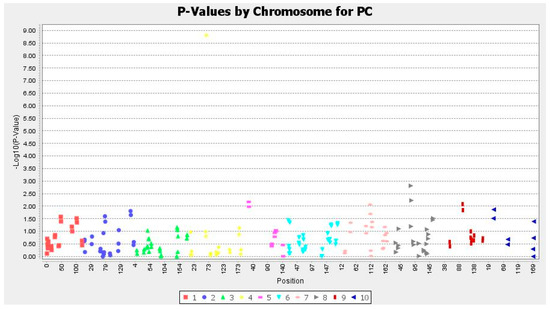
Figure 5.
Manhattan plot created based on generalized linear model (GLM) for protein content of 28 maize and wild accessions using SSR markers.
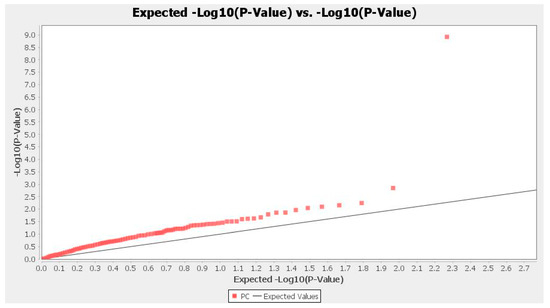
Figure 6.
QQ plot of the analysis for protein content in maize and wild accessions based on GLM analysis using TASSEL software.
4. Discussion
In major crops, genetic diversity for nutritional quality attributes has been limited due to continued selective breeding for yield and yield components to increase production and productivity [43]. In addition to natural selection, domestication and changing environmental conditions have evolved plants to be more adaptive as well as generate the variability for plant architectural, stress adaptation and kernel compositional traits simultaneously. Overall, this continued process has resulted in increased nutritional composition of some traits while drastically reducing others [44]. Maize is a notable example that has discerned domestication effects such as loss of seed coat in cultivated maize in comparison to its wild ancestor teosinte [45]. When it comes to kernel composition in teosinte, starch accumulation is lesser with elevated protein concentration, whereas vice versa in cultivated maize [7]. During domestication, dramatic changes occurred that led maize kernels to lose their stony fruit case. Worldwide, researchers have tried to comprehend the theoretical scenario of this process, and teosinte glume architecture1 appears to be associated with it [46]. Paulis and Wall have analyzed the responsible amino acids that influence protein accumulation in teosinte, tripsacum and maize lines and noted that protein accumulation is more than double in wild progenitors in comparison to cultivated maize [8]. Among these wild accessions, tripsacum (29.3%) has higher protein content than teosinte (28.7%). Similarly, Flint-Garcia et al. also compared kernel traits of maize inbred lines, local heirlooms and teosinte and observed that the teosinte accessions have protein contents ranging from 26.49% to 30.72% [7].
Interestingly, coix (Coix lacryma-jobi) is closely related to cereal crops including Zea and Sorghum genera and possesses higher protein content than cultivated maize. Our results also comparatively correspond with the observations of Venkateswarlu and Chaganti [16] and Ottoboni et al. [17] wherein coix reportedly had 20% protein content. Recently, Feng et al. highlighted the medicinal and edible properties of coix and indicated the presence of significantly higher amount of protein [47]. Among 28 accessions evaluated in this study, teosinte and coix accessions were noted to have higher protein content than the maize inbred lines. Accessions of Zea mays subsp. parviglumis, mexicana, diploperennis and nicaraguensis possess higher protein content than cultivated species, and our findings are in alignment with earlier reported studies. Significant differences in the protein content among wild relatives and cultivated species are probably due to the loss of favorable alleles during their domestication and selective breeding that led to the evolution of modern high-yielding maize cultivars [7,34]. Re-domestication of such wild allelic forms may be of great significance in maize biofortification. Teosinte, the wild progenitor of maize, therefore can serve as a crucial resource for domestication, evolution and genetics studies of maize kernel composition [48,49,50]. Most of these wild species are not extensively investigated especially for kernel quality parameters, and their potential remains untapped. Thus, wild genetic resources offer a strong foundation for the introgression, cloning, and transformation of agronomically favorable genes and QTLs from wild to cultivated species [51].
Molecular markers are key to locate genomic regions that control key traits of economic interest and have been very effective in studying and demonstrating the genetic variation present in wild species [52], and an allele-specific precision strategy can be employed to harvest genetic variability in the existing breeding programs [53]. Population structure analysis in our study indicated four sub-populations with some sort of admixtures with Z. mays subsp. parviglumis and Z. mays subsp. mexicana among all 28 evaluated accessions. Based on the population structure analysis, Fukunaga et al. have also noticed parviglumis–mexicana admixtures in annual teosinte plants grown in the Balsas River valley [54]. Weber et al. conducted an association mapping to identify major regulatory genes involved in variation between teosinte and maize that were collected from the Balsas River valley and found 10 significant associations between five candidate genes for plant and inflorescence architecture [33]. Subsequently, the same research group increased the number of individuals and marker density for more traits compared to the previous study and found similar results [55]. With high mapping resolution, the ability to sample many alleles and the use of naturally existing populations, association mapping is a valuable technique to investigate gene functions that are involved in the genetic architecture of complex characteristics [55,56,57]. In the case of protein content, Jadhav et al. studied marker–trait association in chickpeas and reported two significant QTLs that are linked to TR26.205 and CaM1068.195 markers present on chromosomes 3 and 5, respectively [58].
In maize, numerous QTLs have been identified for protein content, but only a few reports are available on marker–trait association in maize, especially using teosinte [32]. This investigation analyzed teosinte accessions to determine marker–trait associations for protein content and to localize the genomic regions affecting protein content on different chromosomes based on GLM. In the case of MLM, we did not find marker–trait associations, which is likely due to low population size and the limited number of markers utilized for genotyping affecting marker density and the likelihood of detecting any significant association between a marker and concerned trait. Additionally, other research has also reported marker–trait associations in maize for protein, starch, oil [24] and kernel zinc and iron content [59]. Similarly, kernel compositional marker–trait associations for oil and protein content have been studied using genome-wide association in soybean [60] and chickpea (Jadhav et al., 2015, [58]; Karaca et al., 2019, [61]) and for iron and zinc content [62] in lentils. Apart from kernel constituents of oil and protein, marker–trait associations have also extensively been studied for agronomical [63], inflorescence and leaf architecture [64] and grain-quality traits [25] in diverse maize breeding populations. In this study, markers umc1294, umc1171, phi091, umc2182 and bnlg292 exhibited significant associations in genomic regions localized on the 4, 5, 7, 8 and 9 chromosome bins, respectively, that influenced the kernel protein content. Guo et al. also observed QTL on chromosome 5 between markers umc1221–umc2026, which encode protein content accumulation [65]. The teosinte high protein 9 (Thp9) QTL that encodes the enzyme asparagine synthetase 4 located on chromosome 9 has been reported in Zea mays ssp. parviglumis [32]. Several studies that screened wild relatives have shown significant variation for total protein content well beyond that observed in cultivated maize varieties. Hence, the availability of wide variation in protein content among wild species creates opportunities to investigate mechanisms involved in domestication and alleles responsible for the diversification of protein content and allelic diversity, which could likely be exploited for trait introgression in the future. Potential marker–trait association will be very useful to discover useful alleles and their locations in the genome. Wild relatives to the likes of teosinte and tripsacum will be very resourceful to unravel significant marker–trait associations and generate the genetic variability much needed for improved grain quality and overall maize improvement.
5. Conclusions
Teosinte and coix are proven rich allelic sources for higher protein content and serve as potential genetic resources for trait improvement that are hitherto untapped and not fully explored for enhancing protein content in modern maize. The extensive evaluation of 28 accessions comprising teosinte, maize breeding lines and coix was utilized to study SSR marker and protein content association using GLM. Studied accessions were grouped into four diverse subpopulations based on population structure analysis. SSR markers umc1294, umc1171, phi091, umc2182 and bnlg292 distributed across chromosomes 4, 5, 7, 8 and 9, respectively, showed significant association for protein content. The results may be of great significance in pre-breeding as well as in the breeding of maize lines for higher kernel protein content after marker validation. This research has laid the foundation to further develop the mapping population for identifying genomic regions associated with protein content variation; hence, teosinte accessions identified with higher protein content can be further used to develop such a mapping population. Additional research will be required in order to validate these associations as well as to unravel the genetic architecture of complex characteristics and the precise function of key regulatory genes. The findings also suggest that selection in wild species played an important role during domestication for protein content, and that existing genetic variability present in teosinte will be efficiently utilized in the near future for enhancing protein content in maize kernels.
Author Contributions
Conceptualization, N.K.S. and V.S.; methodology, N.K.S. and V.S.; validation, N.K.S.; formal analysis, N.K.S. and V.S.; investigation, N.K.S., V.S., S.S., N.P., P.G., S.V. and S.K.; resources, N.K.S., V.S., J.P.J., S.R.J. and A.N.N.; data curation, N.K.S. and V.S.; writing—original draft preparation, V.S.; writing—review and editing, V.S., S.S., N.K.S., N.P., P.G., S.V., S.K., J.P.J., S.R.J. and A.N.N.; visualization, N.K.S., V.S. and A.N.N.; supervision, N.K.S.; project administration, N.K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
All the data are provided in the presented manuscript.
Acknowledgments
We are grateful to ICAR-AICRP Maize and G.B.P.U.A. and T., Pantnagar for providing resources and facilities for the investigation. ICAR Senior Research Fellowship to the senior author is duly acknowledged. We would also like to acknowledge the support of the European Union’s Horizon 2020 research and innovation program, project PlantaSYST (SGACSANo. 739582 under FPA No. 664620) and the European Regional Development Fund through the Bulgarian “Science and Education for Smart Growth” Operational Programme (project BG05M2OP001-1.003-001-C01).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Strable, J.; Scanlon, M.J. Maize (Zea mays): A Model Organism for Basic and Applied Research in Plant Biology. Cold Spring Harb. Protoc. 2009, 10, pdb-emo132. [Google Scholar] [CrossRef]
- Tian, F.; Stevens, N.M.; Buckler, E.S., IV. Tracking footprints of maize domestication and evidence for a massive selective sweep on chromosome 10. Proc. Natl. Acad. Sci. USA 2009, 106, 9979–9986. [Google Scholar] [CrossRef] [PubMed]
- Gaut, B.S.; Doebley, J.F. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 1997, 94, 6809–6814. [Google Scholar] [CrossRef]
- Wei, F.; Coe, E.D.; Nelson, W.; Bharti, A.K.; Engler, F.; Butler, E.; Kim, H.; Goicoechea, J.L.; Chen, M.; Lee, S.; et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007, 3, e123. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Vigouroux, Y.; Goodman, M.M.; Sanchez, G.J.; Buckler, E.; Doebley, J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 2002, 99, 6080–6084. [Google Scholar] [CrossRef]
- Mammadov, J.; Buyyarapu, R.; Guttikonda, S.K.; Parliament, K.; Abdurakhmonov, I.Y.; Kumpatla, S.P. Wild relatives of maize, rice, cotton, and soybean: Treasure troves for tolerance to biotic and abiotic stresses. Front. Plant Sci. 2018, 9, 886. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Bodnar, A.L.; Scott, M.P. Wide variability in kernel composition, seed characteristics, and zein profiles among diverse maize inbreds, landraces, and teosinte. Theor. Appl. Genet. 2009, 119, 1129–1142. [Google Scholar] [CrossRef] [PubMed]
- Paulis, J.W.; Wall, J.S. Comparison of the protein compositions of selected corns and their wild relatives, teosinte and Tripsacum. J. Agric. Food Chem. 1977, 25, 265–270. [Google Scholar] [CrossRef]
- Wang, L.; Xu, C.; Qu, M.; Zhang, J. Kernel amino acid composition and protein content of introgression lines from Zea mays ssp. mexicana into cultivated maize. J. Cereal Sci. 2008, 48, 387–393. [Google Scholar] [CrossRef]
- Arora, R.K. Job’s-tears (Coix lacryma-jobi)-a minor food and fodder crop of northeastern India. Econ. Bot. 1977, 31, 358–366. [Google Scholar] [CrossRef]
- Wester, P.S. Notes on Adlay. Philip. Agric. Rev. 1920, 13, 217–222. [Google Scholar]
- Schaffhausen, R.V. Adlay or Job’s tears—A cereal of potentially greater economic importance. Econ. Bot. 1952, 6, 216–227. [Google Scholar] [CrossRef]
- Woo, J.H.; Li, D.; Wilsbach, K.; Orita, H.; Coulter, J.; Tully, E.; Kwon, T.K.; Xu, S.; Gabrielson, E. Coix seed extract, a commonly used treatment for cancer in China, inhibits NFκB and protein kinase C signaling. Cancer Biol. Ther. 2007, 6, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.O.; Yun, S.J.; Jung, B.; Lee, E.H.; Hahm, D.H.; Shim, I.; Lee, H.J. Hypolipidemic effects of crude extract of adlay seed (Coix lachrymajobi var. mayuen) in obesity rat fed high fat diet: Relations of TNF-α and leptin mRNA expressions and serum lipid levels. Life Sci. 2004, 75, 1391–1404. [Google Scholar] [CrossRef]
- Kellogg, E.A.; Watson, L. Phylogenetic studies of a large data set. I. Bambusoideae, Andropogonodae, and Pooideae (Gramineae). Bot. Rev. 1993, 59, 273–343. [Google Scholar] [CrossRef]
- Venkateswarlu, J.; Chaganti, R.S.K. Job’s tears (Coix lacryma-jobi L.). ICAR Tech. Bull. 1973, 44, 54–59. [Google Scholar]
- Ottoboni, L.M.; Leite, A.; Targon, M.L.; Crozier, A.; Arruda, P. Characterization of the storage protein in seed of Coix lacryma-jobi var. Adlay. J. Agric. Food Chem. 1990, 38, 631–635. [Google Scholar] [CrossRef]
- Wang, J.; Liu, L.; Ball, T.; Yu, L.; Li, Y.; Xing, F. Revealing a 5,000-y-old beer recipe in China. Proc. Natl. Acad. Sci. USA 2016, 113, 6444–6448. [Google Scholar] [CrossRef]
- Fu, Y.H.; Yang, C.; Meng, Q.; Liu, F.; Shen, G.; Zhou, M.; Ao, M. Genetic diversity and structure of Coix lacryma-jobi L. from its world secondary diversity center, Southwest China. Int. J. Genom. 2019, 2019, 9815697. [Google Scholar] [CrossRef]
- Liu, H.; Shi, J.; Cai, Z.; Huang, Y.; Lv, M.; Du, H.; Gao, Q.; Zuo, Y.; Dong, Z.; Huang, W.; et al. Evolution and domestication footprints uncovered from the genomes of Coix. Mol. Plant 2020, 13, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.L. Molecular markers and marker-assisted breeding in plants. Plant Breed. Lab. Fields 2013, 3, 45–83. [Google Scholar] [CrossRef]
- Park, J.H.; Suresh, S.; Piao, X.M.; Cho, G.T.; Lee, S.Y.; Baek, H.J.; Lee, C.W.; Chung, J.W. Application of simple sequence repeat (SSR) markers for the discrimination of Korean and Chinese sesame (Sesamum indicum L.) accessions. Plant Breed. Biotech. 2014, 2, 80–87. [Google Scholar] [CrossRef]
- Kumar, B.; Choudhary, M.; Kumar, P.; Kumar, K.; Kumar, S.; Singh, B.K.; Lahkar, C.; Kumar, P.; Dar, Z.A.; Devlash, R.; et al. Population Structure Analysis and Association Mapping for Turcicum Leaf Blight Resistance in Tropical Maize Using SSR Markers. Genes 2022, 13, 618. [Google Scholar] [CrossRef] [PubMed]
- Dudley, J.W.; Dijkhuizen, A.; Paul, C.; Coates, S.T.; Rocheford, T.R. Effects of random mating on marker–QTL associations in the cross of the Illinois high protein × Illinois low protein maize strains. Crop Sci. 2004, 44, 1419–1428. [Google Scholar] [CrossRef]
- Zheng, Y.; Yuan, F.; Huang, Y.; Zhao, Y.; Jia, X.; Zhu, L.; Guo, J. Genome-wide association studies of grain quality traits in maize. Sci. Rep. 2021, 11, 9797. [Google Scholar] [CrossRef]
- Li, Y.; Dong, Y.; Niu, S.; Cui, D.; Wang, Y.; Wei, M.; Li, X.; Fu, J.; Zhang, Z.; Chen, H.; et al. QTL identification of kernel composition traits with popcorn using both F2: 3 and BC2F2 populations developed from the same cross. J. Cereal Sci. 2008, 48, 625–631. [Google Scholar] [CrossRef]
- Wang, G.; Wang, F.; Wang, G.; Wang, F.; Zhang, X.; Zhong, M.; Zhang, J.; Lin, D.; Tang, Y.; Xu, Z.; et al. Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 2012, 24, 3447–3462. [Google Scholar] [CrossRef]
- Wang, G.; Qi, W.; Wu, Q.; Yao, D.; Zhang, J.; Zhu, J.; Wang, G.; Wang, G.; Tang, Y.; Song, R. Identification and characterization of maize floury4 as a novel semidominant opaque mutant that disrupts protein body assembly. Plant Physiol. 2014, 165, 582–594. [Google Scholar] [CrossRef]
- Kim, C.S.; Gibbon, B.C.; Gillikin, J.W.; Larkins, B.A.; Boston, R.S.; Jung, R. The maize Mucronate mutation is a deletion in the 16-kDa γ-zein gene that induces the unfolded protein response 1. Plant J. 2006, 48, 440–451. [Google Scholar] [CrossRef]
- Karn, A.; Gillman, J.D.; Flint-Garcia, S.A. Genetic analysis of teosinte alleles for kernel composition traits in maize. G3 Genes Genomes Genet. 2017, 7, 1157–1164. [Google Scholar] [CrossRef]
- Sood, S.; Flint-Garcia, S.; Willcox, M.C.; Holland, J.B. Mining natural variation for maize improvement: Selection on phenotypes and genes. In Genomics of Plant Genetic Resources; Springer: Dordrecht, The Netherlands, 2014; Volume 1, pp. 615–649. [Google Scholar] [CrossRef]
- Wu, Y.; Huang, Y.; Wang, H.; Zhu, Y.; Huang, X.; Li, S.; Wu, X.; Zhao, Y.; Bao, Z.; Qin, L.; et al. Teosinte high protein 9 enhances the seed protein content and nitrogen utilization efficiency in maize. Nature 2022, 612, 292–300. [Google Scholar] [CrossRef]
- Weber, A.; Clark, R.M.; Vaughn, L.; de Jesus Sánchez-Gonzalez, J.; Yu, J.; Yandell, B.S.; Bradbury, P.; Doebley, J. Major regulatory genes in maize contribute to standing variation in teosinte (Zea mays ssp. parviglumis). Genetics 2007, 177, 2349–2359. [Google Scholar] [CrossRef]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Chhonkar, P.K.; Pande, R.N. Soil reaction in soil, plant, water analysis method: Manual. IARI ICAR New Delhi 1999, 1, 11–13. [Google Scholar]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Anandan, A.; Nagireddy, R.; Sabarinathan, S.; Bhatta, B.B.; Mahender, A.; Vinothkumar, M.; Parameswaran, C.; Panneerselvam, P.; Subudhi, H.; Meher, J.; et al. Multi-trait association study identifies loci associated with tolerance of low phosphorus in Oryza sativa and its wild relatives. Sci. Rep. 2022, 12, 4089. [Google Scholar] [CrossRef]
- Mehta, C.R.; Patel, N.R. IBM SPSS Exact Tests; IBM Corporation: Armonk, NY, USA, 2011; Volume 17, pp. 23–24. Available online: https://www.sussex.ac.uk/its/pdfs/SPSS_Exact_Tests_19.pdf (accessed on 12 July 2023).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Fu, Y.B. Understanding crop genetic diversity under modern plant breeding. Theor. Appl. Genet. 2015, 128, 2131–2142. [Google Scholar] [CrossRef]
- Van Tassel, D.L.; Tesdell, O.; Schlautman, B.; Rubin, M.J.; DeHaan, L.R.; Crews, T.E.; Streit Krug, A. New food crop domestication in the age of gene editing: Genetic, agronomic and cultural change remain co-evolutionarily entangled. Front. Plant Sci. 2020, 11, 789. [Google Scholar] [CrossRef]
- Wang, H.; Nussbaum-Wagler, T.; Li, B.; Zhao, Q.; Vigouroux, Y.; Faller, M.; Bomblies, K.; Lukens, L.; Doebley, J.F. The origin of the naked grains of maize. Nature 2005, 436, 714–719. [Google Scholar] [CrossRef]
- Dorweiler, J.; Stec, A.; Kermicle, J.; Doebley, J. Teosinte glume architecture 1: A genetic locus controlling a key step in maize evolution. Science 1993, 262, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Zhao, Y.; Zhang, Z.; Zhang, S.; Zhang, H.; Yu, M.; Ma, Y. The edible and medicinal value of Coix lacryma-jobi and key cultivation techniques for high and stable yield. Nat. Resour. 2020, 11, 569–575. [Google Scholar] [CrossRef]
- Doebley, J. Molecular evidence and the evolution of maize. Econ. Bot. 1990, 44, 6–27. [Google Scholar] [CrossRef]
- Evans, M.M.; Kermicle, J.L. Teosinte crossing barrier1, a locus governing hybridization of teosinte with maize. Theor. Appl. Genet. 2001, 103, 259–265. [Google Scholar] [CrossRef]
- Sanz-Alferez, S.; San Miguel, P.; Jin, Y.K.; Springer, P.S.; Bennetzen, J.L. Structure and evolution of the Cinful retrotransposon family of maize. Genome 2003, 46, 745–752. [Google Scholar] [CrossRef]
- Nevo, E.; Chen, G. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ. 2010, 33, 670–685. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Zou, C.; Lu, Y.; Xie, C.; Zhang, X.; Prasanna, B.M.; Olsen, M.S. Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 2017, 68, 2641–2666. [Google Scholar] [CrossRef]
- Bohra, A.; Kilian, B.; Sivasankar, S.; Caccamo, M.; Mba, C.; McCouch, S.R.; Varshney, R.K. Reap the crop wild relatives for breeding future crops. Trends Biotechnol. 2022, 40, 412–431. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, K.; Hill, J.; Vigouroux, Y.; Matsuoka, Y.; Sanchez, G.J.; Liu, K.; Buckler, E.S.; Doebley, J. Genetic diversity and population structure of teosinte. Genetics 2005, 169, 2241–2254. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.L.; Briggs, W.H.; Rucker, J.; Baltazar, B.M.; de Jesus Sánchez-Gonzalez, J.; Feng, P.; Buckler, E.S.; Doebley, J. The genetic architecture of complex traits in teosinte (Zea mays ssp. parviglumis): New evidence from association mapping. Genetics 2008, 180, 1221–1232. [Google Scholar] [CrossRef]
- Myles, S.; Peiffer, J.; Brown, P.J.; Ersoz, E.S.; Zhang, Z.; Costich, D.E.; Buckler, E.S. Association mapping: Critical considerations shift from genotyping to experimental design. Plant Cell 2009, 21, 2194–2202. [Google Scholar] [CrossRef]
- Yan, J.; Warburton, M.; Crouch, J. Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci. 2011, 51, 433–449. [Google Scholar] [CrossRef]
- Jadhav, A.A.; Rayate, S.J.; Mhase, L.B.; Thudi, M.; Chitikineni, A.; Harer, P.N.; Jadhav, A.S.; Varshney, R.K.; Kulwal, P.L. Marker-trait association study for protein content in chickpea (Cicer arietinum L.). J. Genet. 2015, 94, 279–286. [Google Scholar] [CrossRef]
- Hindu, V.; Palacios-Rojas, N.; Babu, R.; Suwarno, W.B.; Rashid, Z.; Usha, R.; Saykhedkar, G.R.; Nair, S.K. Identification and validation of genomic regions influencing kernel zinc and iron in maize. Theor. Appl. Genet. 2018, 131, 1443–1457. [Google Scholar] [CrossRef]
- Hwang, E.Y.; Song, Q.; Jia, G.; Specht, J.E.; Hyten, D.L.; Costa, J.; Cregan, P.B. A genome-wide association study of seed protein and oil content in soybean. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef]
- Karaca, N.; Ates, D.; Nemli, S.; Ozkuru, E.; Yilmaz, H.; Yagmur, B.; Kartal, C.; Tosun, M.; Ozdestan, O.; Otles, S.; et al. Genome-Wide Association Studies of Protein, Lutein, Vitamin C, and Fructose Concentration in Wild and Cultivated Chickpea Seeds. Crop Sci. 2019, 59, 2652–2666. [Google Scholar] [CrossRef]
- Khazaei, H.; Podder, R.; Caron, C.T.; Kundu, S.S.; Diapari, M.; Vandenberg, A.; Bett, K.E. Marker–trait association analysis of iron and zinc concentration in lentil (Lens culinaris Medik.) seeds. TPG 2017, 10, plantgenome2017.02.0007. [Google Scholar] [CrossRef]
- Yang, N.; Lu, Y.; Yang, X.; Huang, J.; Zhou, Y.; Ali, F.; Wen, W.; Liu, J.; Li, J.; Yan, J. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet. 2014, 10, e1004573. [Google Scholar] [CrossRef] [PubMed]
- Rice, B.R.; Fernandes, S.B.; Lipka, A.E. Multi-trait genome-wide association studies reveal loci associated with maize inflorescence and leaf architecture. Plant Cell Physiol. 2020, 61, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yang, X.; Chander, S.; Yan, J.; Zhang, J.; Song, T.; Li, J. Identification of unconditional and conditional QTL for oil, protein and starch content in maize. Crop J. 2013, 1, 34–42. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).