Abstract
4-Coumarate: coenzyme A ligase (4CL; EC 6.2.1.12) is an important enzyme in the phenylpropanoid metabolic pathway that controls the biosynthesis of lignin and flavonoids. In this study, to identify the function of the Ag4CL3 gene of celery, the Ag4CL3 gene was cloned from celery cv. “Nanxuan Liuhe Ziqin”. Sequence analysis results showed that the Ag4CL3 gene contained an open reading frame (ORF) with a length of 1688 bp, and 555 amino acids were encoded. The Ag4CL3 protein was highly conserved among different plant species. Phylogenetic analysis demonstrated that the 4CL proteins from celery and carrot belonged to the same clade. The Ag4CL3 protein was mainly composed of 31.89% α-helixes, 18.02% extended strands, 6.67% β-turns, and 43.42% random coils, and the signal peptide was unfound. A total of 62 phosphorylation sites and a class-I superfamily of adenylate-forming domains were found. As the growth time increased, the plant height and stem thickness also increased, and the petiole lignin content increased and became lignified gradually. The relative expression levels of the Ag4CL3 gene in “Nanxuan Liuhe Ziqin” petioles were higher than those in other tissues, with the highest level occurring 70 d after sowing. The lignin contents in the transgenic Arabidopsis thaliana lines hosting the Ag4CL3 gene were higher than those in the WT. In this study, the overexpression of Ag4CL3 led to the significant upregulation of lignin biosynthesis gene expression in transgenic A. thaliana plants, except for AtPAL, AtCCR, and AtLAC. This study speculates that Ag4CL3 genes are related to lignin synthesis in A. graveolens.
1. Introduction
Celery (Apium graveolens L.) is a widely cultivated Apiaceae vegetable worldwide [1], and its leaves and petioles are the main edible tissues. The medicinal value and rich nutrients of celery [2,3], such as dietary fiber [4], anthocyanin [5,6], and apigenin [7,8,9], have been reported. Dietary fiber is a polysaccharide that is not easily digested and includes cellulose, hemicellulose, and lignin [10].
Lignin is an important secondary metabolite and is the second largest biopolymer in the world. In particular, lignin accounts for 30% of the organic carbon content in the biosphere [11]. Moreover, lignin is a complex phenolic polymer that is composed of p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol, which are three alcohol monomers [12]. Its structure is relatively stable and it is one of the main components that make up the skeleton of woody and herbaceous plants. Monomers in the chemical composition of lignin have three major types: p-hydroxyphenyl (H) lignin, formed by the polymerization of p-hydroxyphenylpropane structural monomers; guaiacyl (G) lignin, formed by the polymerization of guaiacyl propane structural monomers; and syringal (S) lignin, formed by the polymerization of syringal propane structural monomers [13].
Lignin is an insoluble dietary fiber that naturally exists in vegetables and fills the cellulose frame. It is an important physiological function that enhances the mechanical strength of plants, assists in the transport of water in tissues, and improves the resistance of plants to stress [12]. The lignin content dynamically changes during plant growth and development, the vegetable tissue gradually becomes solid and rough, and the tissue becomes lignified and not easy to chew, which seriously reduces vegetable quality and affects taste [14].
The 4-Coumarate acid: coenzyme A ligase (4CL; EC 6.2.1.12) is related to lignin biosynthesis [15], which can catalyze six phenolic acids, namely, coumaric acid, cinnamic acid, caffeic acid, ferulic acid, 5-hydroxy ferulic acid, and sinapic acid, to generate corresponding CoA lipids: cinnamoyl CoA, coumaryl CoA, caffeyl CoA, feruloyl CoA, 5-hydroxyl feruloyl CoA, and sinapol CoA. These CoA lipids are precursors of lignin synthesis, and coumarin coenzyme A is a common precursor of lignin and flavonoid synthesis [16]. Therefore, 4CL is at the turning point of the phenylpropanoid metabolic pathway to form different types of products, which is related to the trend of the lignin-specific pathway [17].
In previous research, 4CL genes have been cloned and studied in a large variety of plants, where they comprise small gene families in most cases [18]. For example, parsley (Petroselinum crispum) has two 4CL genes [19], tobacco (Nicotiana tabacum) and mulberry (Morus alba) have three 4CL genes [20,21], the Arabidopsis genome [22] and soybean (Glycine max) [23] have four 4CL genes, and the rice (Oryza sativa) genome has five 4CL genes [24]. Phylogenetic analysis shows that 4CLs in dicotyledonous plants can be divided into two categories: class I and class II. Class-I 4CLs are mainly involved in the biosynthesis of lignin, whereas class-II 4CLs are often involved in pathways other than lignin in the phenylpropanoid pathway. For example, in Arabidopsis, At4CL1 and At4CL2 mainly participate in the synthesis of lignin, whereas At4CL3 mainly affects the synthesis of flavonoids [25]. Similar to the situation in Arabidopsis, Pt4CL1 is highly expressed in the xylem of Populus euphratica, which is mainly involved in lignin biosynthesis; Pt4CL2 is expressed in the epidermis of leaves and stems, and participates in the biosynthesis of phenolic substances, such as flavonoids [26].
In celery, the function of the Ag4CL gene in lignin biosynthesis is unclear. In this study, we cloned the Ag4CL3 gene from celery and detected its expression patterns in different tissues and growth stages of celery. Transgenic A. thaliana plants overexpressing Ag4CL3 were obtained to examine the lignin level, and then comparison of lignin content between transgenic and wild type Arabidopsis. This study further identified the roles of Ag4CL3 in lignin biosynthesis, laying a foundation for future studies on the regulation of lignin synthesis and breeding new cultivars.
2. Materials and Methods
2.1. Plant Material and Experimental Design
Celery cv. “Nanxuan Liuhe Ziqin”, Columbia wild type A thaliana ecotype (WT), and transgenic A thaliana were grown in pots that contained a mixture of soil, vermiculite, and perlite at the Institute of Horticulture, Guizhou Academy of Agricultural Sciences (106.67° E, 26.51° N). The celery and A thaliana plants were grown at 25/18 °C (day/night) for 16/8 h in a phytotron with a relative humidity of 70~75%, and the light intensity was 300 μmol m−2·s−1 during daytime. The petioles of celery were collected 30 d (seedling stage), 50 d (growth period), and 70 d (commercial stage) after sowing. The roots, leaves, and petioles of celery were collected 70 d (commercial stage) after sowing. All the samples were immediately frozen in liquid nitrogen and then stored at −80 °C for RNA extraction. WT (wild type) and Ag4CL3-OE lines of A. thaliana were grown on a chat/vermiculite/perlite mixture. Each experiment was performed in three biological replicates.
2.2. Determination of Lignin Content
The lignin contents of the purple celery petioles and A. thaliana rosette leaves were extracted and measured in accordance with previous research methods [27,28]. The sample was ground in liquid nitrogen with a mortar and pestle, approximately 1 g of the sample was ground in 99.7% ethanol, and the mixture was centrifuged at 14,000 rpm·min−1 for 20 min. The sediment was collected and air-dried at room temperature overnight. Approximately 10 mg of the dried sediment was weighed and transferred to a 2 mL clean centrifuge tube. Next, 1 mL of 2 M HCl and 0.1 mL of thioglycolic acid were added. The tube with the mixture was incubated at 100 °C for 8 h, cooled on ice, and then centrifuged at 14,000 rpm/min for 20 min at 4 °C. The sediment was washed with 1 mL of deionized water and dissolved in 1 mL of 1 M NaOH. It was incubated at 25 °C for 18 h, and centrifuged at 14,000 rpm/min for 20 min, and the supernatant was transferred to a new 2 mL centrifuge tube. About 1 mL of concentrated HCl was added, and the mixture was kept at 4 °C for 6 h to precipitate the lignin thioglycolic acid. After centrifugation at 14,000 rpm/min for another 20 min, the sediment was dissolved in 1 mL of 1 M NaOH. Finally, the 1 M NaOH solution was used as a blank control, and the absorbance value was measured at 280 nm.
2.3. Total RNA Extraction and cDNA Synthesis
The total RNA of celery and A. thaliana was extracted using a total RNA extraction kit (Huayueyang, Beijing, China), according to the instructions. Then, the RNA was converted into cDNA using a HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, Nanjing, China), based on the manufacturer’s instruction.
2.4. Bioinformatics Analysis
The 4CL3 protein sequences of other species were downloaded from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/) (accessed on 24 November 2022). The primers were designed and obtained by Primer Premier 6.0. BioXM software, which was used to analyze the nucleotide and encoded amino acid sequences of the Ag4CL3 gene. The secondary and tertiary structures of the Ag4CL3 protein were predicted and established using SOPMA software (http://pbil.ibcp.fr/) (accessed on 24 November 2022) and a SWISS models server (https://swissmodel.expasy.org/) (accessed on 24 November 2022), respectively. The conserved functional domain of the Ag4CL3 protein was predicted by the CDD of NCBI. To investigate the phylogenetic relationships of the 4CL proteins, MEGA7.0 software was used to construct a phylogenetic tree using the maximum likelihood method, Jones-Taylor-Thornton (JTT) model, Gamma Distributed (G). According to the 4CL classification of Arabidopsis thaliana, Glycine max, and Populus tremuloides, Ag4CL3 was classified and predicted.
2.5. Overexpression Vector Construction and A. thaliana Transformation
The putative Ag4CL3 gene sequence was retrieved from the celery genome and transcriptome database [29,30]. The full length ORF (open reading frame) of Ag4CL3 was amplified with specific primers (Ag4CL3-KpnI-F: 5′-GCGGGTCGACGGTACCATGCCAAGTC-TCAGCCAATC-3′; Ag4CL3-KpnI-R: 5′-TAGACATATGGGTACCTTATAATCTTGCTG-AGGCTCC-3′). The PCR product was cloned into pCAMBIA2301 and sequenced. The recombinant plasmid (35S: Ag4CL3) was introduced into Agrobacterium tumefaciens strain GV3101 via the electroporation method. The floral dip method was used for the Agrobacterium-mediated transformation of A. thaliana [31]. Transgenic A. thaliana was initially screened on MS medium containing kanamycin (50 mg·L−1), and then PCR amplification and sequencing were conducted using Ag4CL3 recombinant primers.
2.6. Real-Time Quantitative PCR Analysis
Real-time quantitative PCR (RT-qPCR) was conducted to detect the expression level of Ag4CL3 in the seedling stage and different tissues of “Nanxuan Liuhe Ziqin”. Premier 6.0 software was used to design primers (Ag4CL3-qF: 5′-ACTCTTCAGGCACTACTGGACGA-3′, Ag4CL3-qR: 5′-CAGCATAAAAAAACC-AAACACAT-3′). The AgActin gene was used as an internal standard [32]. The specific primers of the samples of WT and transgenic A. thaliana plants for the lignin biosynthesis gene were also designed by Premier 6.0 software (Table 1). AtActin was used as an internal reference gene [33].

Table 1.
The specific primers of transgenic A. thaliana plants about lignin-biosynthesis-gene.
The ChamQ SYBR ColorMaster Mix (Vazyme, Nanjing, China) and Bio-Rad IQ5 re-al-time PCR system (Bio-Rad, Hercules, CA, USA) were used for qPCR. The qPCR used a 20 μL system, including 2 μL of cDNA, 0.5 μL of forward and reverse primers, 10 μL of SYBR Premix Ex Taq, and 7 μL of ddH2O. Three biological replicates were performed for each sample. The reaction condition for qPCR is 95 °C for 5 min, 40 cycles at 95 °C for 10 s, 60 °C for 30 s, and melting curve analysis (65 to 95 °C, increasing by 0.5 °C every 5 s). The gene expression level was calculated by 2−ΔΔCT [34].
2.7. Statistical Analysis
All data in the text were obtained from the average of three biological repeats. Data significant differences were analyzed using SPSS 25 software by one-way ANOVA at 0.05 levels.
3. Results
3.1. Changes of Lignin Contents
As the growth time increased, the plant height and stem thickness also increased (Figure 1A–D and Table 2). Lignin content is an important index that influences the aging of celery. During the growth stage, the lignin content of the petioles gradually increased, and the lignin content of petioles at 70 d of growth increased to 222.25 mg·g−1 (Figure 1D).
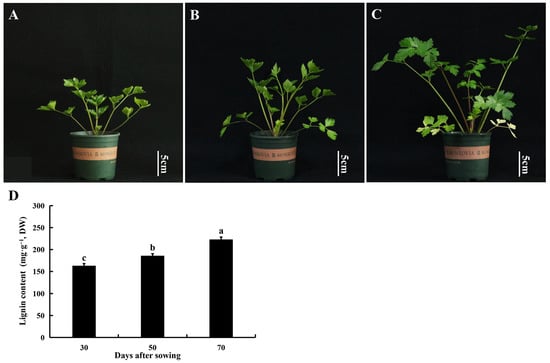
Figure 1.
Morphological changes and lignin content of “Nanxuan Liuhe Ziqin” petioles during growth. (A) 30 d (seedling stage) after sowing; (B) 50 d (vigorous growth period) after sowing; (C) 70 d (commercial stage) after sowing; (D) Lignin content of celery during growth. Data are expressed as the means ± standard deviation (SD) of three replicates. Different lowercase letters indicate significant differences at 0.05 levels.

Table 2.
Change of morphology of celery growth stage.
3.2. Analysis of the Ag4CL3 Sequence
The Ag4CL3 gene cDNA sequence was successfully cloned from “Nanxuan Liuhe Ziqin” (GenBank OP156995) and contained an ORF length of 1668 bp, encoding 555 amino acids (Figure 2A). The theoretical isoelectric point was 6.89, and the protein molecular weight was 59.85 kDa. The secondary structure mainly consisted of 31.89% α-helixes, 18.02% extended strands, 6.67% β-turns, and 43.42% random coils (Figure 2B). The tertiary structure composition is consistent with its secondary structure composition (Figure 2C). The Ag4CL3 protein has a class-I superfamily of adenylate-forming domains. It has an active site, an AMP binding site, putative CoA binding site, and acyl-activating enzyme (AAE) conserved motif (Figure 2D). In order to investigate the phylogenetic relationships of 4CL proteins, a phylogenetic tree was constructed using the maximum likelihood method for a total of 10 amino acid sequences of 4CLs from A. thaliana, G. max, and P. tremuloides (Figure 3). The results showed that Ag4CL3 successfully clustered with At4CL1, At4CL2, Gm4CL1, Gm4CL2, and Pt4CL1, indicating that Ag4CL3 belonged to class I. Moreover, the functional studies of class I and class II genes in A. thaliana, G. max, and P. tremuloides showed that they were involved in lignin synthesis and flavonoid synthesis. Therefore, it is speculated that Ag4CL3 plays a pivotal role in lignin biosynthesis in the celery.
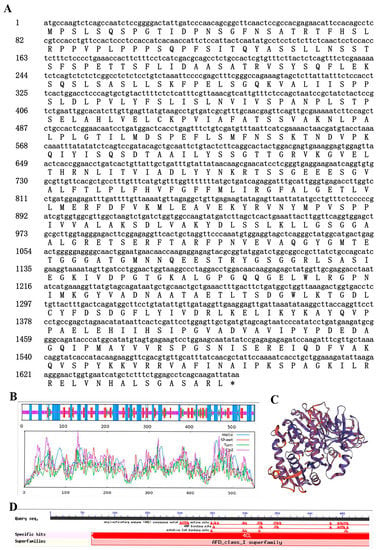
Figure 2.
Bioinformatics analysis of Ag4CL3. (A) Nucleotide and encoded amino acid sequence of Ag4CL3 gene, *: Stop codon; (B) The secondary structure model of the Ag4CL3 protein; (C) The tertiary structure model of the Ag4CL3 protein; (D) Prediction of conserved functional domain of theAg4CL3 protein.
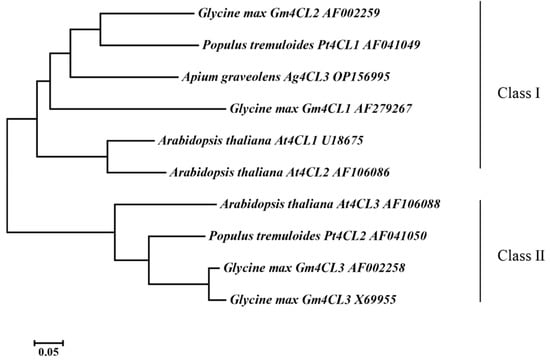
Figure 3.
Phylogenetic tree of 4CL3 proteins from celery and other plant species.
3.3. Ag4CL3 Gene Expression Analysis
RT-qPCR was used to analyze the relative expression levels of the Ag4CL3 gene in the different tissues of “Nanxuan Liuhe Ziqin”. At 70 d after sowing, the Ag4CL3 gene was expressed in the roots, leaves, and petioles of “Nanxuan Liuhe Ziqin”, but the relative expression was significantly different. The relative expression of the Ag4CL3 gene was highest in petioles, second in roots, and lowest in leaves (Figure 4A). In the seedling stage (30 d after sowing), the growth period (50 d after sowing), and the commercial stage (70 d after sowing), the relative expression of the Ag4CL3 gene in the petioles showed remarkable differences. The highest relative expression of the Ag4CL3 gene occurred at the commercial stage (70 d after sowing) (Figure 4B).
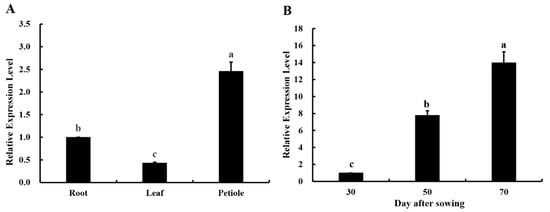
Figure 4.
Comparison of the relative expression of the Ag4CL3 gene in petioles at different growth stages (A) and in different tissues (B) of A. graveolens “Nanxuan Liuhe Ziqin”. Data are expressed as the means ± standard deviation (SD) of three replicates. Different lowercase letters indicate significant differences at 0.05 levels.
3.4. Identification of Transgenic A. thaliana and Overexpression of the Ag4CL3 Upregulated the Lignin Content in Arabidopsis
To research the function of the Ag4CL3 gene, transgenic A. thaliana lines were created via Agrobacterium-mediated transformation. The transgenic A. thaliana lines (L1, L2, and L3) were screened on MS medium containing kanamycin. Moreover, approximately 1600 bp PCR products were observed in the transgenic lines L1, L2, and L3, based on PCR amplification, but they were not detected in the WT plants (Figure 5A). The results indicated that Ag4CL3 was successfully transferred into A. thaliana, and three OE lines harboring the Ag4CL3 gene were obtained. According to the lignin content determination, the transgenic A. thaliana lines exhibited higher lignin accumulation than the WT (Figure 5B). The lignin contents in the WT and transgenic A. thaliana lines were 106.57, 237.22, 250.10, and 316.55 mg·g−1 DW (dry weight), respectively. The Ag4CL3 gene was expressed significantly higher in transgenic A. thaliana than in WT plants (Figure 5C). These results indicate that the Ag4CL3 gene was introduced, and that the successful overexpression of Ag4CL3 upregulated the lignin level in transgenic A. thaliana.
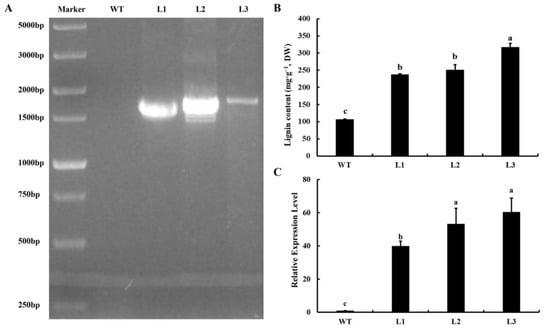
Figure 5.
Identification of transgenic A. thaliana using (A) PCR amplification; (B) Comparison of lignin content between WT and transgenic A. thaliana lines (L1, L2, L3); (C) Ag4CL3 gene expressed in WT and transgenic A. thaliana lines (L1, L2, L3). Data are expressed as the means ± standard deviation (SD) of three replicates. Different lowercase letters indicate significant differences at 0.05 levels.
3.5. Expression of Lignin Biosynthesis Related Genes in A. thaliana Plants
To investigate further the effect of Ag4CL3 on lignin biosynthesis, the expression of other lignin biosynthesis genes was investigated in transgenic A. thaliana (Figure 6). The relative expression levels of AtPAL and AtCCR of the WT plants were higher than those of the transgenic A. thaliana plants, but the expression levels of the remaining genes in WT plants were lower than those in transgenic A. thaliana plants (L1, L2, L3). Furthermore, the expression levels of AtHCT in transgenic line L2 were higher than those in the WT and other transgenic lines. This result showed that gene expression in the transgenic A. thaliana plants was significantly upregulated, except for AtPAL, AtCCR, and AtLAC. Therefore, the overexpression of Ag4CL3 can affect the biosynthesis of lignin in A. thaliana.
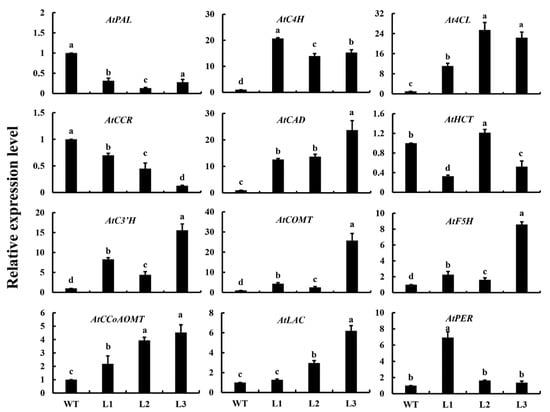
Figure 6.
Expression profiles of lignin-related structural genes in A. thaliana plants. Data are expressed as the means ± standard deviation (SD) of three replicates. Different lowercase letters indicate significant differences at 0.05 levels.
4. Discussion
Celery is a popular vegetable crop in China and around the world, and its leaf blades and petioles contain rich nutrients that humans need. Celery is rich in dietary fiber and contains more lignin than other vegetables. The biosynthesis of lignin plays an important role in the growth and development of plants that not only “affects” the taste and flavor of vegetable crops, but also supports plant structure and promotes disease resistance in plants, such as carrots [35], Toona sinensis [36], tea [13], and maize [37]. Appropriate amounts of lignin support green and straight celery with a delicious taste, which is beneficial for improving the quality and yield of celery. In our study, we found that as the lignin content of celery petioles increased, the celery tissues became lignified and inedible. Therefore, studying the regulation of lignin synthesis during the growth and development of celery is crucial for the quality of celery.
Regarding the lignin metabolism pathway, the first step is the formation of aromatic amino acids through the shikimic acid pathway, and the next step is hydroxycinnamic acids and hydroxycinnamoyl CoA compounds, which are synthesized from aromatic amino acids through deamination, hydroxylation, and methylation. The genes involved in lignin biosynthesis have been validated in multiple species, such as Eucalyptus [38], Acacia auriculiformis, Acacia mangium [39], and Chinese fir [40]. With many years of research, the key enzymes involved in regulating lignin biosynthesis pathways currently include phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT), p-coumaroyl 3′-hydroxylase (C3′H), caffeoyl-CoA O-methyItransferase (CCoAOMT), ferulate 5-hydroxylase (F5H), caffeic acid O-methyItransferase (COMT), laccase (LAC), and peroxidase (PER) [41,42,43].
4CL is a key enzyme involved in lignin biosynthesis, which catalyzes the conversion of 4-cinnamic acid to 4-cinnamoyl CoA, and participates in the biosynthesis of other secondary metabolites [44]. 4CL has been confirmed to exist in various plants, and its expression is tissue specific. Five 4CLs were found in rice [45], of which Os4CL2 was specifically expressed in anthers and strongly activated by ultraviolet radiation, indicating that it may be involved in the formation of flavonoids. By contrast, the remaining 4CLs (Os4CL1, Os4CL3, Os4CL4, Os4CL5) were involved in lignin formation [46]. Twelve Pg4CLs were identified in the pomegranate genome [47], and Pg4CL5 successfully clustered with At4CL1, At4CL2, and At4CL4, indicating that Pg4CL5 belonged to class I, which was involved in lignin synthesis; Pg4CL2 clustered with At4CL3 and belonged to class II, which was related to flavonoid synthesis.
Antisense inhibits the expression of Os4CL3 in rice; the plants become dwarfed, and the content of coumaric acid and ferulic acid increases, whereas the lignin content decreases, resulting in plant anther dysplasia. Moreover, the fertility and yield of transgenic rice decreased [24,48]. In Fraxinus mandsshurica [49], the Fm4CL2-overexpressing (OE-Fm4CL2) tobacco showed increased lignin content and decreased hemicellulose content. In addition, silencing Gh4CL7 in upland cotton resulted in a decrease of approximately 20% in lignin content compared with the control group, whereas overexpressing Gh4CL7 in transgenic A. thaliana OE lines increased the lignin content by approximately 10% [50].
Previous studies have shown that changes in lignin content are closely associated with the expression of lignin biosynthesis genes [51,52]. In the lignin biosynthesis pathway, PAL, C4H, 4CL, CCR, CAD, HCT, C3′H, COMT, F5H, CCoAOMT, LAC, and PER are structural genes that directly affect lignin formation through enzyme catalysis. PAL and C4H are not specifically involved in lignin synthesis, but they are involved in the synthesis of non-lignin phenolic compounds, such as flavonoids and salicylic acid. PAL and C4H gene expression was inhibited, their activity was controlled, and the precursor required for lignin monomer synthesis upstream of the lignin synthesis pathway was reduced, thereby affecting lignin synthesis [53]. The overexpression or inhibition of the CCR gene not only affects transgenic plant lignin content, but also affects its normal growth and development [54,55,56,57]. The activity of the CAD gene was inhibited in certain plants, and the lignin content remained unchanged [58,59]. The overexpression of the F5H gene in tobacco, poplar, and Arabidopsis significantly increased S-lignin biosynthesis, whereas G-lignin biosynthesis was significantly inhibited [60,61,62]. Reducing the COMT gene caused the lignin content to decrease in transgenic alfalfa [63] and poplar [64]. In transgenic alfalfa, the activity of the CCoAOMT gene was associated with G-lignin content [65]. The inhibition of the C3H gene in transgenic alfalfa did not significantly change the lignin content, but the H-lignin content increased by approximately 65% relative to the S-lignin and G-lignin contents [66,67]. Silencing AtHCT gene expression inhibited lignin synthesis, and the height of transgenic A. thaliana plants was significantly reduced [68]. The overexpression of 4CL genes can affect the expression of downstream genes in lignin, such as CCR, CAD, HCT, C3′H, COMT, F5H, CCoAOMT, LAC, and PER. In this study, the overexpression of Ag4CL3 led to a significant upregulation of lignin biosynthesis gene expression in transgenic A. thaliana plants, except for AtPAL, AtCCR, and AtLAC.
In our investigation, we cloned the Ag4CL3 gene sequence approximately 1668 bp fragment successfully from Apium graveolens. Moreover, during the growth stages of celery, the expression level of Ag4CL3 gradually increased in the petioles and leaf blades, which is consistent with the increasing lignin accumulation at the three stages. Therefore, this gene may be involved in the biosynthetic pathway of lignin. This finding is consistent with the expression results in Populus tremuloide [26] and Nicotiana tabacum [69]. To further discuss the function of the Ag4CL3 gene, we overexpressed the Ag4CL3 gene and measured the lignin content of three A. thaliana OE lines and WT. Compared with the WT, the overexpression of Ag4CL3 upregulated the lignin content in three transgenic A. thaliana lines. Meanwhile, the overexpression of Ag4CL3 led to the lignin biosynthesis gene expression in the transgenic A. thaliana plants being significantly upregulated, except for AtPAL, AtCCR, and AtLAC. The overexpression of Ag4CL3 can possibly improve the activity of 4-Coumarate: coenzyme A ligase, which is involved in the lignin biosynthesis pathway in A. thaliana. With the published celery genome and transcriptomic data, we found five 4CL genes in celery. In this study, only the Ag4CL3 gene was studied, and the function of the remaining four genes needs further investigation.
5. Conclusions
4-Coumarate: coenzyme A ligase is an important enzyme in the lignin biosynthesis pathway. In this study, we cloned the Ag4CL3 gene sequence successfully from Apium graveolens. The Ag4CL3 gene contains an ORF with a length of 1688 bp, 555 amino acids. The overexpression of Ag4CL3 led to the lignin biosynthesis gene expression in the transgenic A. thaliana plants being almost significantly upregulated, except for AtPAL, AtCCR, and AtLAC. This study speculates that Ag4CL3 genes are related to lignin synthesis in A. graveolens.
Author Contributions
Conceptualization, X.-L.Z., S.-H.Z. and G.-F.T.; Methodology, X.-L.Z., S.-H.Z. and Q.Z.; Software, X.-L.Z. and Q.Z.; Validation, X.-L.Z. and G.-F.T.; Formal analysis, S.-H.Z., Q.L., K.W. and Z.-F.C.; Data curation and writing—original draft preparation, X.-L.Z. and G.-F.T.; Writing—review and editing, G.-F.T.; Supervision, G.-F.T.; Project administration, X.-L.Z. and G.-F.T.; Visualization, X.-L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by Guizhou Province Basic Research Program ([2019]1307); Guizhou Academy of Agricultural Sciences Support Project [Qian Nongkeyuan Science and Technology Innovation No. (2022) 07]; Project of Guizhou Provincial Department of Science and Technology (No. Qiankehe Fuqi [2022] 005); Vegetable System Project of Guizhou (GZCYTX2023-0101); and Construction of Guiyang Vegetable Germplasm Resources Research Center (Zhuke contract [2021] No. 5-1).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fu, N.; Wang, P.Y.; Liu, X.D.; Shen, H.L. Use of EST-SSR markers for evaluating genetic diversity and fingerprinting celery (Apium graveolens L.) cultivars. Molecules 2014, 19, 1939–1955. [Google Scholar] [CrossRef] [PubMed]
- Hostetler, G.L.; Riedl, K.M.; Schwartz, S.J. Effects of food formulation and thermal processing on flavones in celery and chamomile. Food Chem. 2013, 141, 1406–1411. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Hou, X.L.; Wang, F.; Tan, G.F.; Xu, Z.S.; Xiong, A.S. Advances in the research of celery, an important Apiaceae vegetable crop. Crit. Rev. Biotechnol. 2018, 38, 172–183. [Google Scholar] [CrossRef]
- Duan, A.Q.; Tao, J.P.; Jia, L.L.; Tan, G.F.; Liu, J.X.; Li, T.; Chen, L.Z.; Su, X.J.; Feng, K.; Xu, Z.S.; et al. AgNAC1, a celery transcription factor, related to regulation on lignin biosynthesis and salt tolerance. Genomics 2020, 112, 5254–5264. [Google Scholar] [CrossRef]
- Feng, K.; Liu, J.X.; Duan, A.Q.; Li, T.; Yang, Q.Q.; Xu, Z.S.; Xiong, A.S. AgMYB2 transcription factor is involved in the regulation of anthocyanin biosynthesis in purple celery (Apium graveolens L.). Planta 2018, 248, 1249–1261. [Google Scholar] [CrossRef]
- Feng, K.; Xing, G.M.; Liu, J.X.; Wang, H.; Tan, G.F.; Wang, G.L.; Xu, Z.S.; Xiong, A.S. AgMYB1, an R2R3-MYB factor, plays a role in anthocyanin production and enhancement of antioxidant capacity in celery. Veg. Res. 2021, 1, 2–12. [Google Scholar] [CrossRef]
- Tan, G.F.; Ma, J.; Zhang, X.Y.; Xu, Z.S.; Xiong, A.S. AgFNS overexpression increase apigenin and decrease anthocyanins in petioles of transgenic celery. Plant Sci. 2017, 263, 31–38. [Google Scholar] [CrossRef]
- Yan, J.; Yu, L.; He, L.; Zhu, L.; Xu, S.; Wan, Y.; Wang, H.; Wang, Y.; Zhu, W. Comparative transcriptome analysis of celery leaf blades identified an R2R3-MYB transcription factor that regulates apigenin metabolism. J. Agric. Food Chem. 2019, 67, 5265–5277. [Google Scholar] [CrossRef]
- Wang, H.; Liu, J.X.; Feng, K.; Li, T.; Duan, A.Q.; Liu, Y.H.; Liu, H.; Xiong, A.S. AgMYB12, a novel R2R3-MYB transcription factor, regulates apigenin biosynthesis by interacting with the AgFNS gene in celery. Plant Cell Rep. 2021, 41, 139–151. [Google Scholar] [CrossRef]
- Dhingra, D.; Michael, M.; Rajput, H.; Patil, R.T. Dietary fibre in foods: A review. J. Food Sci. Tech. 2012, 49, 255–266. [Google Scholar] [CrossRef]
- Liu, Q.Q.; Luo, L.; Zheng, L.Q. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, J.M.; Chapple, C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002, 5, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Teng, R.M.; Wang, W.L.; Wang, Y.; Shen, W.; Zhuang, J. Identification of genes revealed differential expression profiles and lignin accumulation during leaf and stem development in tea plant (Camellia sinensis (L.) O. Kuntze). Protoplasma 2019, 256, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Thimm, J.C.; Burritt, D.J.; Sims, I.M.; Newman, R.H.; Ducker, W.A.; Melton, L.D. Celery (Apium graveolens) parenchyma cell walls: Cell walls with minimal xyloglucan. Physiol. Plant 2002, 116, 164–171. [Google Scholar] [CrossRef]
- Wang, D.D.; Bai, H.; Chen, W.Q.; Lu, H.; Jiang, X.N. Identifying a cinnamoyl coenzyme a reductase (CCR) activity with 4-coumaric acid: Coenzyme a ligase (4CL) reaction products in Populus tomentosa. J. Plant Biol. 2009, 52, 482–491. [Google Scholar] [CrossRef]
- Beuerle, T.; Pichersky, E. Enzymatic synthesis and purification of aromatic coenzyme a esters. Anal. Biochem. 2002, 302, 305–312. [Google Scholar] [CrossRef]
- Schneider, K.; Hovel, K.; Witzel, K.; Hamberger, B.; Schomburg, D.; Stuible, H.P. The substrate specificity-determining amino acid code of 4-coumarate: CoA ligase. Proc. Natl. Acad. Sci. USA 2003, 100, 8601–8606. [Google Scholar] [CrossRef]
- Saballos, A.; Sattler, S.E.; Sanchez, E.; Foster, T.P.; Xin, Z.G.; Kang, C.H.; Pedersen, J.F.; Wermerris, W. Brown midrib2 (Bmr2) encodes the major 4-coumarate: Coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J. 2012, 70, 818–830. [Google Scholar] [CrossRef]
- Cukovica, D.; Ehlting, J.; VanZiffle, J.A.; Douglas, C.J. Structure and evolution of 4-coumarate: Coenzyme a ligase (4CL) gene families. Biol. Chem. 2001, 382, 645–654. [Google Scholar] [CrossRef]
- Kajita, S.; Hishiyama, S.; Tomimura, Y.; Katayama, Y.; Omori, S.J. Structural characterization of modified lignin in transgenic tobacco plants in which the activity of 4-coumarate:coenzyme A ligase is depressed. Plant Physiol. 1997, 114, 871–879. [Google Scholar] [CrossRef]
- Wan, C.H.; Yu, J.; Cai, Y.X.; Zhu, P.P.; Liu, C.Y.; Zhao, A.C.; Lv, R.H.; Li, M.J.; Xu, F.X.; Yu, M.D. Characterization and functional analysis of 4-coumarate:coa ligase genes in mulberry. PLoS ONE 2016, 23, e0157414. [Google Scholar]
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four isoforms of Arabidopsis 4-coumarate: CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 2015, 169, 2409–2421. [Google Scholar] [CrossRef] [PubMed]
- Christian, L.; Britta, M.; Judith, F.; Annette, U.; Friedrich, L.; Harald, M.; Jurgen, E. Divergent members of a soybean (Glycine max L.) 4-coumarate:coenzyme A ligase gene family: Primary structures, catalytic properties, and differential expression. Eur. J. Biochem. 2002, 269, 1304–1315. [Google Scholar]
- Gui, J.S.; Shen, J.H.; Li, L.G. Functional characterization of evolutionarily divergent 4-coumarate: Coenzyme a ligases in rice. Plant Physiol. 2011, 157, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.S.; Chen, W.J.; Deng, F.; Yasuhiro, Y. Differential properties of 4-coumarate: CoA ligase related to growth suppression by chalcone in maize and rice. Plant Growth Regul. 2005, 46, 169–176. [Google Scholar] [CrossRef]
- Hu, W.J.; Kawaoka, A.; Tsai, C.J.; Lung, J.; Osakabe, K.; Ebinuma, H.; Chiang, V.L. Compartmentalized expression of two structurally and functionally distinct 4-coumarate: CoA ligase genes in aspen (Populus tremuloides). Proc. Natl. Acad. Sci. USA 1998, 95, 5407–5412. [Google Scholar] [CrossRef]
- Cai, C.; Xu, C.J.; Li, X.; Ferguson, L.; Chen, K.S. Accumulation of lignin in relation to change in activities of lignification enzymes in loquat fruit flesh after harvest. Postharvest Biol. Technol. 2006, 40, 163–169. [Google Scholar] [CrossRef]
- Cervilla, L.M.; Rosales, M.A.; Rubio-Wilhelmi, M.M.; Sánchez-Rodríguez, E.; Blasco, B.; Ríos, J.J.; Romero, L.; Ruiz, J.M. Involvement of lignification and membrane permeability in the tomato root response to boron toxicity. Plant Sci. 2009, 176, 545–552. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Li, M.Y.; Jiang, Q.; Xu, Z.S.; Liu, J.X.; Xiong, A.S. CeleryDB: A genomic database for celery. Database 2018, 2018, bay070. [Google Scholar] [CrossRef]
- Li, M.Y.; Feng, K.; Hou, X.L.; Jiang, Q.; Xu, Z.S.; Wang, G.L.; Liu, J.X.; Wang, F.; Xiong, A.S. The genome sequence of celery (Apium graveolens L.), an important leaf vegetable crop rich in apigenin in the Apiaceae family. Hortic. Res. 2020, 7, 9. [Google Scholar] [CrossRef]
- Zhang, X.R.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Wang, F.; Jiang, Q.; Wang, G.L.; Tian, C.; Xiong, A.S. Validation and comparison of reference genes for qPCR normalization of celery (Apium graveolens) at different development stages. Front. Plant Sci. 2016, 17, 313. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Q.; Dong, C.; Li, X.; Du, J.C.; Qian, M.; Sun, X.D.; Yang, Y.P. A novel Ap2/ERF transcription factor from Stipa purpurea leads to enhanced drought tolerance in Arabidopsis thaliana. Plant Cell Rep. 2016, 35, 2227–2239. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Huang, Y.; Zhang, X.Y.; Xu, Z.S.; Wang, F.; Xiong, A.S. Transcriptome-based identification of genes revealed differential expression profiles and lignin accumulation during root development in cultivated and wild carrots. Plant Cell Rep. 2016, 35, 1743–1755. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhong, X.L.; Cai, X.; Zhu, S.H.; Meng, P.H.; Zhang, J.; Tan, G.F. Comparative Physiological analysis of lignification, anthocyanin metabolism and correlated gene expression in red Toona sinensis buds during cold storage. Agronomy 2023, 13, 119. [Google Scholar] [CrossRef]
- Cardinal, A.J.; Lee, M.; Moore, K.J. Genetic mapping and analysis of quantitative trait loci affecting fiber and lignin content in maize. Theor. Appl. Genet. 2003, 106, 866–874. [Google Scholar] [CrossRef]
- Harakava, R. Genes encoding enzymes of the lignin biosynthesis pathway in Eucalyptus. Genet. Mol. Biol. 2005, 28, 601–607. [Google Scholar] [CrossRef]
- Wong, M.M.L.; Cannon, C.H.; Wickneswari, R. Identification of lignin genes and regulatory sequences involved in secondary cell wall formation in Acacia auriculiformis and Acacia mangium via de novo transcriptome sequencing. BMC Genom. 2011, 12, 342. [Google Scholar] [CrossRef]
- Huang, H.H.; Xu, L.L.; Tong, Z.K.; Lin, E.P.; Liu, Q.P.; Cheng, L.J.; Zhu, M.Y. De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genom. 2012, 13, 648. [Google Scholar] [CrossRef]
- Riboulet, C.; Guillaumie, S.; Méchin, V.; Bosio, M.; Pichon, M.; Goffner, D.; Lapierre, C.; Pollet, B.; Lefevre, B.; Martinant, J.P.; et al. Kinetics of phenylpropanoid gene expression in maize growing internodes: Relationships with cell wall deposition. Crop Sci. 2009, 49, 211–223. [Google Scholar] [CrossRef]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.G.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef]
- Chowdhury, M.E.K.; Choi, B.; Cho, B.K.; Kim, J.B.; Park, S.U.; Natarajan, s.; Lim, H.S.; Bae, H. Regulation of 4CL, encoding 4-coumarate: Coenzyme A ligase, expression in kenaf under diverse stress conditions. Plant Omics 2013, 6, 254–262. [Google Scholar]
- Silber, M.V.; Meimberg, H.; Ebel, J. Identification of a 4-coumarate: CoA ligase gene family in the moss. Physcomitrella Patens Phytochem. 2008, 69, 2449–2456. [Google Scholar] [CrossRef]
- Sun, H.Y.; Li, Y.; Feng, S.Q.; Zou, W.H.; Guo, K.; Fan, C.F.; Si, S.L.; Peng, L.C. Analysis of five rice 4-coumarate:coenzyme A ligase enzyme activity and stress response for potential roles in lignin and flavonoid biosynthesis in rice. Biochem. Biophys. Res. Commun. 2013, 430, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Guo, L.H.; Zhao, Y.J.; Zhao, X.Q.; Yuan, Z.H. Systematic analysis and expression profiles of the 4-Coumarate: CoA Ligase (4CL) gene family in pomegranate (Punica granatum L.). Int. J. Mol. Sci. 2022, 23, 3509. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Su, W.L.; Zhang, H.; Zhan, Y.G.; Zeng, F.S. Fraxinus mandshurica 4-coumarate-CoA ligase 2 enhances drought and osmotic stress tolerance of tobacco by increasing coniferyl alcohol content. Plant Physiol. Biochem. 2020, 155, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.C.; Xiong, X.P.; Zhang, X.L.; Feng, H.J.; Zhu, Q.H.; Sun, J.; Li, Y.J. Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance. Plant Biol. 2020, 20, 125. [Google Scholar] [CrossRef]
- Ali, M.B.; McNear, D.H., Jr. Induced transcriptional profiling of phenylpropanoid pathway genes increased flavonoid and lignin content in Arabidopsis leaves in response to microbial products. BMC Plant Biol. 2014, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Que, F.; Xu, Z.S.; Wang, F.; Xiong, A.S. Exogenous gibberellin enhances secondary xylem development and lignification in carrot taproot. Protoplasma 2017, 254, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zang, C.; Ge, H.; Zhang, J.; Grierson, D.; Yin, X.R.; Chen, K.S. Involvement of PAL, C4H, and 4CL in chilling injury-induced flesh lignification of loquat fruit. HortScience 2017, 52, 127–131. [Google Scholar] [CrossRef]
- Laskar, D.D.; Jourdes, M.; Patten, A.M.; Helms, G.L.; Davin, L.B.; Lewis, N.G. The Arabidopsis cinnamoyl CoA reductase irx4 mutant has a delayed but coherent (normal) program of lignification. Plant J. 2006, 48, 674–686. [Google Scholar] [CrossRef]
- Chabannes, M.; Ruel, K.; Yoshinaga, A.; Chabbert, B.; Jauneau, A.; Joseleau, J.P.; Boudet, A.M. In situ analysis of lignins in transgenic tobacco reveals a differential impact of individual transformations on the spatial patterns of lignin deposition at the cellular and subcellular levels. Plant J. 2001, 28, 271–282. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, A.; Holt, K.; Piquemal, J.; Grima-Pettenati, J.; Boudet, A.; Pollet, B.; Lapierre, C.; PetitConil, M.; Schuch, W.G.; Halpin, C. Improved paper pulp from plants with suppressed cinnamoyl-coa reductase or cinnamyl alcohol dehydrogenase. Transgenic Res. 2002, 11, 495–503. [Google Scholar] [CrossRef]
- Lapierre, C.; Pollet, B.; Petitconil, M.; Toval, G.; Romero, J.; Pilate, G.; Leple´, J.C.; Boerjan, W.; Ferret, V.; Nadai, V.D.; et al. Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid o-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol. 1999, 119, 153–163. [Google Scholar] [CrossRef]
- Halpin, C.; Knight, M.E.; Foxon, G.A.; Campbell, M.M.; Boudet, A.M.; Boon, J.J.; Chabbert, B.; Tollier, M.T.; Schuch, W.G. Manipulation of lignin quality by downregulation of cinnamyl alcohol dehydrogenase. Plant J. 1994, 6, 339–350. [Google Scholar] [CrossRef]
- Chabannes, M.; Barakate, A.; Lapierre, C.; Marita, J.M.; Ralph, J.; Pean, M.; Danoun, S.; Halpin, C.; GrimaPettenati, J.; Boudet, A.M. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 2001, 28, 257–270. [Google Scholar] [CrossRef]
- Franke, R.; Mcmichael, C.M.; Meyer, K.; Shirley, A.M.; Cusumano, J.C.; Chapple, C. Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 2000, 22, 223–234. [Google Scholar] [CrossRef]
- Sibout, R.; Baucher, M.; Gatineau, M.; Van Doorsselaere, J.; Mila, I.; Pollet, B.; Maba, B.; Pilate, G.; Lapierre, C.; Boerjan, W.; et al. Expression of a poplar cDNA encoding a ferulate-5-hydroxylase/coniferaldehyde 5-hydroxylase increases S lignin deposition in Arabidopsis thaliana. Plant Physiol. Biochem. 2002, 40, 1087–1096. [Google Scholar] [CrossRef]
- Huntley, S.K.; Ellis, D.; Gilbert, M.; Chapple, C.; Mansfield, S.D. Significant increases in pulping efficiency in C4H-F5H-transformed poplars: Improved chemical savings and reduced environmental toxins. J. Agric. Food Chem. 2003, 51, 6178–6183. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.J.; Chen, F.; Inoue, K.; Blount, J.W.; Dixon, R.A. Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl coa 3-O-methyltransferase in transgenic alfalfa: Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 2001, 13, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Functions of BMPs, Runx2, and osterix in the development of bone and cartilage. Nihon Rinsho 2005, 63, 1671–1677. [Google Scholar]
- Zhong, R.Q.; Herbert Morrison, W.S.; Himmelsbach, D.; Poole, F.L.; Ye, Z.H. Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 2000, 124, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Ralph, J.; Akiyama, T.; Kim, H.; Lu, F.; Schatz, P.F.; Marita, J.M.; Ralph, S.A.; Reddy, M.S.; Chen, F.; Dixon, R.A. Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J. Biol. Chem. 2006, 281, 8843–8853. [Google Scholar] [CrossRef]
- Reddy, M.S.; Chen, F.; Shadle, G.; Jackson, L.; Aljoe, H.; Dixon, R.A. Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc. Natl. Acad. Sci. USA 2005, 102, 16573–16578. [Google Scholar] [CrossRef]
- Besseau, S.; Hoffmann, L.; Geoffroy, P.; Lapierre, C.; Pollet, B.; Legrand, M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 2007, 19, 148–162. [Google Scholar] [CrossRef]
- Lee, D.; Douglas, C.J. Two divergent members of a tobacco 4-coumarate: Coenzyme A ligase (4CL) gene family (cDNA structure, gene inheritance and expression, and properties of recombinant proteins). Plant Physiol. 1996, 112, 193–205. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).