Abstract
The wheat (Triticum aestivum) microbiome is essential to its growth and adaptation under the current climatic crisis. Wheat breeding programs are often mainly focused on obtaining more resistant cultivars; thus, plant genotype-by-microbiome interactions have gained attention. In this sense, local wheat cultivars represent a unique opportunity to examine how bacterial communities are recruited and support plant growth under field conditions. In this study, we explored the diversity, community structure, and potential functions of root-associated bacterial communities of four Chilean wheat (Triticum aestivum) cultivars under field conditions through Illumina MiSeq. Analyses showed that Proteobacteria was the most abundant phylum in root endosphere (51.1 to 74.4%) and rhizosphere samples (39.3 to 44.9%) across wheat cultivars. Significant differences (p ≤ 0.05) in alpha and beta diversity were observed in root endosphere and rhizosphere samples, independently of wheat genotypes. Potassium was identified as the main factor driving the rhizosphere microbiomes of wheat. A higher proportion of shared operational taxonomic units (OTUs) were found in rhizosphere (mainly Pseudomonas, Flavobacterium, and Janthinobacterium) compared with root endosphere (dominated by Delftia, Acinetobacter, Stenotrophomonas, Kaistobacter) samples across all cultivars. Analyses of larger predicted functional activities revealed that chemoheterotrophy and aerobic chemoheterotrophy were more observed in the root endosphere environment, whereas among the minor functions, nitrogen cycling was the more predicted trait, related to rhizosphere samples. A co-occurrence analysis revealed complex bacterial interactions in wheat cultivars’ niche microbiomes identifying three (Comamonadaceae, Enterobacteraceae, Micrococcaceae) and four (Corynebacteraceae, Dermabacteraceae, Xanthomonadaceae, Staphylococcaceae) families as keystone taxa for the root endosphere and rhizosphere, respectively. It is suggested that such findings on the differences in root microbiomes associated with wheat cultivars under field conditions would help to develop new cultivars with abilities to recruit specific bacterial communities.
1. Introduction
Wheat (Triticum aestivum) is one of the most consumed crops worldwide, thus increasing its productivity through the efficient application of fertilizers, water, and pesticides is the major challenge in the current agricultural scenario. Wheat microbiomes are recognized as a crucial component of the next green revolution, as they harbor the potential to improve the resilience and sustainability of agroecosystems under several stressors, including the current climatic crisis [1]. Nutrient acquisition, protection against soil-borne diseases, and stress tolerance have shown to be modulated by microbiomes in the roots and the rhizosphere of wheat plants [2,3]. In this sense, most bacteria associated with crop roots reside in the bulk soil [4], being influenced by field management (e.g., input of agrochemicals, tillage, crop rotations, etc.). This has been observed to generate a reduced capacity for plants to recruit and establish optimal root-associated bacterial communities [5]. Moreover, edaphic factors, including water stress, soil pH, and temperature, also impact the root microbiome structure, diversity, and functionality [6,7]. On the other hand, wheat cultivars, germplasm, and phenological stage also determine the associated bacterial communities, with a concomitant effect on their functional diversity and shaping the assembly of root-associated bacterial communities [5,8]. Moreover, significant changes have been suggested to have occurred during the process of breeding in the ability of wheat to assemble bacterial and fungal communities [9]. For example, the bacteria colonization rate in wheat cultivars was faster than that in wheat ancestors.
Bacteria associate with plants’ root systems through the colonization of the rhizosphere (soil influenced by root exudates), the rhizoplane (attaching to the surface of roots), or through insertion into the endosphere (making their way into plant tissues) [10]. The selection of stress-resistant wheat cultivars has often been suggested to induce changes in the root architecture and physiology, potentially influencing the composition of bacterial communities and their ability to colonize root systems [11]. Such effects have been attributed to changes in root exudation, an important source of organic substrates available to microorganisms in the rhizoplane and rhizosphere [12]. Additionally, modern wheat cultivars selected under similar fertilization regimes will recruit different bacterial communities [13]. These findings have also been observed in wheat plants grown under contrasted fertilization regimens and soil chemical properties, where the cultivars played the most important role in determining the diversity and structure of bacterial communities among rhizosphere and root endosphere samples [14,15]. However, studies have also described that cultivar influences on bacterial community composition are less than those observed for soil type and environmental factors [16,17]. For example, wheat cultivars grown in soils from Cameroon, Senegal, France, and Italy presented bacterial communities more influenced by the management and soil type rather than plant genotypes or ploidy level [18]. As both factors exert varying influence, a specific assessment for each particular cultivar and environment is required.
Advances in high-throughput sequencing (HTS) and microbial-specific databases have opened the possibility to unravel the network interactions among various members and/or groups of bacterial communities allowing us to identify keystone taxa that contribute to their stability and/or responsiveness [13,19,20,21]. For example, Simonin et al. [18] identified that <3% (103 bacteria, 2 archaea, 41 fungi and 31 protists) of all taxa identified in the rhizosphere of eight wheat cultivars were found to be part of the core microbiome. Despite this, the authors described this group as representing ~50% of the total abundance in those communities. As such, members of Bradyrhizobium, Massilia, Variovorax genera have been recognized as some keystone taxa, highly abundant on modern wheat roots, independently of soil origin and land use history [22]. In other study, endophytic co-occurrence networks were integrated by members of Flavobacterium, Pseudomonas, and Janthinobacterium genera for all tested wheat plants [23].
In view of the shared bacteria observed and given that variation is modulated by either the cultivar or field and environmental conditions, local knowledge about the factors influencing the composition and assembly of bacterial communities associated with wheat cultivars under field conditions is essential. Therefore, we explored the variation in the diversity, community structure, and potential functions of rhizosphere and root endosphere bacterial communities of three locally bred (Fena, Patras, Rocky) and a reference imported (Joker, Netherlands) wheat (Triticum aestivum) cultivars under field conditions. The selected cultivars were resistant to yellow- and red rust, as well as powdered mildew. However, Septoria susceptibility was present on two cultivars (Fena and Patras). Additionally, using a co-occurrence network analysis, functional microbial indicators were predicted from bacterial communities associated with the rhizosphere and the root endosphere.
2. Material and Methods
2.1. Rhizosphere and Wheat Cultivars Plant Collection
Wheat plants along with their rhizosphere samples were collected on 20 September in 2018 from experimental farming plots located in Lautaro, La Araucanía region (38°32′47.5′′ S, 72°27′43.6′′ W), Chile. The farm was in a typical oceanic climate; it had a humid temperate climate with cool summers and winters, with an annual average precipitation of 145 mm, and a daily average temperature of 58.5 °F. The sampled soils were characterized as to contain low phosphorus (P) and high organic matter contents; the averages of extractable P and organic matter in a 0–20 cm layer were 26 mg kg−1 and 9.3%, respectively (Table S1).
Four wheat cultivars (Triticum aestivum cv. Fena, Patras, Joker, and Rocky) corresponding to the anthesis stage were sampled. Briefly, three representative healthy specimens with similar heights and diameters and their adjacent soils (20 cm topsoil layer) were randomly taken in a 10 m transect by using the clean spade to remove intact roots from soils. Collected plants and soils were transferred into plastic bags, stored on ice coolers and immediately transported to the Applied Microbial Ecology Laboratory (EMAlab) of Universidad de La Frontera, Temuco, Chile. Both sample types were stored at −80 °C until DNA extraction. Wheat cultivars were grown in an Andisol under rotation with oat (Avena sativa) and rapeseed (Brassica napus) since 2012. Before our sampling, the experimental plots were used for rapeseed experiments. At sowing, our sampled plants were subjected to a fertilization program with 140 kg of urea ha−1 and treated with commercial pre-emergence herbicides at doses of 1 L ha−1 (12% w/v of flufenacet, flurtamone, and diflufenican). Data from the physicochemical characterization of each soil sample (reported by Rilling et al., [24]) were used to determine the relationship between soil factors and the composition of rhizobacterial communities between four wheat cultivars under field conditions. Detailed information on the physicochemical parameters of the soil samples can be found in Table S1.
2.2. DNA Extraction
Total genomic DNA was extracted from rhizosphere and root endosphere samples. Rhizosphere samples (~0.25 g) of each cultivar sample were obtained from the soil adhered to roots through vigorous shaking and processed with DNeasy PowerSoil DNA isolation kits (QIAGEN N.V., Venlo, The Netherlands) in triplicate. Samples were lysed via a bead-beating protocol, according to the kit manufacturer instructions. For endosphere samples, ~0.25 g of root tissue was initially vigorously washed with sterile distilled water (SDW), and rhizoplane surfaces were sterilized as described by Barra et al. (2016). Briefly, root surfaces were washed in 70% (v/v) ethanol for 3 min, immersed in 2.5% (v/v) sodium hypochlorite (NaOCl) for 5 min, rinsed three times with SDW, and macerated with 1 mL of SDW via a sterile mortar and pestle. Total DNA was extracted from 0.25 mL of surface sterile root homogenate using a Quick-DNATM Plant/Seed Miniprep Kit (Zymo Research, Seattle, WA, USA), according to the manufacturer instructions. The extracts’ DNA concentrations were measured using a Qubit4TM (Thermo Fisher Scientific, Waltham, MA, USA) using broad-range DNA assays. In parallel, RNA and protein assay kits were used on those samples to prevent contamination during the extractions.
2.3. Library Preparation and 16S rRNA Amplicon Sequencing
The libraries and sequencing of bacterial communities associated with the root endosphere and rhizosphere of wheat cultivars were carried out by using Illumina MiSeq as follows: the V4 hypervariable region of the 16S rRNA gene for each biological replicate’s extracted DNA sample was amplified using primer set 515F (5′- GTG CCA GCM GCC GCG GTA A -3′) and 806R (5′- GGA CTA CHV GGG TWT CTA AT -3′). The primers were coupled to specific Illumina Inc. adapters, and the dual indexing method was used for the amplicon library construction [12]. Libraries were pooled to 4 nM, denatured, loaded into Illumina MiSeqTM v3 sequencing kit, and sequenced for 301 cycles in Illumina MiSeq (Illumina, Inc., San Diego, CA, USA) with the support of the University of Minnesota Genomics Center (UMGC, Minneapolis, MN, USA). Raw sequence files were uploaded to NCBI SRA database under bioproject no. PRJNA867668.
2.4. Bioinformatic Processing of the Sequences and Statistical Analysis of the Data
Primers and adapters were removed from raw sequence reads via trimming and processing using SHI7, preserving high-quality reads (QC > 35) [25]. Trimmed sequences were aligned into OTUs, UCHIME was used for the removal of chimera [25], and sequences with >97% similarity were grouped. Nonmicrobial sequence reads (e.g., chloroplasts and mitochondria) were removed via QIIME [20], while data were rarefied to 7000 reads for the biodiversity analysis. Taxonomy assignation was performed using NINJA-OPS and the Silva database (version 123) [26,27]. Richness (OTUs observed, abundance-based, Goods coverage, Chao1, and jackknife) and diversity (Shannon index, Simpson index, and q-stat) values were also calculated via Mothur [28]. The relative abundance taxonomy for the most and lesser observed taxa was graphed in Microsoft Excel. Shared OTUs among rhizobacterial communities across samples were also observed via VennDiagram in R 4.2.1 [29]. Functional traits predictions were performed using the provided database and scripts of the Functional Annotation of Prokaryotic Taxa (FAPROTAX) project [30].
For the co-occurrence network analysis, root endosphere and rhizosphere data were agglomerated to family level and “Spearman” correlation matrixes were constructed for the families with the highest variance among samples, using |0.6| as the lowest accepted correlation (p < 0.05) under the R “NetCoMi” package [31]. The identified clusters, degree, closeness, betweenness, and eigenvector centralities on those matrixes were used for the identification of hubs (or putative keystone taxa) (p < 0.01). Networks were then plotted using “ggplot2” and “igraph” modules in R [32]. In parallel, microbial indicators at the genus level were predicted under the multipatt function using the indicspecies [33] package in R as described by Zhang et al. [12] (p < 0.01).
3. Results
3.1. Alpha Diversity of Root-Associated Bacterial Communities
The average alpha diversity indexes (±SD) obtained from root endosphere and rhizosphere bacterial communities associated to wheat cultivars (Fena, Rocky, Patras, and Joker) are summarized in Table 1. Similar coverage percentages were obtained among wheat cultivars for each plant niche analyzed (p ≤ 0.05). However, the coverage percentage was significantly higher in rhizosphere samples compared to those observed in root endosphere samples for all cultivars analyzed. On average, significant differences in the number of total OTUs at 97% similarity were observed among cultivars, where Patras and Rocky had the highest values (1554 and 936 OTUs on root endosphere and rhizosphere samples, respectively). Moreover, the root endosphere associated to Fena and Patras cultivars showed significant higher OTUs in comparison to their rhizosphere samples, with values ranging from 769 to 130 and 1554 to 472 OTUs, respectively. On the other hand, the abundance-based coverage estimates (ACE) and Chao1 indexes were significant different among wheat cultivars, higher values were attributed to the root endosphere from Patras (1883 and 1894, respectively) and to rhizosphere samples from Rocky (1034 and 1059, respectively) (p ≤ 0.05). By contrast, Fena and Patras showed significantly higher (p ≤ 0.05) ACE and Chao1 indexes from the root endosphere compared to those observed in their rhizosphere samples. For the alpha diversity, significant differences in the Shannon index were observed between the root endosphere from Patras (5.3) and Rocky (4.1); however, among rhizosphere samples, both Rocky and Patras showed a higher diversity compared with the other cultivars analyzed (6.2 to 5.9, respectively). Differences in the Shannon index were also evidenced between plant compartments, for example rhizosphere samples from Rocky (6.2) and the root endosphere from Fena (4.6) showed a greater diversity compared with their respective root endosphere (4.1) and rhizosphere (3.9) samples. Despite this, differences in Simpson indexes were not detected from bacterial communities associated with the root endosphere among wheat cultivars, ranging between 0.022 and 0.071. However, a greater diversity was found in rhizosphere samples from Fena cultivars (0.031) compared to those obtained in the rhizosphere samples from Rocky, Patras, and Joker as revealed by Simpson indexes of 0.003, 0.004, and 0.010, respectively (ANOVA, p < 0.05).

Table 1.
Coverage and alpha diversity of bacterial communities in the root endosphere and rhizosphere of wheat cultivars.
3.2. Taxonomic Affiliation of Root-Associated Bacterial Communities
The taxonomic assignment of 16S rRNA sequences of bacterial communities revealed that members of the phylum Proteobacteria were the most abundant among wheat cultivars in both plant compartments, with values of 51.1 to 74.4 and 39.3 to 44.9% for root endosphere and rhizosphere samples, respectively (Figure 1A; Table S2). After Proteobacteria, root endosphere bacteria communities in the four cultivars were dominated by Bacteriodetes (15 to 30.2%), followed by Actinobacteria (6.7 to 13%) and Firmicutes (2.1 to 2.7%) but only in Fena, Rocky, and Joker cultivars, while Patras presented a low relative abundance in this niche for Firmicutes (0.3%). Patras and Joker presented higher abundances of Acidobacteria (0.9 to 1.4%), Verrucomicrobia (1.1 to 2.8%) and Chloroflexi (1.1 to 2.5%). The relative abundances of various bacterial phyla in the rhizosphere samples were different from those in the endosphere, an indication for the differences between niches. In general, Bacteroidetes was less abundant in the rhizosphere than in the root endosphere across cultivars (6.0–11%). Actinobacteria were the second most abundant phyla (10.8–16.1%) in Rocky, Patras, and Joker. For Fena, Firmicutes (25.4%) were more abundant than for other cultivars (14.7, 7.6, and 4.9% for Joker, Patras, and Rocky, respectively). In addition, Acidobacteria phyla were also higher in Rocky (14.5%) and Patras (14.1%) compared to those observed in Joker (6.7%) and Fena (1.3%) cultivars. Members of phyla Gemmatimonadetes (1.8 to 3.7%), Chloroflexi (2.5 to 3.5%), Verrucomicrobia (2.2 to 3.5%), and Planctomycetes (2.7%) were also observed in rhizosphere samples from Joker, Patras, and Rocky cultivars (Figure 1A; Table S2).
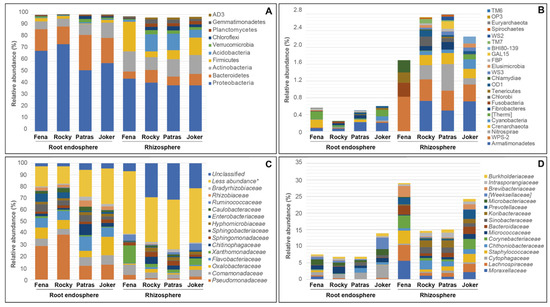
Figure 1.
Bacterial community composition in the root endosphere and rhizosphere samples for different wheat cultivars. Bars represent average relative abundance values from each cultivar sequenced (in triplicate) for more (A,C) and less abundant (B,D) representative taxa at the phylum and family level, respectively. Relative abundance values can be found in Tables S2–S5. Raw values can be found in Table S12. * p ≤ 0.05.
In relation with less abundant taxa, the relative abundances showed a broad taxonomic diversity among samples (Figure 1B; Table S3). There were more bacterial groups in rhizosphere samples compared to those present in the root endosphere samples and they were mainly attributed to members of the phyla Armatimonadetes (0.5 to 0.7%), followed by Eremiobacterota (formerly WPS-2) (0.4 to 0.8%), and Nitrospirae (0.1 to 0.6%) present in the rhizosphere of Joker, Patras, and Rocky. Interestingly, the minor relative abundances associated with the rhizosphere of Fena cultivars were for other rare taxa, including members of the phyla Fusobacteria (0.32%), Chlamydiae (0.29%), and Tenericutes (0.24%). In the root endosphere, Fena cultivars also had larger proportions of Armatimonadetes (0.06 to 0.21%) and Thermi (0.01 to 0.19%), with a small percentage associated with Crenarchaeota and Cyanobacteria.
At the family level, a greater relative abundance of taxa in the rhizosphere samples were attributed to less abundant and unclassified taxa, with relative abundances ranging from 38.4 to 53.6% and from 6.8 to 31.1%, respectively (Figure 1C; Table S4). In addition, members of bacterial families Xanthomonadaceae (2.4 to 15.2%) and Comamonadaceae (3.5 to 8.8%) were also abundant in all rhizosphere samples. On the other hand, the root endosphere samples showed a higher relative abundance of Pseudomonadaceae (11.8 to 38.7%), less abundant taxa (14.9 to 30.5%), followed by Oxalobacteraceae (5.3 to 15.7%) and Flavobacteriaceae (7.5 to 12.3%). Members of the family Pseudomonadaceae were found to be more abundant in the endosphere samples, but not in the rhizospheres independent of wheat cultivars. With respect to less abundant families, a higher diversity was observed in rhizosphere samples compared to root endosphere, where larger relative abundances were associated with Fena and Joker cultivars in comparison with other wheat varieties (Figure 1D; Table S5). Moreover, the rhizosphere samples showed a greater abundance of Moraxellaceae (5.6 to 0.8%) and Lachnospiraceae (4.7 to 0.8%) followed by Staphylococcaceae (3.9 to 0.7%) and Corynebacteriaceae (3.8 to 0.5%).
3.3. Structure of Root-Associated Bacterial Communities and Their Relationship with Soil Parameters
The root-associated community structure similarities from our four wheat cultivars determined using a principal coordinate analysis (PCoA) are shown in Figure 2A. A clear separation was observed between rhizosphere and root endosphere bacterial communities associated with the wheat cultivars, where the first two axes explained 29% and 13% of the total variation in the rhizosphere and root endosphere, respectively. The PERMANOVA analysis confirmed the differences in the bacterial community structure associated with the plant niche (p < 0.001) (Table S6). In this sense, rhizosphere bacteria communities showed more dissimilarities within cultivars when compared to those in the root endosphere samples; however, no clustering was identified by PCoA in response to wheat cultivars (Fena, Joker, Patras, and Rocky). The PERMANOVA analysis showed that wheat cultivars exerted no statistical influence (p < 0.075) on both rhizosphere and root endophytic bacterial communities when all the samples were analyzed (Table S6). The relationship between the physicochemical parameters of soil and bacterial communities revealed that the potassium (K) content was the only factor with a significant effect (R = 0.84; p < 0.005) on the structure of the rhizobacterial communities between the four evaluated cultivars (Figure 2B; Table S7).
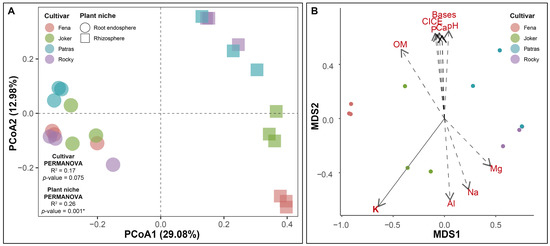
Figure 2.
PCoA plots using Bray–Curtis similarity metrics indicate a greater dissimilarity between the root endosphere and rhizosphere samples (PCoA 1) compared to that among wheat cultivars bacterial community distance matrixes (PCoA) (A), and the influence of soil physicochemical data upon the rhizosphere (MDS) (B). Data points represent the mean values for the rhizosphere or endosphere niches of individual samples belonging to a wheat cultivar. * = p-value under 0.05.
3.4. Shared OTUs and Predicted Functions of Root-Associated Bacterial Communities
A Venn diagram analysis showed a greater number of OTUs shared between root endosphere samples (294; 13.3%) than those observed in rhizosphere samples (65; 2.65%) among all wheat cultivars evaluated (Figure 3A,B and Tables S9 and S10). The main shared OTUs observed in all root endosphere samples were assigned to phyla Proteobacteria (70.9%), Bacteroidetes (19.8%), and Actinobacteria (7.5%). Among these, families Pseudomonaceae, Flavobacteraceae, Oxalobacteraceae, Comamonadaceae, and Sphingobacteraceae represented the major fraction at 31.4, 10.6, 10.3, 7.8, and 5.2%, respectively. At the genus level, the most abundant taxa among shared OTUs were identified as Pseudomonas (30.6%), Flavobacterium (10.6%), Janthinobacterium (8.6%), and one OTU recognized as Oxalobacteraceae but without genus assignation (6.2%). For the rhizosphere, the Proteobacteria, Actinobacteria, and Firmicutes represented 53.4, 21.7, and 10.2% of shared phyla among all cultivars, respectively (Table S9). In depth, at the family level, there was less diversity but more homogeneity, with Xanthomonadaceae (12.2%), Comamonadaceae (9.6%), Sphingomonadaceae (6.5%), Pseudomonaceae (6.5%), Moraxellaceae (5.9%), Bradyrhizobiaceae (5.9%), and Ruminococcaceae (4.1%) being the most observed. At the genus level, Delftia (7.9%), Acinetobacter (5.9%), Stenotrophomonas (5.8%), Kaistobacter (5.7%) were the most abundant. Complementarily, two unassigned OTUs genera classified as Xanthomonadaceae (6.2%) and Pseudomonaceae (5.3%) were also identified (Table S9).
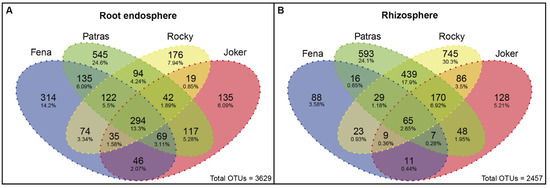
Figure 3.
Venn diagrams showing numbers of unique and shared bacterial OTUs for the root endosphere (A) and rhizosphere (B) niches of the four wheat cultivars. The percentages are relative to the total OTU number observed for both niches. The OTUs shared between all cultivars can be found in Tables S9 and S10.
On the other hand, higher numbers of unique OTUs were observed in the root endosphere from Patras (545; 24.6%) and Fena (314; 14.2%) cultivars, whereas fewer unique OTUs were observed in Rocky and Joker cultivars (176; 7.9% and 135; 6.0%, respectively) (Figure 3A). Moreover, rhizosphere samples showed the greater amount of unique OTUs from Rocky (745; 30.3%) and Patras (593; 24.1%) in comparison with Jocker (128; 5.2%) and Fena (88; 3.5%) cultivars (Figure 3B).
Predicted bacterial functional traits for the taxa in the root endosphere and rhizosphere were more numerous for the root endosphere than rhizosphere (Figure 4, Table S8). For all cultivars, chemoheterotrophy (44.2 to 22.8%) and aerobic chemoheterotrophy (39.7 to 15.9%) were the most abundant metabolic groups, independent of the plant niche or cultivar. A larger proportion of metabolic function assignments was related with fermentation and animal parasites or symbionts in the rhizosphere samples compared to those from the root endosphere samples, with values ranging from 6.6 to 7.1% and from 2.8 to 0.6%, respectively. Among the lesser abundant predicted traits, a majority were assigned to nitrogen cycling processes such as nitrate reduction, nitrogen fixation, nitrate respiration, nitrogen respiration, ureolysis, and nitrification (Figure 4, Table S8). Contrastingly, although similar functions were predicted, only ~5% of the root endosphere functionality was explained.
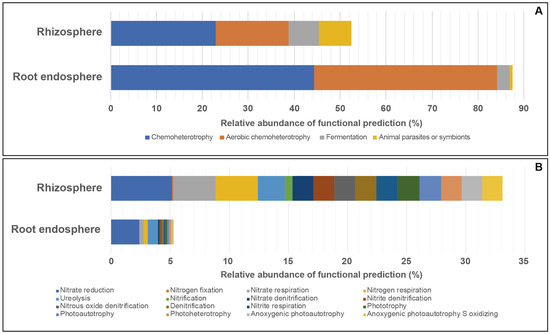
Figure 4.
Major (A) and minor (B) proportions of predicted soil-related functional traits from the root endosphere and rhizosphere for all sampled wheat cultivars. Traits unrelated to soil functionality were classified as others. Bars represent the average relative abundance value for each cultivar and niche sampled. Figure data can be found in Table S8.
3.5. Co-Occurrence Network and Microbial Indicators for Root-Associated Bacterial Communities
A network analysis was conducted to determine the co-occurrence patterns and putative keystone taxa of root-endosphere- and rhizosphere-associated bacterial communities across the cultivars (Figure 5). Despite both networks sharing several taxa, differences in terms of composition, number of clusters, dispersion, number of nodes (or modules), and the connections between them (e.g., Enterobacteraceae and Oxalobacteraceae influence on both networks) were observed.
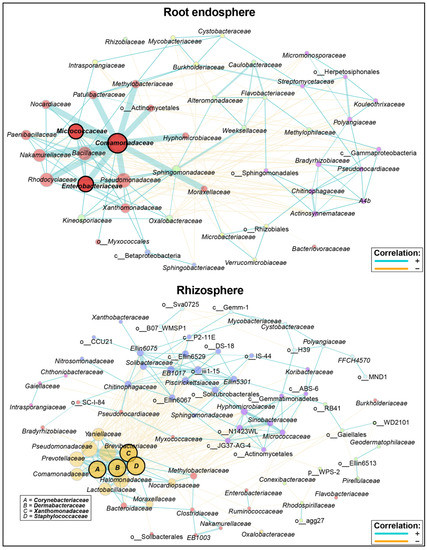
Figure 5.
Co-occurrence networks for rhizosphere and root endosphere niches on studied wheat cultivars. Connected families represent significant positive or negative correlation (p ≥ 0.05). Edge stops represent correlation between two nodes, while thickness and proximity represent the correlation strength. Node color represent statistic cluster association. Node sizes were defined by eigenvector values. Highlighted taxa names represent identified putative keystone taxa based on betweenness, closeness, degree, and eigenvector values (p ≥ 0.05). Node labels containing “o__” or “c__” were not classified at the family level and thus are represented with the nearest taxonomic level assigned.
For example, the root endosphere network was composed of fifty (50) families identified among five (5) main clusters, with all the nodes and clusters directly or indirectly connected through positive correlations. The root endosphere network was described as 0.48, 0.18, 50.5, and 0.04 for the clustering coefficient, modularity, positive edges (%), and natural connectivity, respectively (Table S11). Nodes within clusters were highly connected; however, connections to other clusters were limited. Among those, the largest connected cluster (LCC) was formed by strong correlations among 13 families (Comamonadaceae, Micrococcaceae, Enterobacteraceae, Pseudomonadaceae, Moraxellaceae, Hyphomicrobiaceae, Methylobacteraceae, Patulibacteraceae, Nocardiaceae, Paenibacillaceae, Nakamurellaceae, Rhodocyclaceae, and Bacillaceae), with the Comamonadaceae family as the central node.
The rhizosphere network was composed of seventy-seven (77) families, and eight (8) clusters (Figure 5). Results showed that not all clusters were connected by direct or indirect positive correlations, while node connections within clusters were less disperse than those in root endosphere. The rhizosphere network presented values of 0.44, 0.22, 48.9, and 0.04 for the clustering coefficient, modularity, positive edges (%), and natural connectivity, respectively, indicating a limited connectivity between nodes and clusters compared to that in the root endosphere (Table S11). This was evident from the highly connected cluster consisting of Corynebacteraceae, Dermabacteraceae, Xanthomonadaceae, Staphylococcaceae, Halomonadaceae, Lactobacillaceae, Comamonadaceae, Prevotellaceae, Pseudomonaceae, Yaniellaceae, Brevibacteraceae, Moraxellaceae, and Nocardiopsaceae, with strong correlations. Thus, the weight of the cluster was evenly distributed between Corynebacteraceae, Dermabacteraceae, Xanthomonadaceae, and Staphylococcaceae families as represented by node size (“eigenvector”) (Figure 5).
In terms of hubs (or putative keystone taxa), the root endosphere network showed three families (Comamonadaceae, Enterobacteraceae, Micrococcaceae), while the rhizosphere network revealed four families (Corynebacteraceae, Dermabacteraceae, Xanthomonadaceae, Staphylococcaceae). The relative abundance of the sum of hub families indicated 11.7 and 11.6% of the total assigned taxonomies for each niche (Table S11).
On the other hand, the microbial indicator analyses using the taxonomic assignments from the phylum to genus level revealed the associations between abundance taxon and plant compartments (Table 2). Notably, more indicator taxa were identified in the root endosphere than those observed in rhizosphere samples. For example, only 15 taxa could be classified at the genus level, belonging to Proteobacteria, Verrucomicrobia, Actinobacteria, and Firmicutes phyla. Among them, Chthoniobacter (R = 0.84) and Variovorax (R = 0.83), followed by Arthrospira (R = 0.77) and Steroidobacter (R = 0.70) were significantly abundant in the root endosphere (p ≤ 0.01). In contrast, only one taxon identified as Blautia (R = 0.81) presented a greater abundance in rhizosphere samples.

Table 2.
Microbial indicators (genus level) in the root endosphere and rhizosphere of wheat.
4. Discussion
Root-associated bacterial communities play a crucial role in crops’ fitness and development, as well as their adaptation and tolerance to environmental stresses [34]. The taxonomic composition and functions of plant bacterial communities are influenced by many environmental factors, including soil type and plant host genotypes [35]. The breeding of wheat cultivars is mainly focused on sustaining productivity through the development of plant genotypes to resistance to disease and pests, with little attention to root traits and their potential to recruit a beneficial bacterial community [36]. Thus, given the importance of bacterial communities for increasing crop productivity [37], additional studies are required to understand the impact of wheat breeding on beneficial bacteria recruitment across the below- and above-ground plant compartments. In this study, we examined the composition and predicted functionality of root-associated bacterial communities from four wheat cultivars (Fena, Patras, Rocky, and Joker) grown under field conditions on an Andisol soil. The DNA metabarcoding analysis of 16S rRNA gene libraries (V4 region) from plant rootsrevealed a greater difference in bacterial richness (revealed by the number of OTUs) in the endosphere, with the most abundant OTUs associated with Patras (Table 1). In addition, the Shannon index was higher in the rhizosphere than in the root endosphere in all treatments with the exception of the Fena genotype. However, the Simpson index associated with the rhizosphere revealed a lower diversity in comparison with the root endosphere in three over four cultivars analyzed. It is widely known that bacterial communities associated with the root endosphere are less diverse than those observed from rhizosphere samples mainly from the plant-based selection pressures [34,38]. Similar to the results reported by Rilling et al. [24] and Zhang et al. [34], the diversity and functions of the bacterial community associated with different wheat cultivars under field conditions were higher in the rhizosphere, when compared with other plant niches. Donn et al. [39] found that genotype (varietal) and crop sequence effects accounted for only a small amount of total variation in bacterial communities and the most dominant factors that influenced wheat-root-associated populations included the crop development stage, plant age, and how tightly the soil was attached to the roots i.e., tightly bound vs. loosely bound rhizosphere soil. Despite this, studies conducted by Zheng et al. [40] demonstrated significant interactive effects between the wheat variety, location, and growth stage on the bacterial community assembly in field conditions. The study site and endosphere–rhizosphere compartments have been recognized as main factors that describe a high percentage of the differences among the prokaryotic community [18,41,42].
Based on a taxonomic annotation, Proteobacteria was the most abundant phylum in both plant niches, independently of the wheat cultivar analyzed (Figure 1A, Table S2). This phylum is recognized as the most abundant in soil samples [43,44,45] and other plant niches, particularly the endosphere [34,46] and rhizosphere [47,48] probably because of the copiotrophic nature of many members of this phylum. In addition, the Bacteroidetes phylum was highly abundant in the root endosphere, whereas members of the Firmicutes and Actinobacteria phyla were more abundant in rhizosphere samples across all wheat cultivars. These proportions were also observed for the shared OTUs among all cultivars (Table S9). Bacteroidetes has been reported as a common predominant bacterial taxon in the endospheric tissue of crops such as canola, wheat, and barley [15,49]. A recent study has indicated that members of Bacteroidetes encode a novel and unique constitutive alkaline phosphatase PafA; therefore, their presence as core microbiota in crops could be important for phosphorus mobilization [50]. Several other studies have shown that Actinobacteria and Firmicutes are the dominant phyla in the rhizosphere of diverse wheat cultivars [11,51]. These phyla are considered as K-strategists characterized by their low growth rates and high persistency in soils, even under low nutrient availability [52]. Previous studies have reported that the slow-growing bacteria (K-strategists) are predominant in the rhizosphere of mature roots, whereas fast-growing bacteria (r-strategists) are dominant in young immature roots [53]. At the family level, Pseudomonadaceae was described as the dominant group observed in both the root endosphere and rhizosphere of the analyzed cultivars. Donn et al. [39] found that members belonging to the Family Pseudomonadaceae and Oxalobacteraceae predominated on young roots during early vegetative growth but were significantly reduced on older and senescing roots. In this sense, Pseudomonadaceae associated with roots were observed in modern wheat cultivars “Lewjain Eltan”, and “Hill 81” [54,55]. Predominantly, Pseudomonas spp. are highly prevalent in the rhizosphere of winter and spring wheat cultivars, harboring several plant-growth-promoting traits [56]. This can be explained by some Pseudomonas strains harboring accessory genes that confer a specific adaptability to colonize and persist in the rhizosphere via active chemotaxis toward root exudates, biofilm formation, motility, and their versatility to use a wide range of carbon sources [57].
Interestingly, our results revealed significant differences in the bacterial community structures associated with the root endosphere compared with rhizosphere samples (Figure 2A; Table S6). Niche differentiation in the bacterial community composition has been previously described in many crops, such as wheat, barley, rice, and oats [38,58,59,60], as well as in native plants [12,34]. Through the rhizosphere, plants establish a selective barrier for endosphere microbes, which can colonize the root interior, move inside, and occupy the intracellular space of the cortex and vascular bundle [61]. The wheat genotype selection also triggers and modulates the bacterial community structure in root–soil interface and plant tissue [62]. Despite the latter, our results show that for these wheat cultivars the bacterial community structures for both niches were not significantly influenced. Simonin et al. [18] also reported that wheat varieties did not exert a significant effect on the rhizobacterial microbiome in eight wheat cultivars; rather, the environmental factors such as soil type and management had a greater impact. In contrast, Dilla-Ermita et al. [16] established a growth chamber and field assay with eight wheat cultivars using the same soil, revealing that the most influential factor in rhizosphere communities was the cultivar rather than the environment. These findings have also been observed from the endosphere microbiome of wheat roots, which differed between ancient and modern wheat categories and among the type of cultivars as well [63]. On the other hand, changes in the bacterial endophyte community have also been related to stress conditions such as drought and salinity rather than with the wheat genotypes [7]. For this, our results showed that only K was a significant factor influencing the structure of the bacterial community in the rhizosphere samples (Figure 3B; Table S7). Yu et al. [64] indicated that available and total K can influence bacterial community composition, not only in the bulk soil but also in communities in the rhizosphere. Recently, it has been reported that the application of incremental amounts of K fertilization shapes communities’ functionality in tobacco root systems [65]. Similarly, the addition of K2O has been observed to modify the root exudations of cotton, changing the composition of its root bacterial communities and reducing the biotic stress induced by Verticillium spp. [66].
Our results showed that higher proportions of OTUs observed were shared among wheat cultivars, particularly those obtained from rhizosphere samples. These findings are inconsistent with studies showing an OTU distribution of endophytes among different wheat varieties [7]. In addition, the bacterial communities in both niches indicated a similar proportion of the total communities at the phylum level but still differed in terms of deeper species composition (family and genus levels). For the root endosphere, the dominant shared OTUs in all root endosphere communities (Pseudomonas, Flavobacterium, and Janthinobacterium) were consistent with those observed in the wheat endosphere (root, shoot, and leaves) by Kuzniar et al. [67]. Similarly, the shared rhizosphere OTUs were dominated by Delftia, Kaistobacter, Acinetobacter, Stenotrophomonas and two non-identified Xanthomonadaceae, and Pseudomonaceae bacteria, also described as dominant taxa in soybean- and wheat-root-associated communities [68]. Complementarily, the same taxa were identified as key members of Triticum durum kernels during storage [69]. The Delftia and Acinetobacter species harbor versatile metabolic pathways; thus, their persistence in the rhizosphere may be associated with their ability to convert various substrates derived from root turnover processes [70,71]. The findings in this study and some supporting evidence in the literature indicate that most wheat cultivars might recruit those taxa describe above despite differences in cultivar, soil, or the environment.
On the other hand, specific families identified in the root endosphere and rhizosphere co-occurrence network were statistically confirmed to be microbial indicators, at the genus level, for each niche. For the root endosphere, the genera identified were Variovorax, Bosea, Nocardia, Luteolibacter, and Prosthecobacter, members of the families Comamonadaceae, Verrucomicrobiaceae, Hyphomicrobiaceae, Nocardiaceae, and Verrucomicrobiaceae, respectively (Table 2). In terms of the hubs observed in the network, links with Comamonadaceae might be related to the genus Variovorax, identified in the microbial indicator analyses. Strains of Variovorax such as V. paradoxus have been shown to be a metabolically versatile plant-growth-promoting endophyte in a number of crops including through their ability to enhance the host plant’s stress tolerance to abiotic and biotic stresses and disease resistance [72,73]. We hypothesize a similar influence for the genus Bosea detected as a microbial indicator influencing the Hyphomicrobiaceae node present in the network. In a prior study, a strain of this taxon was isolated from the same samples and confirmed to harbor the nifH gene in the root endosphere of the same wheat cultivars evaluated in this study [24], supporting the nitrogen cycle functionality predictions observed. Additionally, the detection of Yaniellaceae (phylum Actinobacteria) as one of the hubs in the network analyses, as well as a microbial indicator, suggests that members of this family, some of them being known halotolerant bacteria, as key taxa relevant for the rhizosphere of wheat.
Finally, when relating the hub families with the communities’ predicted functionality, the Enterobacteraceae, Comamonadaceae, Hyphomicrobiaceae, Xanthomonadaceae, Corynebacteraceae, and Staphylococcaceae families have been related to nitrogen cycling across diverse ecosystems [74], which could explain the large proportion of nitrogen cycle activities predicted by OTUs associated with wheat cultivars (Figure 4B; Table S8). Therefore, our results suggest that members of these families may contribute to the microbiome stability and could be pivotal functional taxa associated with nitrogen metabolisms in the root microbiome. It is widely known that microbiomes can affect plant performance positively and negatively through their plant-growth-promoting mechanisms. Several reports have indicated that the changes in the abundance of some specific diazotrophic taxa such as Azospirillum, could affect crops N availability [75,76]. Similarly, members of the genus Variovorax (Comamonadaceae) have been shown to induce stress tolerance genes in wheat, promoting plant growth and yield under water-stress conditions [73]. Although the underlying role of keystone taxa in improving the microbiome stability and functions still need to be resolved, our findings about such bacterial taxa would improve the insight into the bacterial community recruitment by wheat cultivars under field conditions. This new knowledge, in our opinion, is pivotal to developing well-adapted wheat cultivars with a greater fitness and sustainability.
5. Conclusions
In this study, significant differences in the diversity, richness, and abundance of bacterial OTUs were observed between the root endosphere and rhizosphere, independently of wheat cultivars. A higher proportion of OTUs were shared across the rhizosphere in comparison with root endosphere samples, suggesting that plants recruit specific but different bacterial communities according to the plant compartment. In addition, potassium (K) was determined as the one of the main factors driving the rhizosphere bacterial components of wheat in these soils. A major proportion of functional groups predicted in the root endosphere communities were mainly attributed to chemoheterotrophy and aerobic chemoheterotrophy, whereas nitrogen-cycling-related functional predictions were less abundant in the rhizosphere samples. The co-occurrence network analysis revealed the complexity of the root–bacteria associations in wheat cultivars, highlighting the Enterobacteraceae, Comamonadaceae, Hyphomicrobiaceae, Xanthomonadaceae, Corynebacteraceae, Dermabacteraceae, and Staphylococcaceae families as the hubs or keystone taxa. The findings improve the current knowledge about the recruitment of specific bacterial communities in the two plant niches for different wheat cultivars under field conditions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13051392/s1. Table S1: Physicochemical parameters of sampled soils; Table S2: Relative abundance for most abundant Phyla; Table S3: Relative abundances for less abundant Phyla; Table S4: Relative abundance for most abundant Families; Table S5: Relative abundances for less abundant Families; Table S6: PERMANOVA (Adonis), PERMANOVA (QIIME2), and ANOSIM beta diversity tables; Table S7: Physicochemical parameters PCoA explanation values; Table S8: Functional traits prediction; Table S9: Shared OTUs relative abundances; Table S10: Shared OTUs taxonomy; Table S11: Network centralities and keystone taxa; Table S12: Raw OTUs relative abundance; Table S13: Alpha diversity indexes values per replicate.
Author Contributions
J.I.R. and N.G.I. performed the sampling and experimentation procedures. J.J.A. and J.I.R. designed the experiments and produced the first manuscript draft. Q.Z. and J.I.R. performed the bioinformatic and statistical analyses. Q.Z., V.V.S.R.G., M.A.J., J.M., J.I.R. and J.J.A. contributed to the interpretation of the findings and reviewed and edited the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by The National Fund for Scientific and Technological Development (FONDECYT) project no. 1201386 and 1221228 (M.A.J. and J.J.A.), FONDECYT Postdoctorado no. 3210594 (J.I.R.) by Chile National Research and Development Agency (ANID), Dirección de Investigación Universidad de La Frontera (DIUFRO) DI21-0044 (J.J.A.), and Millennium Science Initiative Program—ICN2021_044 (J.J.A.). The participation of V.V.S.R.G. was supported by CSIRO Ag & Food, Australia through the MOSH-Future Science Platform.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences reported are available in the NCBI GenBank database (http://ncbi.nlm.nih.gov: accessed on 9 August 2022 under PRJNA867668 code).
Conflicts of Interest
The authors declare having no conflict of interest for all data and experiments comprised in this manuscript.
References
- Arora, N.K.; Fatima, T.; Mishra, I.; Verma, S. Microbe-based Inoculants: Role in Next Green Revolution. In Environmental Concerns and Sustainable Development; Shukla, V., Kumar, N., Eds.; Springer: Singapore, 2020. [Google Scholar] [CrossRef]
- Acuña, J.J.; Campos, M.; de la Luz Mora, M.; Jaisi, D.P.; Jorquera, M.A. ACCD-producing rhizobacteria from an Andean Altiplano native plant (Parastrephia quadrangularis) and their potential to alleviate salt stress in wheat seedlings. Appl. Soil Ecol. 2019, 136, 184–190. [Google Scholar] [CrossRef]
- Durán, P.; Jorquera, M.; Viscardi, S.; Carrion, V.J.; Mora MD, L.L.; Pozo, M.J. Screening and characterization of potentially suppressive soils against Gaeumannomyces graminis under extensive wheat cropping by Chilean indigenous communities. Front. Microbiol. 2017, 8, 1552. [Google Scholar] [CrossRef]
- Lagos, L.; Maruyama, F.; Nannipieri, P.; Mora, M.L.; Ogram, A.; Jorquera, M. A Current overview on the study of bacteria in the rhizosphere by modern molecular techniques: A mini-review. J. Soil Sci. Plant Nutr. 2015, 15, 504–523. [Google Scholar] [CrossRef]
- Abdullaeva, Y.; Ratering, S.; Ambika Manirajan, B.; Rosado-Porto, D.; Schnell, S.; Cardinale, M. Domestication Impacts the Wheat-Associated Microbiota and the Rhizosphere Colonization by Seed- and Soil-Originated Microbiomes, Across Different Fields. Front. Plant Sci. 2022, 12, 806915. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, X.; Qin, Q.; Dinkins, R.D.; Zhu, H. The impacts of domestication and breeding on nitrogen fixation symbiosis in legumes. Front. Genet. 2020, 11, 00973. [Google Scholar] [CrossRef]
- Žiarovská, J.; Medo, J.; Kyseľ, M.; Zamiešková, L.; Kačániová, M. Endophytic bacterial microbiome diversity in early developmental stage plant tissues of wheat varieties. Plants 2020, 9, 266. [Google Scholar] [CrossRef]
- Rheault, K.; Lachance, D.; Morency, M.J.; Thiffault, É.; Guittonny, M.; Isabel, N.; Martineau, C.; Séguin, A. Plant genotype influences physicochemical properties of substrate as well as bacterial and fungal assemblages in the rhizosphere of Balsam Poplar. Front. Microbiol. 2020, 11, 575625. [Google Scholar] [CrossRef]
- Kinnunen-Grubb, M.; Sapkota, R.; Vignola, M.; Nunes, I.M.; Nicolaisen, M. Breeding selection imposed a different selective pressure on the wheat root-associated microbiome. FEMS Microbial. Ecol. 2020, 96, fiaa196. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Kavamura, V.N.; Robinson, R.J.; Hughes, D.; Clark, I.; Rossmann, M.; Melo, I.S.D. Wheat dwarfing influences selection of the rhizosphere microbiome. Sci. Rep. 2020, 10, 1452. [Google Scholar] [CrossRef]
- Zhang, Q.; Acuña, J.J.; Inostroza, N.G.; Duran, P.; Mora, M.L.; Sadowsky, M.J.; Jorquera, M.A. Niche Differentiation in the composition, predicted function, and co-occurrence networks in bacterial communities associated with antarctic vascular plants. Front. Microbiol. 2020, 11, 1036. [Google Scholar] [CrossRef]
- Harman, G.; Khadka, R.; Doni, F.; Uphoff, N. Benefits to plant health and productivity from enhancing plant microbial symbionts. Front. Plant Sci. 2021, 11, 610065. [Google Scholar] [CrossRef]
- Cangioli, L.; Mancini, M.; Napoli, M.; Fagorzi, C.; Orlandini, S.; Vaccaro, F.; Mengoni, A. Differential response of wheat rhizosphere bacterial community to plant variety and fertilization. Int. J. Mol. Sci. 2022, 23, 3616. [Google Scholar] [CrossRef] [PubMed]
- Cordero, E.J.; de Freitas, J.R.; Germida, J.J. Bacterial microbiomes associated with the rhizosphere, root interior, and aboveground plant organs of wheat and canola at different growth stages. Phytobiomes J. 2021, 5, 442–451. [Google Scholar] [CrossRef]
- Dilla-Ermita, C.J.; Lewis, R.W.; Sullivan, T.S.; Hulbert, S.H. Wheat genotype-specific recruitment of rhizosphere bacterial microbiota under controlled environments. Front. Plant Sci. 2021, 12, 718264. [Google Scholar] [CrossRef] [PubMed]
- Wipf, H.M.L.; Coleman-Derr, D. Evaluating domestication and ploidy effects on the assembly of the wheat bacterial microbiome. PLoS ONE 2021, 16, e0248030. [Google Scholar] [CrossRef]
- Simonin, M.; Dasilva, C.; Terzi, V.; Ngonkeu, E.L.; Diouf, D.; Kane, A. Influence of plant genotype and soil on the wheat rhizosphere microbiome: Evidences for a core microbiome across eight African and European soils. FEMS Microbiol. Ecol. 2020, 96, fiaa067. [Google Scholar] [CrossRef]
- Qiu, L.; Kong, W.; Zhu, H.; Zhang, Q.; Banerjee, S.; Ishii, S.; Sadowsky, M.J.; Gao, J.; Feng, C.; Wang, J.; et al. Halophytes increase rhizosphere microbial diversity, network complexity and function in inland saline ecosystem. Sci. Total Environ. 2022, 831, 154944. [Google Scholar] [CrossRef]
- Qiu, L.; Zhang, Q.; Zhu, H.; Reich, P.B.; Banerjee, S.; van der Heijden, M.G.; Sadowsky, M.J.; Ishii, S.; Jia, X.; Shao, M.; et al. Erosion reduces soil microbial diversity, network complexity and multifunctionality. ISME J. 2021, 15, 2474–2489. [Google Scholar] [CrossRef]
- Tian, L.; Lin, X.; Tian, J.; Ji, L.; Chen, Y.; Tran, L.S.; Tian, C. Research advances of beneficial microbiota associated with crop plants. Int. J. Mol. Sci. 2020, 21, 1792. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Yin, C.; Hulbert, S.; Paulitz, T.C. Core rhizosphere microbiomes of dryland wheat are influenced by location and land use history. Appl. Environ. Microbiol. 2020, 86, e02135-19. [Google Scholar] [CrossRef] [PubMed]
- Kuźniar, A.; Włodarczyk, K.; Grządziel, J.; Goraj, W.; Gałązka, A.; Wolińska, A. Culture-independent analysis of an endophytic core microbiome in two species of wheat: Triticum aestivum L. (cv. ‘Hondia’) and the first report of microbiota in Triticum spelta L. (cv. ‘Rokosz’). System. Appl. Microbiol. 2020, 43, 126025. [Google Scholar] [CrossRef] [PubMed]
- Rilling, J.I.; Acuña, J.J.; Sadowsky, M.J.; Jorquera, M.A. Putative nitrogen-fixing bacteria associated with the rhizosphere and root endosphere of wheat plants grown in an Andisol from southern Chile. Front. Microbiol. 2018, 9, 2710. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghalith, G.A.; Hillmann, B.; Ang, K.; Shields-Cutler, R.; Knights, D. SHI7 is a self-learning pipeline for multipurpose short-read DNA quality control. Msystems 2018, 24, e00202-17. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghalith, G.A.; Montassier, E.; Ward, H.N.; Knights, D. NINJA-OPS: Fast accurate marker gene alignment using concatenated ribosomes. PLoS Comput. Biol. 2016, 12, e1004658. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence Analysis. Appl. Environ. Microbiol. 2011, 77, 3219–3226. [Google Scholar] [CrossRef]
- Schloss, P.D. Reintroducing mothur: 10 years later. Appl. Environ. Microbiol. 2020, 86, e02343-19. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Peschel, S.; Müller, C.L.; von Mutius, E.; Boulesteix, A.L.; Depner, M. NetCoMi: Network construction and comparison for microbiome data in R. Brief. Bioinform. 2021, 22, bbaa290. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. Int. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- De Cáceres, M.; Sol, D.; Lapiedra, O.; Legendre, P. A framework for estimating niche metrics using the resemblance between qualitative resources. Oikos 2011, 120, 1341–1350. [Google Scholar] [CrossRef]
- Zhang, Q.; Acuña, J.J.; Inostroza, N.G.; Mora, M.L.; Radic, S.; Sadowsky, M.J.; Jorquera, M.A. Endophytic bacterial communities associated with roots and leaves of plants growing in Chilean extreme environments. Sci. Rep. 2019, 9, 4950. [Google Scholar] [CrossRef]
- Brachi, B.; Filiault, D.; Whitehurst, H.; Darme, P.; Le Gars, P.; Le Mentec, M.; Morton, T.C.; Kerdaffrec, E.; Rabanal, F.; Anastasio, A.; et al. Plant genetic effects on microbial hubs impact host fitness in repeated field trials. Proc. Natl. Acad. Sci. USA 2022, 119, e2201285119. [Google Scholar] [CrossRef]
- Germida, J.; Siciliano, S. Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol. Fertil. Soils 2001, 33, 410–415. [Google Scholar] [CrossRef]
- Suman, A.; Govindasamy, V.; Ramakrishnan, B.; Aswini, K.; SaiPrasad, J.; Sharma, P.; Pathak, D.; Annapurna, K. Microbial Community and Function-Based Synthetic Bioinoculants: A Perspective for Sustainable Agriculture. Front. Microbiol. 2022, 12, 805498. [Google Scholar] [CrossRef]
- Richardson, A.E.; Kawasaki, A.K.; Condron, L.M.; Ryan, P.R.; Gupta, V.V.S.R. Root Microbiome Structure and Microbial Succession in the Rhizosphere. In Rhizosphere Biology: Interactions between Microbes and Plants; Gupta, V.V.S.R., Sharma, A.K., Eds.; Springer Nature Singapore Pte Ltd.: Singapore, 2021; pp. 109–128. [Google Scholar]
- Donn, S.; Kirkegaard, J.A.; Perera, G.; Richardson, A.E.; Watt, M. Evolution of bacterial communities in the wheat crop rhizosphere. Environ. Microbiol. 2014, 17, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Gong, X. Niche differentiation rather than biogeography shapes the diversity and composition of microbiome of Cycas panzhihuaensis. Microbiome 2019, 7, 152. [Google Scholar] [CrossRef]
- Poudel, R.; Jumpponen, A.; Kennelly, M.M.; Rivard, C.L.; Gomez-Montano, L.; Garrett, K.A. Rootstocks shape the Rhizobiome: Rhizosphere and endosphere bacterial communities in the grafted tomato system. Appl. Environ. Microbiol. 2019, 85, 2. [Google Scholar] [CrossRef]
- Kawasaki, A.; Donn, S.; Ryan, P.R.; Mathesius, U.; Devilla, R.; Jones, A.; Watt, M. Microbiome and Exudates of the Root and Rhizosphere of Brachypodium distachyon, a Model for Wheat. PLoS ONE 2016, 11, e0164533. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the Dominant Soil Bacterial Taxa in Libraries of 16S rRNA and 16S rRNA Genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 623–631. [Google Scholar] [CrossRef]
- Bissett, A.; Fitzgerald, A.; Meintjes, T.; Mele, P.M.; Reith, F.; Dennis, P.G.; Breed, M.F.; Brown, B.; Brown, M.V.; Brugger, J.; et al. Introducing BASE: The biomes of Australian soil environments soil microbial diversity database. GigaScience 2016, 5, 21. [Google Scholar] [CrossRef]
- Cordero, J.; de Freitas, J.R.; Germida, J.J. Bacterial microbiome associated with the rhizosphere and root interior of crops in Saskatchewan, Canada. Can. J. Microbiol. 2020, 66, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, S.; Jiang, Z.; Wu, Y.; Huang, X. Gradient of microbial communities around seagrass roots was mediated by sediment grain size. Ecosphere 2022, 13, e3942. [Google Scholar] [CrossRef]
- Astorga-Eló, M.; Zhang, Q.; Larama, G.; Stoll, A.; Sadowsky, M.J.; Jorquera, M.A. Composition, Predicted Functions and Co-occurrence Networks of Rhizobacterial Communities Impacting Flowering Desert Events in the Atacama Desert, Chile. Front. Microbiol. 2022, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Robertson-Albertyn, S.; Alegria Terrazas, R.; Balbirnie, K.; Blank, M.; Janiak, A.; Szarejko, I.; Chmielewska, B.; Karcz, J.; Morris, J.; Hedley, P.E.; et al. Root Hair Mutations Displace the Barley Rhizosphere Microbiota. Front. Plant. Sci. 2017, 8, 1094. [Google Scholar] [CrossRef] [PubMed]
- Lidbury, I.D.; Borsetto, C.; Murphy, A.R.; Bottrill, A.; Jones, A.M.; Bending, G.D.; Hammond, J.P.; Chen, Y.; Wellington, E.M.; Scanlan, D.J. Niche-adaptation in plant-associated Bacteroidetes favours specialisation in organic phosphorus mineralisation. ISME J. 2021, 15, 1040–1055. [Google Scholar] [CrossRef]
- Xu, T.; Jiang, W.; Qin, D.; Liu, T.; Zhang, J.; Chen, W.; Gao, L. Characterization of the microbial communities in wheat tissues and rhizosphere soil caused by dwarf bunt of wheat. Sci. Rep. 2021, 11, 5773. [Google Scholar] [CrossRef] [PubMed]
- Van Elsas, J.D.; Pratama, A.A.; de Araujo, W.L.; Trevors, J.T. Microbial Interactions in Soil. In Modern Soil Microbiology, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2006; pp. 177–210. [Google Scholar]
- Chiarini, L.; Bevivino, A.; Dalmastri, C.; Nacamulli, C.; Tabacchioni, S. Influence of plant development, cultivar and soil type on microbial colonization of maize roots. App. Soil Ecol. 1998, 8, 11–18. [Google Scholar] [CrossRef]
- Mahoney, A.K.; Yin, C.; Hulbert, S.H. Community structure, species variation, and potential functions of rhizosphere-associated bacteria of different winter wheat (Triticum aestivum) cultivars. Front. Plant Sci. 2017, 8, 132. [Google Scholar] [CrossRef]
- Okubara, P.A.; Kornoely, J.P.; Landa, B.B. Rhizosphere colonization of hexaploid wheat by Pseudomonas fluorescens strains Q8r1-96 and Q2-87 is cultivar-variable and associated with changes in gross root morphology. Biol. Control. 2004, 30, 392–403. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Gałązka, A. Biodiversity in the Rhizosphere of Selected Winter Wheat (Triticum aestivum L.) Cultivars—Genetic and Catabolic Fingerprinting. Agronomy 2020, 10, 953. [Google Scholar] [CrossRef]
- Costa-Gutierrez, S.B.; Adler, C.; Espinosa-Urgel, M.; de Cristóbal, R.E. Pseudomonas putida and its close relatives: Mixing and mastering the perfect tune for plants. Appl. Microbiol. Biotechnol. 2022, 106, 3351–3367. [Google Scholar] [CrossRef]
- Nuccio, E.E.; Starr, E.; Karaoz, U.; Brodie, E.L.; Zhou, J.; Tringe, S.G.; Malmstrom, R.R.; Woyke, T.; Banfield, J.F.; Firestone, M.K.; et al. Niche differentiation is spatially and temporally regulated in the rhizosphere. ISME J. 2020, 14, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Dishetty, N.K.; Bhatnagar, S.; Eisenc, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef]
- Mauger, S.; Ricono, C.; Mony, C.; Chable, V.; Serpolay, E.; Biget, M.; Vandenkoornhuyse, P. Differentiation of endospheric microbiota in ancient and modern wheat cultivar roots. Plant-Environ. Interact. 2021, 2, 235–248. [Google Scholar] [CrossRef]
- Yu, Z.; Liang, K.; Huang, G.; Wang, X.; Lin, M.; Chen, Y.; Zhou, Z. Soil Bacterial Community Shifts Are Driven by Soil Nutrient Availability along a Teak Plantation Chronosequence in Tropical Forests in China. Biology 2021, 15, 1329. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cong, P.; Kuang, S.; Tang, L.; Li, Y.; Dong, J.; Song, W. Long-term excessive application of K2SO4 fertilizer alters bacterial community and functional pathway of tobacco-planting soil. Front. Plant Sci. 2022, 13, 1005303. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Li, Y.; Zhang, X.; Yu, K.; Huo, Y.; Zhu, J.; Wang, Y.; Zhou, Z.; Ali, S.; Tang, Q.; et al. Effects of potassium application on soil ecological resistance to Verticillium wilt of cotton (Gossypium hirsutum L.). Arch. Agron. Soil Sci. 2022, 68, 488–502. [Google Scholar] [CrossRef]
- Kuźniar, A.; Włodarczyk, K.; Grządziel, J.; Woźniak, M.; Furtak, K.; Gałązka, A.; Dziadczyk, E.; Skórzyńska-Polit, E.; Wolińska, A. New Insight into the Composition of Wheat Seed Microbiota. Int. J. Mol. Sci. 2020, 21, 4634. [Google Scholar] [CrossRef]
- Rascovan, N.; Carbonetto, B.; Perrig, D.; Díaz, M.; Canciani, W.; Abalo, M.; Alloati, J.; González-Anta, G.; Vazquez, M.P. Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci. Rep. 2016, 6, 28084. [Google Scholar] [CrossRef]
- Gaglio, R.; Cirlincione, F.; Di Miceli, G.; Franciosi, E.; Di Gerlando, R.; Francesca, N.; Settanni, L.; Moschetti, G. Microbial dynamics in durum wheat kernels during aging. Int. J. Food Microbiol. 2020, 324, 108631. [Google Scholar] [CrossRef]
- Ubalde, M.C.; Braña, V.; Sueiro, F.; Morel, M.A.; Martínez-Rosales, C.; Marquez, C.; Castro-Sowinski, S. The versatility of Delftia sp. isolates as tools for bioremediation and biofertilization technologies. Curr. Microbiol. 2012, 64, 597–603. [Google Scholar] [CrossRef]
- Jung, J.; Park, W. Acinetobacter species as model microorganisms in environmental microbiology: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 2533–2548. [Google Scholar] [CrossRef]
- Han, J.I.; Choi, H.K.; Lee, S.W.; Orwin, P.M.; Kim, J.; LaRoe, S.L.; Kim, T.G.; O’Neil, J.; Leadbetter, J.R.; Lee, S.Y.; et al. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J. Bacteriol. 2011, 193, 1183–1190. [Google Scholar] [CrossRef]
- Chandra, D.; Srivastava, R.; Gupta, V.V.; Franco, C.M.; Paasricha, N.; Saifi, S.K.; Tuteja, N.; Sharma, A.K. Field performance of bacterial inoculants to alleviate water stress effects in wheat (Triticum aestivum L.). Plant. Soil 2019, 441, 261–281. [Google Scholar] [CrossRef]
- Maier, S.; Kratz, A.M.; Weber, J.; Prass, M.; Liu, F.; Clark, A.T.; Abed, R.M.; Su, H.; Cheng, Y.; Eickhorst, T.; et al. Water-driven microbial nitrogen transformations in biological soil crusts causing atmospheric nitrous acid and nitric oxide emissions. ISME J. 2022, 16, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Pankievicz, V.; Plucani, D.; Amaral, F.; Ane, J.M.; Gary, S. Diazotrophic Bacteria and Their Mechanisms to Interact and Benefit Cereals. Mol. Plant-Microbe Interact. MPMI 2021, 34, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Friman, V.P.; Li, L.; Xu, Q.; Guo, J.; Guo, S.; Shen, Q.; Ling, N. Meta-analysis of diazotrophic signatures across terrestrial ecosystems at the continental scale. Environ. Microbiol. 2022, 24, 2013–2028. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).