Abstract
High yield and superior quality are the main objectives of rice breeding and research. While innovations in rice breeding have increased production to meet growing demand, the universal issue of balancing high yield and susperior quality has led to a lack of focus on improving rice quality. With rising living standards, improving rice quality has become increasingly important. Rice grain quality is a complex trait influenced by both genetic and environmental factors, with four primary aspects: milling quality, appearance quality, eating and cooking quality, and nutritional quality. While different populations have varying demands for rice quality, the core traits that contribute to rice quality include grain shape and chalkiness in terms of appearance, as well as endosperm composition that influences cooking and sensory evaluation. Researchers have made substantial advancements in discovering genes/QTLs associated with critical traits including appearance, aroma, texture, and nutritional properties. Markers derived from these genetic discoveries have provided an efficient tool for marker-assisted selection to improve rice quality. Thus, this review focuses on elite genes and their applications in breeding practices to quickly develop superior quality rice varieties that meet various market demands.
1. Introduction
Rice (Oryza sativa L.) is one of the most important staple crops in the world. High yield and superior quality have always been the main goals of rice breeding and basic research. In recent decades, through technological innovations such as dwarf breeding, heterosis utilization, and super rice breeding, rice production has increased significantly, basically meeting the growing demand for rice consumption. However, the contradiction between high yield and superior quality of crops is a universal scientific problem, and there has been insufficient attention given to rice quality in the actual rice variety breeding process. With the gradual improvement in people’s living conditions and consumption levels, improving the quality of rice is becoming increasingly important [1]. The primary focus of current basic research and genetic improvement in rice quality is to develop new superior quality rice varieties that meet diverse market demands and facilitate rapid improvement of rice quality.
Rice quality is a comprehensive trait that encompasses various fundamental characteristics of the product from rice production to consumption after processing. Rice quality is a complex trait influenced by genetic and environmental factors. Although different populations have different focuses on rice quality, rice breeders, producers, and consumers generally have a consistent definition of rice quality, which generally includes four aspects: milling quality, appearance quality, eating and cooking quality, and nutritional quality [2,3,4]. Each aspect of quality can be described by a series of corresponding quantifiable indicators. Milling quality is an important aspect that refers to the ability of rice to withstand the processing procedure, characterized by brown rice recovery (BR) and head rice recovery (HR), whereas appearance quality is primarily concerned with grain length (GL), grain width (GW), the length/width ratio, chalkiness, and grain translucency. Eating and cooking quality (ECQ) mainly reflects the properties and taste of cooked rice and is associated with three starch-related traits, namely, amylose content (AC), gel consistency (GC), and gelatinization temperature (GT). Meanwhile, rice nutritional quality (NQ) is determined by the content and quality of proteins, lipids, minerals, and other beneficial substances that can contribute to human health. While this article does not focus on discussing individual rice quality indicators, several excellent reviews or chapters in books published in recent years can help people gain a more comprehensive understanding of rice quality [5,6,7,8].
Different populations show diverse demands for rice quality. Milling quality is defined as the percentage of the marketable product obtained during the milling process, which begins with brown rice and ends with unbroken white rice. Obviously, milling quality is of greater concern to rice producers. BR and HR are important indicators of rice quality evaluation standards in different countries and are of interest to breeders [6]. The properties exhibited by rice grains following processing are known as the appearance quality, which includes grain shape, chalkiness, and translucency, and directly affects the commercial value of rice. Rice with a uniform shape, less chalkiness, good translucency, and a glossy appearance is more attractive to consumers [9]. Eating and cooking quality refers to all the physical and chemical characteristics and sensory properties of rice during the cooking process and eating. Similar to appearance quality, ECQ is a fundamental indicator of rice quality that consumers can directly perceive. While consumers in various regions may have distinct preferences for rice ECQ, superior quality rice generally exhibits intact grains that are clean and fragrant, soft and elastic but not sticky, palatable, and can maintain their texture when cold [6,10]. Rice is mainly composed of starch, with very low amounts of other nutrients including protein, amino acids, fat, vitamins, and mineral elements. However, additional nutrients in rice have received increasing attention due to their potential importance for maintaining human health.
In general, rice quality traits are determined by a combination of genetic and environmental factors. Core traits that contribute to rice quality include grain shape and chalkiness in terms of appearance, as well as the starch composition that influences cooking and sensory evaluation. The quality of rice grain is closely correlated with rice seed characteristics, such as the pericarp, seed coat, aleurone, starchy endosperm, and embryo [11]. Recently, many quantitative trait loci (QTLs) and critical genes responsible for rice quality traits have been identified, and useful molecular markers have been developed to improve rice quality. Notably, researchers have made significant progress in identifying and characterizing the genes responsible for crucial traits such as appearance (shape and chalk), aroma, texture, and nutritional properties. Here, we mainly discuss some elite genes and their applications in breeding practice.
2. Genetics of Grain Shape
Rice grain appearance is an essential and extensively researched quality trait that is usually determined by grain shape, chalkiness, and transparency. Grain shape is is defined by its length, width, and length/width ratio, and it is a crucial target trait for yield, appearance, domestication, and breeding [12,13,14,15,16,17]. In Indica rice, slender grains are preferred, while Japonica rice is characterized by short and round grains. Over the past few decades, geneticists have identified numerous QTLs related to grain shape in rice, but only a few have been cloned (Figure 1).
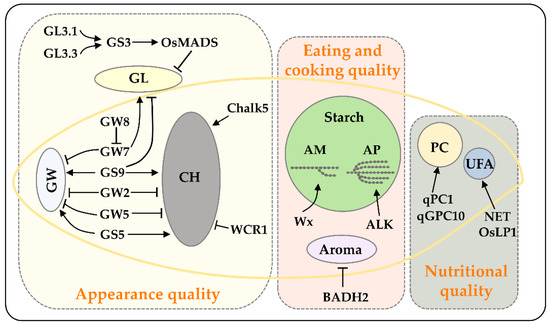
Figure 1.
The major genes controlling rice grain quality. This figure shows some extensively studied genes/QTLs related to the core quality traits including appearance quality, eating and cooking quality, and nutritional quality. Genetic dissection of these genes has enabled the development of functional markers to accelerate breeding programs for improving rice quality. GL, grain length; GW, grain width; CH, chalkiness; AM, amylose; AP, amylopectin; PC, protein content; UFA, unsaturated fatty acids.
2.1. Grain Length
The first QTL that has been cloned for grain length in rice was GS3 (Grain Size on Chromosome 3). GS3 was identified from a NIL population of Minghui 63 (large grain) and Chuan 7 (small grain) and it negatively regulates grain length. By comparing the GS3 sequences from rice varieties with different grain lengths, it was found that the long-grain varieties had a cysteine codon (TGC) to a termination codon mutation in the second exon of the GS3 gene, which resulted in premature termination and a 178-aa truncation of the C-terminus of the protein and led to larger grains [18]. This mutation was strongly selected in both Japonica and Indica rice varieties, providing an explanation for the prevalence of long grain in Indica rice genotypes [19]. GS3 has four functional domains (OSR in the N terminus, a transmembrane domain, TNFR/NGFR, and VWFC in the C terminus), and different alleles of GS3 formed due to natural variation [20]. The GS3 gene is a key regulator of grain size in rice, with different alleles exhibiting distinct effects on grain length. The wild-type GS3 allele (GS3-1) has a medium-length grain shape, which is the genotype for the majority of medium-grain Indica varieties. In most temperate Japonica varieties, such as Nipponbare, the GS3-2 allele has minimal impact on grain length due to a single amino acid difference at the C-terminus compared to that of GS3-1. In contrast, the GS3-3 allele in the Minghui 63 cultivar results in longer grains by eliminating OSR function. Similarly, the GS3-4 allele in the Chuan 7 cultivar leads to extremely short grains by the loss of functional domains at the C-terminus. It was later discovered that GS3 is a G protein γ subunit gene that regulates grain length through the G protein pathway [21,22]. GS3 negatively regulates rice grain length by competitively binding to G protein β subunits with two other γ subunits, DEP1 and GGC2, which inhibits the function of DEP1 and GGC2 and leads to shorter grains [22].
The GL3.1/qGL3 locus encodes a protein phosphatase kelch (PPKL) family member and functions by negatively modulating the longitudinal cell number in grain glumes. This locus is closely linked to GS3 and acts as an enhancer gene, enhancing the effect of GS3 on grain length [23,24,25]. Furthermore, it has been discovered that the GS3 locus displays epistatic interaction with GL3.3, a gene encoding a kinase similar to GSK3/SHAGGY. Plants carrying both gs3 and gl3.3 genotypes exhibit significantly longer grains compared to those carrying only one of the genotypes [26].
2.2. Grain Width
GW2 (Grain Width on Chromosome 2) was the first QTL cloned for rice grain width, which encodes a RING-type E3 ubiquitin ligase [27]. In exon 4 of the GW2 gene, a 1 bp deletion introduces a premature stop codon, leading to a truncated protein and a large-grain phenotype. GW2 negatively regulates cell division by directing its substrate to proteasomes for regulated proteolysis. The loss function of gw2 results in increased cell numbers in the spikelet hull, thereby, enhancing grain width, weight, and yield.
qSW5 (Seed Width on Chromosome 5)/GW5 (Grain Width on Chromosome 5) is a major QTL that controls rice grain width and has been mapped to a genomic region of 2263 bp and 21 kb, respectively [28,29]. The former two studies identified a 1212 bp deletion in this locus as a determinant of wide-grain phenotype. Further investigations revealed that the true gene of GW5 encodes a membrane-localized protein with an IQ calmodulin-binding motif domain and it is located downstream of the 1212 bp deletion [30,31]. GW5 participates in the brassinosteroid pathway by regulating grain growth through inhibition of the kinase activity of GSK2 on OsBZR1 and DLT [31]. Natural variation of GW5 has been found in different haplotypes, including a 1212 bp deletion in most Japonica rice varieties, a 950 bp deletion in most wide-grain Indica varieties, and no deletion in most narrow-grain Indica varieties [30]. This InDel marker can be used to identify functional alleles of GW5/qSW5 for wide and narrow grains. A recent functional analysis revealed that the transcription factor OsSPL12 promoted the expression of GW5 by directly binding to the 1212 bp region in most Indica varieties. This suggests that OsSPL12 may have co-evolved with GW5 to determine the differences in grain shape between Indica and Japonica rice [32].
GS5 (Grain Size on Chromosome 5) is another major QTL in rice that controls grain size by positively regulating grain width, filling, and weight, and it has no other significant effect. It encodes a putative serine carboxypeptidase, and higher expression of GS5 is correlated with larger grain size. The promoter region of GS5 shows the natural variation that is linked to grain width, thereby, contributing to the diverse grain size of rice [33]. Through the identification of two SNPs located between the wide-grain allele GS5-1 and the narrow-grain allele GS5-2 in the upstream region of the gene, differential expression of GS5 during the development of young panicles is attributed to these SNPs. Moreover, enhanced expression of GS5 competitively inhibits the interaction between OsBAK1-7 and OsMSBP1 by occupying the extracellular leucine-rich repeat (LRR) domain of OsBAK1-7. This, in turn, prevents OsBAK1-7 from undergoing endocytosis by interacting with OsMSBP1 and explains the positive association between grain size and GS5 expression [34].
2.3. Grain Length/Width
Grain length and width are negatively correlated, and this requires a mechanism to balance these two traits. Indeed, GW8 and GW7 loci have been proposed to interact and form a module for regulating grain shape. GW8 (Grain Width on Chromosome 8) is a positive regulator of grain width which encodes the OsSPL16 protein that is responsible for cell proliferation [35]. A 10 bp deletion in the OsSPL16 promoter in Basmati rice is associated with the formation of a slenderer grain. GW8 directly binds to the GW7 (Grain Width on Chromosome 7) promoter and represses its expression [36]. GW7, a locus also known as GL7 (Grain Length on Chromosome 7)/SLG7 (Slender Grain on Chromosome 7), is a positive regulator for the grain length/width ratio encoding a TONNEAU1-recruitment motif (TRM) protein homologous to Arabidopsis LONGIFOLIA proteins [36,37,38]. Copy number variations (CNVs) at the GW7 locus lead to the upregulation of GW7, inducing slender grains with higher grain appearance without loss in yield. Thus, the GW8-GW7 module represents a promising strategy for improving rice grain quality. In addition, GS9 (Grain Shape Gene on Chromosome 9) negatively regulates the length/width ratio of rice grains by regulating horizontal cell division and vertical cell elongation [39]. Introducing the gs9 null allele into elite rice cultivars produces significantly more slender rice grains than those with normal GS9 allele and with no changes in grain thickness or weight, suggesting its potential application in the breeding of rice varieties with optimized grain shape [39,40].
2.4. The Utilization of Grain Shape Genes in Genetic Improvement of Rice Grain Quality
The rapid progress in rice genetics and functional genomics has facilitated the development of numerous molecular markers. Marker-assisted selection (MAS) has become a widely used approach in conventional plant breeding programs. MAS is widely used in various breeding techniques such as backcross breeding, forward selection, reverse selection, gene pyramiding, and background screening. By incorporating genes related to grain shape, rice yields and appearance quality can be significantly improved [41] (Table 1).
Sequencing and association analyses of natural variant populations have confirmed that GS3 is the most effective and crucial gene that controls rice grain length. The premature termination of the gs3 mutation is advantageous for designing functional CAPS molecular markers to detect the long- and short-grain genotypes of GS3 [42,43]. Utilizing the five molecular markers developed for GS3, a chromosome segment harboring the recessive gs3 allele has been successfully introgressed into a leading cultivar, Kongyu 131, resulting in the development of new varieties with longer grains (a 12.05% increase in grain length) [44].


Table 1.
Elite alleles in the genetic improvements of rice grain quality by MAS.
Table 1.
Elite alleles in the genetic improvements of rice grain quality by MAS.
| Trait | Allele | Variation | Function Description | Reference |
|---|---|---|---|---|
| Grain shape | gs3 | Loss of function mutation in exon 2 | Increased grain length to produce slender grains. | [35,36,44,45,46] |
| GW7TFA | SNPs and Indels in the promoter | Pyramiding of GW7TFA and gs3 produced much longer grains and increased grain yield. | [36] | |
| gw8 | 10-bp deletion in the promoter | Pyramiding of gw8 and gs3 increased grain length with no yield loss. | [35] | |
| OsMADSlgy3 | An insertion–deletion polymorphism in the splice site of the intron 7/exon 8 junction | Pyramiding of lgy3 and gs3 increased grain length and yield. | [45] | |
| dep1 | A 625 bp deletion in exon 5 | Increased grain length in Japonica rice. | [46] | |
| gl3.1 | C-A at 1092 bp and C-T at 1495 bp | Increased grain length and yield in hybrid rice. | [24] | |
| GSE5ZJB | No deletion | Produced longer but smaller grains with lower chalkiness. | [47] | |
| gs9 | 3 bp Insertion in exon 1 and 4 SNPs in exon 4; 7 kb insertion at 311 bp | Increased grain length. | [39,40] | |
| Chalkiness | chalk5 | 2 SNPs in the promoter | Reduced grain chalkiness in Indica rice. | [48] |
| WCR1A | A functional SNP (A/G) at −1696 bp in the promoter | Reduced grain chalkiness in Japonica rice. | [49] | |
| Eating and cooking quality | Wxmq | G-A in exon 4 | Decreased the AC to 10~15% to produce semi-glutinous rice. | [50] |
| Wxb | G-T at splicing site in intron 1 | Decreased the AC to 10~15%. | [51,52,53] | |
| Wxmw | A-C in exon 6 | Decreased the AC to 14% and improved endosperm transparency. | [54] | |
| ALKb | 3 SNPs in exon 8 | Decreased the AC and GT. | [55] | |
| badh2-E7 | 8 bp Deletion and 3 SNPs in exon 7 | Produced rice variety with aroma. | [55] | |
| Protein content | qPC-1Habataki | Not clear | Decreased the protein content to enhance ECQ. | [56,57] |
The combination of genes involved in grain shape is a valuable approach for producing rice varieties with high yield and superior quality. The large number of genes that regulate grain shape, as well as their wide genetic variation, allow breeders to create diverse rice grain shapes. For example, the pyramiding of gw8 and gs3 into a short-grain variety (HJX74) has produced long-grain shaped rice similar to Basmati 385. Similarly, the development of a new elite Indica variety, Huabiao1, with elongated grains, was achieved by pyramiding these two genes, which substantially improved grain quality [35]. Furthermore, the CNVs and InDel in the GW7 locus can be used to create functional molecular markers without any significant adverse effects, allowing for the design of superior-quality rice varieties. The new hybrid Indica rice varieties, Taifengyou55 and Taifengyou208, developed through QTL pyramiding of the GW7TFA (an allelic variant of the GW7 from a cytoplasmic male sterility line TaifengA) and gs3 alleles, have substantially improved grain quality [36]. Pyramiding of OsMADS1lgy3 (a key effector downstream of GS3 to regulate grain length) and gs3 alleles in the Liangyoupeijiu genetic background has been shown to enhance overall grain yield with improved grain quality [45].
Indica rice is known for having longer grains and better appearance quality compared to Japonica rice. In recent years, improving the grain shape and appearance quality of Japonica rice has been a major focus. The development of the Jiahe series serves as an exemplary case of improving Japonica rice grain shape [17]. The Jiahe series of rice varieties are characterized by enhanced appearance quality and elongated grain shape, which can be attributed to the incorporation of dep1 and gs3 into the breeding program. DEP1 (Dense and Erect Panicle 1) encodes another atypical Gγ subunit that is similar to GS3 [46]. The successful integration of gs3, GW7TFA, and dep1 led to the development of Jiahe 218, a novel long-grain Japonica rice variety with improved grain shape and quality. Derivatives of Jiahe, such as Zhongjia 8 and Jiafengyou 2, also carry gs3 and dep1 and have both good grain shape and quality.
In addition, some other beneficial alleles that control grain shape have been utilized. The qgl3 allele has been applied in breeding elite rice varieties, and crossing between 9311 and NIL-qgl3 with three commercial photo-thermosensitive male sterile lines has been shown to increase grain length and yield without negatively impacting grain quality [24]. The introgression of the gs9 allele, which has slender grains due to the loss of function, has also been used to improve the appearance quality of the Japonica variety Wuyungeng 27 which carries dep1-1 [39,40]. Moreover, pyramiding multiple favorable genes has proven effective in significantly enhancing the appearance quality of rice grains. Rational allelic combinations of the GW5NIP-GS39311-GW79311-GW8NIP-LGY39311 module simultaneously improve grain yield and quality [58], offering promising implications for rice improvement in the future. A recent genetic analysis of grain shape-related genes in the Guangdong Simiao rice varieties, a widely cultivated series of rice varieties in southern China, has identified GS3, GW5, GL7, and GS5 as crucial genes. The study found that the combination of GS3allele3-GW5allele2-GL7allele2-GS5allele2 is the most predominant allelic combination associated with grain shape in this rice variety series [59].
3. Genetics of Grain Chalkiness
Grain chalkiness is one of the most important traits in rice grain quality since it results in an opaque endosperm phenotype caused by the loose and irregular distribution of starch granules and protein bodies [60]. This undesirable trait affects the appearance, milling, cooking, and nutritional quality of rice, thereby, decreasing its marketability and commercial value [10]. Since the formation of chalkiness is highly susceptible to environmental conditions, especially temperature, and the complexity of measuring the chalkiness trait, the majority of major QTLs that impact this trait are challenging to identify. For naturally occurring chalkiness, only two QTLs have been successfully cloned so far (Figure 1).
Chalk5 (Chalk on Chromosome 5) is the first major QTL that has been cloned in rice and is responsible for positively regulating grain chalkiness [48]. It was identified from a double-haploid population created from a cross between two Indica varieties, H94 (with low chalkiness) and Zhenshan 97 (with high chalkiness). Chalk5 encodes a vacuolar H+-translocating pyrophosphatase, and its increased expression leads to an elevation in vacuole H+ concentration, which disturbs endomembrane pH homeostasis and protein formation, causing the formation of air gaps. This abnormal spatial arrangement of storage substances eventually leads to an increase in grain chalkiness [48]. Two conserved SNPs in the Chalk5 promoter result in the high chalkiness of Zhenshan 97, which carries a naturally highly expressed allele of Chalk5, partly accounting for the differences in its mRNA level [48]. Chalk5 is considered to be the major QTL affecting the chalkiness variation in most Indica rice varieties.
The WCR1 (White-Core Rate 1) gene, encoding an F-box protein, is a recently discovered gene that negatively regulates grain chalkiness [49]. It reduces chalkiness by inhibiting the accumulation of reactive oxygen species (ROS) and delaying programmed cell death (PCD) in the endosperm. Additionally, a functional SNP (A/G) located at −1696 bp in the WCR1 promoter region has been associated with both WCR phenotype and nucleotide variation in Asian-cultivated rice accessions. This functional SNP can affect transcription factor binding sites, and molecular markers developed based on this SNP can be used to efficiently identify individuals carrying desirable alleles during selection, potentially speeding up the breeding process by allowing more efficient screening of large populations [49].
Through the process of long-term breeding practices, it has been discovered that slender-grain rice varieties generally possess better appearance qualities such as low chalkiness, while wide-grain varieties tend to have poorer quality. Studies have also shown that grain shape and chalkiness are often co-related, with cloned grain shape QTLs frequently exhibiting chalkiness as a co-effect [61]. The correlation between grain shape and chalkiness has been confirmed, with certain QTLs such as gw2 [27] and GS2 [62] showing a significant increase in chalkiness while enhancing grain width/weight, and others such as GL7/GW7 [36,37], gw8 [35], and gs9 [39] reducing chalkiness percentage by increasing grain length and decreasing grain width. A recent study identified that the grain width QTL GW5/GSE5 has pleiotropic effects on both chalkiness and grain shape [47]. Introducing the GSE5ZJB allele to the Indica rice varieties Zhenshan 97B showed lower chalkiness and longer but smaller grains. Therefore, increasing grain length provides an effective strategy for reducing chalkiness.
Unlike the diverse requirements for grain shape, low chalkiness is a common demand in rice breeding. The development of molecular markers for fine-mapped QTLs related to chalkiness is important in breeding practices. Introducing QTLs or genes with significant genetic effects into the Indica and Japonica cultivars can be an effective strategy for producing superior quality rice with a high yield (Table 1). However, high yield and superior quality are often contradictory and difficult to reconcile. The mining of Chalk5, which is tightly linked to GW5 and GS5, has provided genetic and molecular evidence that has helped to overcome this contradiction [48]. On the one hand, in Indica rice, the haplotype block comprising chalk5, gs5, and GW5, which are tightly linked, leads to the formation of slender, low-chalkiness grains. On the other hand, in Japonica rice, the combination of Chalk5, GS5, and gw5 genes results in the development of wide grains that exhibit high chalkiness. Therefore, breeders generally select slender grain seeds to achieve the coordination of high yield and superior quality in Indica rice. Alternatively, the use of WCR1A from the Japonica variety represents a novel way to overcome the negative association between quality and yield for variety improvement, especially for Indica varieties. Introducing WCR1A from the Japonica variety into the widely planted Indica accession 9311 has resulted in lower chalkiness, higher grain length, and increased grain yield [49].
To enhance appearance quality, the primary objectives should be centered on reducing grain chalkiness, enhancing transparency, and generating grains with the appropriate size and shape that fulfill the demands of diverse consumers. The screening and pyramiding of superior alleles of grain shape and chalkiness will be beneficial for producing excellent grain appearance lines [63].
4. Genetics of Eating and Cooking Quality
The eating and cooking quality (ECQ) of the rice grain is a fundamental component that reflects the flavor of rice. Good ECQ is represented by good palatability, an aromatic scent, a white and shiny appearance, a pliable and supple texture, non-stickiness, and no hardening or rebounding after cooling [10]. Artificial taste testing is not accurate for evaluating ECQ; therefore, three classic physicochemical indicators are commonly used as references: amylose content (AC), gel consistency (GC), and gelatinization temperature (GT) [64]. Starch, the primary component of rice, is divided into amylose and amylopectin based on distinct molecular structures, and amylose is considered the most significant determinant of rice quality [65]. Rice with moderate AC (14~20%) has a soft and fluffy texture which is preferred by most consumers, while glutinous rice is best with a low AC (<2%). GC is a crucial parameter for assessing the textural properties of cooked rice and the stickiness of cooled paste for cooked rice flour. A softer GC is often preferred by consumers. In addition, the GT of rice starch is a key determinant of the rice cooking time. Rice with lower gelatinization temperature (<70 °C) requires less water and a shorter time for rice cooking, making it more popular. These characteristics are interrelated and jointly determine the ECQ of rice; AC is negatively correlated with GC and GT value, while GC is positively correlated with the GT value [66,67,68].
The endosperm, which mainly comprises starch and protein, constitutes the primary edible part of the rice seed. An evaluation of rice ECQ is primarily related to the physicochemical characteristics of starch. At the molecular level, genes involved in endosperm starch synthesis and regulation potentially play a significant role in the formation of rice ECQ [69,70]. For detailed information related to starch biosynthesis, please refer to the excellent review and book chapters published recently [71,72]. Here, we shall concentrate solely on several crucial genes that dictate rice quality. The biosynthesis of starch in the endosperm of cereals is a highly conserved pathway, involving several key enzymes such as adenosine diphosphate (ADP)-glucose pyrophosphorylase (AGPase), granule-bound starch synthase (GBSS), soluble starch synthase (SS), starch-branching enzyme (SBE), and starch debranching enzyme (DBE) [72,73]. AGPase is responsible for the synthesis of ADP glucose (ADPG), which serves as the primary substrate for starch synthesis [70]. ADPG is then transported to the amyloplast to synthesize amylose and amylopectin. In brief, amylose is synthesized mainly by GBSS I, whereas amylopectin is synthesized by a combination of multiple isoforms of SS, SBE, and DBE [70]. The complexity of the amylose and amylopectin composition, which is controlled by multiple isozymes of enzymes, makes it difficult to genetically dissect ECQ [64].
4.1. Wx Determines the Amylose Content
In rice, GBSSⅠ is encoded by the Wx (Waxy) gene, which is mainly responsible for the synthesis of amylose (Figure 1). Variations in allelic forms of Wx determine the amylose content in rice grain and serve as a decisive factor in regulating ECQ [64,74,75]. Wx is located on chromosome 6 and consists of 14 exons and 13 introns [76]. It is specifically expressed in the endosperm of developing rice seeds, with the transcription start site located in the first exon and the translation start codon located in the second exon [77,78]. A 3.3 kb pre-mRNA is first transcribed, which is then spliced to form a mature mRNA of 2.3 kb that is further translated into GBSSⅠ. Glutinous rice and high-AC varieties produce mRNA of 3.3 kb and 2.3 kb, respectively, while the intermediate-AC varieties exhibit a combination of the two [77]. Any changes in the structure of the Wx exons and introns can affect both the expression level and protein function. There are multiple alleles of the Wx gene in rice, each with distinct single nucleotide polymorphisms (SNPs) and varying amylose content (AC) levels [54,79,80,81,82,83,84,85,86]. The Wxa and Wxlv alleles have AC levels greater than 25%, while Wxin has AC levels between 18% and 22%, and Wxb has AC levels between 15% and 18%. Other alleles such as Wxmw, Wxla, Wxmq, Wxmp, Wxop, and Wxhp have AC levels ranging from approximately 10% to 15%. The wx allele has the lowest AC level of around 2%. Two major alleles, Wxa and Wxb, are predominantly distributed in Indica and Japonica rice varieties, respectively, and evolved from the ancestral Wxlv allele that originated from wild rice. The original Wxlv allele underwent mutations to become Wxa in the Indica subspecies and Wxb in the Japonica subspecies. Later, Wxlv mutated to Wxin in Indica, and Wxb mutated to Wxmq in Japonica as a result of artificial mutagenesis [85]. Based on the distinct characteristics of these various Wx alleles, molecular markers can be used to assist in the targeted improvement of rice AC and ECQ. For example, a functional marker for Wxin has been developed to screen lines with intermediate AC, which is identified by a SNP (A to C) in exon 6 that distinguishes high and intermediate amylose varieties [82,87].
In addition to its impact on rice AC, Wx also serves as the major gene for controlling GC [88,89]. The dominant QTL for GC, qGC6, has been confirmed to be located at the Wx locus [90]. GC and AC have been shown to have a significant negative correlation. Therefore, Wx exhibits pleiotropy and acts as a major gene affecting the ECQ of rice.
4.2. ALK and Wx Influence Gelatinization Temperature
The gelatinization temperature (GT) is another important indicator of rice ECQ and is determined by the alkali degeneration (ALK) gene located near the Wx locus on chromosome 6. ALK, the dominant QTL for GT, encodes the SSⅡa enzyme, which plays a critical role in the formation of amylopectin medium-length branching chains [91] (Figure 1). A comparison of the ALK gene sequences in different rice varieties has revealed that two amino acid substitutions within the coding region of the gene are responsible for changes in SSⅡa enzyme activity. These changes affect the synthesis of amylopectin medium-length branching chains, alter the crystal structure of starch granules, and ultimately result in changes in the GT [91,92,93]. Previous studies have identified at least three SNPs in exon 8 of the ALK gene that are tightly associated with GT variation, including positions Ex8-733 bp (A/G), Ex8-864 bp (G/T), and Ex8-865 bp (C/T) [81,94,95]. These SNPs generate three haplotypes, including ALKa (A-GC) and ALKb (G-TT), which control low GT, and ALKc (G-GC), which controls high GT [68,96].
The Wx gene plays a major role in regulating both AC and GC and also has a minor effect on GT. Allelic variations in the Wx gene could significantly affect the activity of SSIIa during grain filling. By pyramiding different haplotypes of Wx and ALK, a better ECQ has been achieved for lower AC, lower GT, and soft GC [97]. ALK also has an impact on AC, GC, and pasting properties. The loss-of-function mutant ssⅡa exhibits higher AC with a reduced GT compared to the wild type [95]. Taken together, the combination of allelic variants of Wx and ALK genes are responsible for most of the phenotypic variations in GT and AC in rice varieties, and these genes are key factors in determining the ECQ of rice.
4.3. Aroma
The aroma of rice is a key attribute of its eating and sensory quality, and it can have a significant impact on market price. Currently, rice varieties that exhibit a distinctive aroma are highly sought after by producers and consumers. Extensive research has revealed the presence of hundreds of volatile compounds in rice, among which 2-acetyl-1-pyrroline (2-AP) is considered to be the principal compound responsible for the fragrance of rice grains. Extensive studies have focused on its genetic regulation. Now, it is well known that fragrance in rice results from the loss of function of the betaine aldehyde dehydrogenase 2 (badh2) gene on chromosome 8 [98,99] (Figure 1), which leads to the enhancement of 2-AP biosynthesis in fragrant rice [100]. The badh2 gene is composed of 15 exons and 14 introns, and its non-functional recessive alleles display a range of variations throughout the sequence. Currently, more than 10 alleles of badh2 have been identified, such as badh2-E7, badh2-E2, badh2-E4-5, badh2-p-50UTR, and badh2-p [98,100,101,102,103,104,105]. An 8 bp deletion in exon 7 (badh2-E7) is the predominant allele in most aromatic varieties including the famous Jasmine and Basmati fragrant rice [98,100], and a haplotype analysis has revealed that the badh2 allele in Indica varieties originated from Japonica rice [102,106]. The nucleotide diversity of badh2 allows the development of specific molecular markers to discriminate aromatic and non-aromatic rice cultivars by PCR amplification [101,103,107], which greatly facilitated the selection and breeding of aromatic rice cultivars.
4.4. Genetic Improvement of Rice Eating and Cooking Quality
As the core of improving rice quality, the goal of ECQ improvement is to obtain rice varieties that possess desirable characteristics such as good palatability, aromatic scent, white and shiny appearance, pliable and supple texture, no stickiness, and no hardening or rebounding after cooling [10]. Fortunately, the relevant genes that determine these traits have been cloned, and their abundant allelic variations have been utilized by breeders for ECQ improvements through marker-assisted selection (MAS) (Table 1).
Most consumers prefer medium-grain rice with a relatively soft texture; therefore, improving the cooking and eating quality of rice usually focuses on reducing amylose content (AC). MAS of the Wx alleles has been widely used to improve grain quality for a long time. Decades ago, a DNA marker for Wxmq was applicated to varietal identification in Japan [79]. Later, several novel Wx alleles were utilized to breed rice varieties with better eating quality in Japan [108,109,110]. For example, the Japonica rice variety Yumepirika, which has a low amylose content of 16.1% due to the presence of the Wx1-1 allele (a 37 bp deletion in intron 10), is an example of a rice variety which known for its sticky texture. It has been developed and assessed as a superior quality rice variety for the Japanese market [109,110]. In China, the Wxb allele is the most widely used gene in the ECQ improvement process, especially in hybrid rice. The poor quality of the widely used female parent of hybrids, Zhenshan 97, was improved by updating the Wx locus, which led to reduced AC and increased GC and GT [51]. Using MAS, the Wxb allele was introgressed into the maintainer lines of Longtefu B and Zhenshan 97B, resulting in the development of low-amylose content maintainers and restorers with improved cooking and eating quality. The allelic combination significantly reduced the AC and improved cooking and eating quality [52]. Similarly, the hybrid variety Xieyou 57 was also improved by the incorporation of the Wxb allele, leading to better characteristics in terms of ECQ parameters [53]. Recently, the Wx genotyping for thirty-six main parents of hybrid rice in China revealed that only Wxa, Wxlv, and Wxb alleles existed in these main parents, and the allelic combination gradually changed to Wxb/Wxb as the quality improved [111]. In addition to Wxb, the Wxmq allele that controls low AC has been successfully used for superior quality rice breeding in China over the past decade. The low-AC rice is also called semi-glutinous rice or soft rice [50]. Consumers prefer this type of rice due to its favorable eating quality, which combines the softness of glutinous rice and the elasticity of non-glutinous rice. When cooked, this rice has a soft texture and excellent ability to puff up. A series of Japonica varieties, such as Nangeng 46, Nangeng 5055, and Nangeng 9108, with low AC (10–15%), have been produced through the combination of high-yield Japonica varieties with the good quality japonica rice “Kantou 194,” which harbors the Wxmq allele [50,112]. It should be noted that the low AC in soft rice does not necessarily mean better quality, as grains with excessively low AC often exhibit dark or dull endosperm appearances and reduced transparency, leading to a decline in overall appearance quality. However, the recently discovered Wxmw allele shows promise as a solution to this issue, as it not only contributes to a relatively low AC of approximately 14% but also improves endosperm transparency, potentially enhancing the appearance quality of soft rice [54].
For aroma improvement, badh2 currently stands as the only target gene in rice breeding programs. To this end, several gene-specific molecular markers, highly correlated with 2-AP levels, have been developed for aroma enrichment by MAS [113,114]. The utilization of CRISPR/Cas9 gene editing has led to the generation of a succession of badh2 null alleles, resulting in the successful cultivation of numerous high-aroma rice cultivars [115,116,117,118]. Nevertheless, it should be noted that the badh2 gene cannot fully account for all the phenotypic variations observed in aromatic rice cultivars. Some of these cultivars, despite not having any functional badh2 gene variant, possess relatively elevated levels 2-AP levels and exhibit distinct aromatic characteristics [10,119]. Furthermore, the 2-AP content varies significantly among different aromatic rice types and cultivars, with some cultivars containing low 2-AP levels [119]. Hence, it is reasonable to speculate that there may exist other genes or chemical constituents that could influence or regulate the aromatic properties of rice and that further analysis of its genetic mechanisms is required for the effective utilization of this trait.
Typically, enhancements in ECQ traits have been achieved through the manipulation of a single gene. However, ECQ is a complex trait, and pyramiding multiple beneficial alleles is necessary for ECQ improvements. Additionally, ALK has also been proven to be a good target for rice ECQ improvement by altering rice amylopectin [96]. In a previous study, Wxa, ALKb, and badh2-E7 were introgressed into a key maintainer line Ⅱ-32B with poor quality using MAS. As a result, it exhibited low AC and GT with aroma as expected [55]. Similarly, it has been proposed that the coordinated expression of ALK and Wx is a feasible approach for rice ECQ improvement [120]. Through rational design, new rice varieties have been developed by introducing favorable alleles from Nipponbare (Wx and ALK) that control good ECQ and alleles from 9311 (GS3 and GW5) that control good appearance traits into the Indica rice variety Teqing. As a result, the appearance, ECQ, and yield were promoted simultaneously [121].
5. Genetics of Nutritional Quality
In addition to starch, brown rice contains other nutrients such as proteins, storage lipids, as well as trace amounts of amino acids, minerals, vitamins, and phytochemicals [122]. Milled rice leaves only endosperm with the removal of embryo and bran layers through polish processing. Therefore, the nutritional quality of rice grain is determined by the major components of the polished grain, which are proteins and lipids [10] (Figure 1).
5.1. Protein
Rice grains have protein as their second main component. Protein content (PC) varies across different rice varieties and is typically found to be between 5% and 16%, with Indica rice generally having a slightly higher content (from 2% to 3%) compared to Japonica rice [123,124,125]. Grain protein in rice is mainly composed of glutelins, prolamins, globulins, and albumins. Among them, glutelin, as the most abundant storage protein, has the highest nutritional value because of its high digestibility and lysine content [126]. Any significant changes in glutelin content will certainly affect rice quality. Protein content is a key factor not only for evaluating the nutritional quality of rice but also for its eating and cooking quality [127]. Previous research indicates that higher protein content generally leads to reduced ECQ, while rice grain with lower protein content generally exhibits better ECQ [128,129,130].
There have been significant efforts aimed at understanding the genetic basis of rice PC. Many QTLs for this trait have been detected [56,131,132,133,134,135,136,137,138,139]. However, PC is very susceptible to environmental conditions, and almost all studies have used phenotyping data from only a single environment. Thus, the detected QTLs for rice PC are frequently inconsistent because of the different environments or populations researchers used. To date, only two major QTLs, qPC1 and qGPC-10, which underlay natural variation and control PC in rice, were map-based cloned and functionally characterized [125,140]. The qPC1, encoding a putative amino acid transporter OsAAP6, controls PC by regulating the synthesis and accumulation of storage proteins and starch [140]. It functions as a positive regulator of PC in rice. By increasing the expression of this gene in rice varieties with low PC, the total amount of amino acids and PC can be increased, which can ultimately improve the nutritional quality of the grain. A genetic variation analysis revealed that two nucleotide changes in the OsAAP6 5′-UTR seem to be associated with PC diversity mainly in Indica cultivars [140]. The qGPC-10/OsGluA2 is another positive PC regulator which encodes a glutelin type-A2 precursor [125]. OsGluA2 enhances PC by increasing the total amount of glutelin content. Similar to qPC1, no variation has been found in the coding regions of this gene, but one SNP was beendetected upstream of the coding regions. This functional SNP could account for the expression level differences between the OsGluA2 alleles and it clarifies two haplotypes, OsGluA2LET and OsGluA2HET. OsGluA2LET is the low PC type which mainly residues in Japonica cultivars, while the high PC type OsGluA2HET is in Indica cultivars [125].
Generally, a high grain protein content is thought to be favorable for nutritional value, while high PC usually leads to densely structured rice grains, and thus, poor palatability [141,142]. Hence, balancing nutrition quality and eating and cooking quality in rice grain is a feasible strategy in breeding practice. Developing rice varieties with desirable ECQ has led to a focus on reducing the protein content in rice grains. For example, many famous commercial varieties with good ECQ usually contain PC less than 7%, such as Koshihikari in Japan and Kongyu131 in northern China [125]. Little progress has been achieved in the breeding improvement of rice PC due to limited gene resources. The identified QTL qPC-1 has been applied to rice quality improvement [56]. When the Habataki allele of qPC-1 was introduced into a Japonica background, it resulted in decreased protein content but improved eating and cooking quality (Table 1). The same allele was also utilized in the genetic improvement of Nangeng 46 to increase its palatability [57]. OsAPP6 and OsGluA2 are the potential candidates known so far for the manipulation of rice PC known. The low expression alleles, OsAPP6 and the OsGluA2LET, are promising target genes for low PC rice breeding through MAS. Still, more valuable gene resources are needed for rice nutritional quality and eating and cooking quality improvement.
5.2. Lipids
Lipids are mainly stored in rice embryo and aleurone, which are the bran layers. The lipid content in rice endosperm is very low. Rice lipids contain a high proportion of triacylglycerols (TAGs) along with a smaller amount of phospholipids (PLs) and free fatty acids [143]. The main fatty acids include palmitic acid, oleic acid (OA), and linoleic acid (LA). Among them, the unsaturated fatty acids, OA and LA, are good for human health because they are essential nutrients that cannot be synthesized by the human body [8,17]. Lipids not only affect rice nutritional quality but also influence the ECQ. Phospholipids and glycolipids can form complexes with amylose and amylopectin, reducing the expansibility of starch and increasing the GT, thereby affecting the texture of cooked rice [144,145]. In addition, the unsaturated fatty acids of milled grain have been reported to contribute to the rice aroma [146]. Fragrant rice varieties generally have a higher concentration of unsaturated fatty acids in their grains. Therefore, increasing the content of unsaturated fatty acids can be a potential target for improving both the rice ECQ and the NQ. Recently, new progress has been achieved in deciphering the genetic basis of oil biosynthesis in rice grains. A GWAS research on oil composition and oil concentration identified natural variants in four genes (PAL6, LIN6, MYR2, and ARA6) involved in oil metabolism and they showed variation among rice subspecies [147]. A rice grain nutrition QTL, NET (Nutrition, Eating, Taste), regulates lipid content and accumulation of nutrient metabolites, such as vitamins, amino acids, and polyphenols, thus, influencing the taste of rice grain [148]. Another GWAS study identified a key glycerolipid-related gene, OsLP1 (diacylglycerol choline phosphotransferase) that contributes to variations in saturated TAG [149]. The allelic variation in the OsLP1 sequence between Indica and Japonica results in different saturated TAG levels. Although there is no evidence for their use in MAS, these genes/QTLs described above showed clear diversities among Indica and Japonica subpopulations and could be used as biomarkers to facilitate breeding for enhanced oil and grain quality in the future.
6. External Factors That Affect Rice Grain Quality
In addition to the main genetic factors related to the variety, the improvement of rice quality is largely influenced by the environment and cultivation practices. For instance, the northeast region of China is a famous production area for superior quality Japonica rice, characterized by both excellent appearance and taste quality. This is largely due to the long growth period, low occurrence of high-temperature weather, and high organic matter content in the soil. Conversely, various factors in the southern region of China, such as short growth periods, extreme weather, and heavy use of fertilizers, are not conducive to improving the quality of rice. Conversely, various factors in the southern region of China, such as short growth periods, the occurrence of extreme weather, and heavy use of fertilizers, are not conducive to improving the quality of rice. As a result, the rice produced in this region often has poor appearance quality and high protein content. Additionally, the expression of fragrance genes is easily influenced by environmental factors such as soil, water, temperature, and light [102,119]. Therefore, many fragrant rice varieties lose their fragrance when grown outside their original production area. Most of the quality traits of rice are quantitative traits that are highly susceptible to environmental influences, which makes it challenging to improve rice quality through conventional methods. Adverse environmental factors, such as drought, low temperature, high temperature, and salinity stress, not only affect rice yield, but also have a significant impact on its appearance quality, eating and cooking quality, milling quality, and nutritional quality [150]. For instance, drought stress at the flowering stage has been shown to induce no significant effect on the appearance and nutritional quality except for increased grain chalkiness [151]. Low temperatures during the flowering period have been reported to improve milling and nutritional qualities but reduce the cooking and eating quality of late-season rice in southern China [152]. Salinity adversely affects the grain quality by decreasing the head rice recovery, amylose content and by increasing the chalky rice rate [153].
6.1. High Temperature Predominantly Affects Rice Quality
Traditionally, drought, salinity, and related stresses could be managed manually through appropriate field management, but temperature cannot be controlled artificially. Temperature stress, especially high temperature (HT), is the primary ecological factor that affects rice quality. During the entire growth period of rice, the grain filling stage has the greatest impact on rice quality, and this stage often coincides with high temperatures. In years with an extreme climate, high temperature can result in poor grain filling, decreased milling quality, and increased chalkiness. Therefore, in recent years, exploring the genetic mechanisms underlying the decline in rice quality caused by high temperatures has been a hot research topic.
During grain filling, HT stress can alter the chemical composition of rice grains, including starch and storage proteins, which, in turn, affects the quality of the rice [154] (Figure 2). Since starch is the main component of the rice grain, most of the research has focused on the effects of HT on starch. HT can reduce the content and change the structure of grain starch, leading to a reduction in amylose content and short-chain amylopectin, an increase in long-chain amylopectin, and an increase in gelatinization temperature [155,156,157]. Genetically, HT-induced impairment of starch accumulation is primarily due to decreased expression of genes involved in starch biosynthesis, particularly GBSSⅠ and BEⅡb [158], and a subsequent decline in their corresponding enzyme activities [159]. The decrease in amylose content in rice endosperm caused by HT has been observed for a long time [160]. The main reason for this symptom is the decrease in Wx/GBSSⅠ activity [158,161,162]. The transcription of the Wx gene is inhibited under HT, but the splicing efficiency of the first intron of the Wx gene is higher in the HT-tolerant variety. This leads to an increase in the percentage of the large isoform of Wx pre-mRNA, which shows higher enzyme activity, and likely stabilizes amylose content at high temperatures [162]. In addition, suppression of the HT-induced OsMADS7 gene in endosperm increases Wx expression and stabilizes amylose content under HT conditions [163] (Figure 2). Apart from GBSSⅠ, the expressions and activities of amylopectin-related enzymes are also influenced by HT, resulting in an altered chain length distribution in amylopectin [158,164,165] (Figure 2).
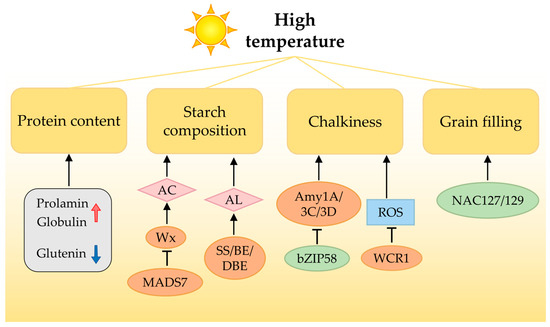
Figure 2.
High temperature (HT) negatively affects rice grain quality. During grain filling, HT can alter the chemical composition of starch and storage proteins in rice grains through various aspects. Moreover, HT-induced chalkiness is the most common and severe symptom during the reproductive stage. Additionally, several transcription factors regulate HT-induced chalkiness. AC, amylose content; AL, amylopectin length; ROS, reactive oxygen species. The up arrow in red and the down arrow in blue represent increase and decrease of protein contents, respectively.
HT may also change the composition of seed storage proteins (Figure 2). At the early filling stage, HT stress can increase the accumulation of all classes of storage proteins, but decrease the accumulation of prolamins during maturation [154]. However, during the later grain-filling stage, the expression of grain protein-related genes decreases under HT stress, which disrupts the normal folding process of storage proteins. This leads to a decrease in the accumulation of 13 kD prolamin and globulin, and an increase in glutenin content in mature rice, which negatively affects the eating quality of rice [126,154,158].
Chalkiness is the most severe symptom caused by HT stress during the grain-filling stage, and HT is considered to be the major climatic factor conferring the chalky trait of rice [166,167]. HT at the milky stage of grain filling has the greatest influence on rice grain chalkiness [166]. The formation of chalkiness under HT is mainly related to changes in grain components, which have been discussed above. Additionally, some other components have been reported to affect the formation of chalkiness in recent years (Figure 2). HT upregulates the expression of α-amylase genes and its enzyme activity during seed maturation [158,168]. Suppressing α-amylase genes expression has been shown to result in a reduction in chalky grain formation under HT [168]. By contrast, the overexpressing of α-amylase genes, such as Amy1A/3C/3D, has been found to result in varying degrees of grain chalkiness [169]. In addition, ROS (reactive oxygen species) is also involved in inducing chalkiness under HT. An increased content of ROS and the expression of NADPH oxidase genes are associated with chalkiness occurrence in grains exposed to HT, accompanied by the increased activity of α-amylase [170]. Furthermore, the F-box protein WCR1 was recently found to reduce grain chalkiness through the MT2b-dependent ROS scavenging pathways in rice endosperm [49] (Figure 2). Additionally, several transcription factors have been reported to regulate HT-induced chalkiness, including OsbZIP58, ONAC127, and ONAC129 [171,172] (Figure 2).
Although there have been some advances in understanding the genetic and molecular mechanisms underlying the decrease in rice quality caused by HT, few natural gene resources have been reported for the genetic improvement of rice quality under HT stress. To date, a few QTLs associated with HT tolerance during the reproductive and grain-filling stages have been identified in rice [162,173,174,175,176], which could be potential genetic resources to facilitate the development of high-temperature-tolerant rice varieties through molecular breeding. Strengthening the breeding of HT-tolerant rice varieties by screening for varietal differences in grain quality when ripened under high temperature is of crucial importance for coping with climate change.
6.2. Field Management Effects on Grain Quality
Field management has relatively minor effects on rice yield and quality compared to genetic and environmental factors. The application of nitrogen fertilizers increases grain yield dramatically but causes a series of problems, including declined rice quality. Numerous studies have examined the impact of nitrogen application on rice quality, and the consensus is that nitrogen application is generally positively correlated with rice milling quality and nutritional quality [177,178], but has negative effects on rice appearance quality [179] and eating and cooking quality [178,180]. However, the appropriate application of nitrogen can maintain and improve rice quality [179,181]. For example, the application of a moderate nitrogen rate (210–260 kg/ha) remarkably increased the milling quality and nutritional quality and resulted in moderate rice eating and cooking quality in a Japonica cultivar [181]. Nitrogen application has the greatest impact on eating and cooking quality, mainly through its influence on protein content and starch properties. High nitrogen levels can increase amylose content, reduce amylopectin branching, and increase protein content, leading to a hard texture of cooked rice and reduced palatability [182,183,184,185]. Therefore, to balance rice yield and quality, the use of chemical fertilizers should be controlled within a reasonable range, and biochar application may provide an alternative solution. The application of biochar has been widely recognized as a beneficial practice for improving rice yield while reducing nitrogen fertilizer usage [184]. More recently, several studies have investigated the potential impact of biochar application on rice grain quality. It has been suggested that biochar application can positively influence various grain quality traits, including milled rice rate, starch viscosity attributes, eating quality, and grain appearance, with the specific effects varying depending on the dosage of biochar used [184,186,187]. Furthermore, the combined application of biochar and a low nitrogen rate (135 kg/ha) has emerged as a promising approach for enhancing both yield and grain quality in a sustainable manner [184].
Environmental conditions and cultural managements belong to the preharvest factors which have significant effects on rice grain quality, while postharvest operations, such as seed drying, storage conditions, and milling processes, have been used to maintain desirable rice grain quality [188]. These postharvest factors play a critical role in determining the commercial quality and value of rice and should receive more attention.
7. Perspectives
7.1. Deepening Basic Research on Rice Quality Traits
In the past decade, significant progress has been made in the functional genomics research of rice quality, including the cloning and molecular regulation of important genes related to rice grain quality traits. However, rice grain quality is composed of multiple traits, and there are universal interactions among different quality traits and between quality and environmental factors. Our understanding of these scientific issues is still insufficient, which greatly limits the genetic improvement of rice quality. In the future, there are still many key scientific questions to be answered in the basic research of rice grain quality.
The fundamental research on rice grain quality aims to address the trade-off between rice yield and quality. Grain shape is a crucial trait that is deeply studied in rice quality research, as it is a key factor in achieving high yields and superior quality simultaneously. Numerous essential genes that control the shape and quality of rice grains, as well as significant biosynthetic and regulatory pathways, have been discovered in the past few decades. Moreover, some varieties have been successfully bred with excellent rice quality and high yield, but most of the favorable genes come from common varieties used in production. Rare genes such as GL3.1, GL7/GW7, and GS2 have been cloned and provide favorable resources for rice quality improvement, but they often come with adverse effects such as increased chalkiness and reduced grain weight. Grain shape is the result of the interaction of multiple genes, but the mechanisms underlying their interactions are still poorly understood. There is still a major challenge in understanding the regulatory mechanisms that control these traits. Specifically, identifying the upstream and downstream components of these pathways and constructing a comprehensive genetic and molecular regulatory network for rice is necessary for a more thorough understanding of grain development and improvement.
Appearance quality is an important trait of rice grain quality. Any rice grain with a transparent endosperm and low chalkiness is considered to be of high quality and will be highly favored by consumers, regardless of its grain shape. Therefore, chalkiness is a major trait and important research direction in rice grain quality improvement and basic research. Chalkiness is related to the genetic factors of the variety itself and is also highly susceptible to high temperature stress. Because Chalk5 and the major-effect gene GS5 of grain width are closely linked in the Indica varieties, appropriately increasing grain length and reducing grain width to maintain grain weight is an effective way to break the unfavorable linkage and reduce chalkiness. Moreover, the exploration of the genetic basis of rice grain quality under high temperature stress is relatively limited, as it is more challenging to measure grain quality than grain yield. It is necessary to screen abundant natural variation germplasm or artificially induced mutants and to explore materials and their excellent genes that are tolerant to high temperature, and thus, solve the problem of HT-induced chalkiness formation.
Aroma is a symbol of premium rice, but there are issues with unclear genetic mechanisms and unstable fragrance in the breeding process of fragrant rice. Some fragrant rice varieties without functional loss of badh2 alleles also have high 2-AP content, indicating that rice may have new functional genes and aromatic substances that control fragrance [119,189]. In the future, it will be important to focus on the collection and identification of fragrant rice resources. These resources can be used to discover new varieties that are not controlled by the badh2 gene. Additionally, it is important to identify fragrant rice that exhibits stable or significant changes in aroma under different environmental conditions. By exploring these resources, it may be possible to discover new aromatic substances and genes, as well as their regulatory mechanisms.
The milling quality of rice, which is mainly measured by brown rice recovery, milled rice recovery, and head rice recovery, is closely related to grain shape and chalkiness. Generally, the milling quality of short and round grains with low chalkiness is higher than that of long and slender grains with high chalkiness [186]. Rice milling quality directly affects the yield of polished rice, but it is easily influenced by factors such as milling methods and storage conditions. Related studies on functional genomic research of milling quality have not received sufficient attention, and most studies are only limited to the initial mapping of related QTLs, with no critical genes being cloned yet. However, a recent GWAS study presented a possible molecular basis of milling quality [190]. Specifically, the study highlighted the pivotal role of the Wx gene in regulating head rice recovery by modulating the amorphous and semi-crystalline layered structure of starch granules. This finding suggested that the amylose content, which is controlled by the Wx gene, is closely linked to milling quality, along with the traditional determinants of grain shape and chalkiness. In the future, more attention should be paid to the identification and functional analysis of key genes related to rice milling quality. By doing so, we can improve efficiency and ensure the stable production of superior quality rice.
7.2. Application of New Technology to Grain Quality Improvement
Map-based gene cloning is a classic and effective method for analyzing the genetic basis of rice quality, but it is time-consuming and cumbersome, and it may be difficult to mine natural gene resources that control quality traits. Valuable variation is present in natural accessions, which is why GWAS approaches have been employed in basic research and breeding practices for superior quality rice [191,192]. Furthermore, the resequencing of 3010 rice accessions has made it possible to use GWAS approaches to identify minor alleles for grain quality from diverse rice collections [193].
Although transgenic technology has played a pivotal role in crop improvement, such as the successful creation of vitamin A-enriched ”Golden Rice” and iron-enriched rice [194,195], policy limitations have restricted the application of this technology in rice breeding. However, recent innovations in CRISPR/Cas technology, such as prime editing and base editing, provide important technical support for crop improvement. Improving rice grain quality through gene-editing technology is a fast and efficient method that can accelerate the process of rice quality improvement. Gene-editing technology is now widely applied to directly generate new rice varieties with improved grain quality by manipulating elite genes, including GS3, GW2, GW5, TGW6, GS9, Wx, OsAAP, badh2, and FAD2-1, indicating that genome editing has significant potential to improve the quality of rice grains [7,112,196].
7.3. Directions of Breeding Rice Variety with Superior Grain Quality
As a staple food, the eating and cooking quality, which determines whether it is delicious or not, is the core of rice quality, while the appearance quality is directly linked to the commercial valueof rice. Thus, breeding rice varieties with both superior appearance and eating quality represents the primary focus of superior quality rice breeding. Long-grain rice, characterized by lower chalkiness, low amylose content, and low protein content, usually presents excellent appearance quality and good taste. As such, recently, breeders in China have devoted considerable attention to long-grain Japonica and super-long-grain Indica rice. The varying preferences for rice taste exist in different populations among various regions and countries, which can be achieved by fine tuning the amylose content through introducing new Wx alleles, starch synthesis-related genes such as SSIIa, SSIIIa, or gene loci that moderately increase AC. The influence of Jasmine rice from Thailand has led to aroma becoming a premium quality trait, with significant economic bonuses for rice farmers. Hence, developing fragrant rice varieties has become an essential aspect that cannot be disregarded.
The quality of the rice grain in conventional Japonica rice is often superior to that of Indica rice. Although hybrid rice has yield advantages, its quality is often inferior to that of conventional rice. Therefore, strengthening the breeding of superior quality conventional and hybrid Indica rice varieties is an important direction for future rice breeding. It is worth noting that hybrid Japonica rice, which has received little attention, has potential advantages in yield and disease resistance over conventional Japonica rice, despite its small planting area. Currently, compared with conventional Japonica rice, hybrid Japonica rice has higher chalkiness and lower head rice rate, and the research and promotion of superior quality hybrid Japonica rice is an important breakthrough for increasing grain production in Japonica rice areas.
Furthermore, with the improvement of living standards and the pursuit of nutrition and health by consumers, the development of functional rice with special nutritional value or special needs for specific populations, such as rice with low glutelin content, high resistant starch, and high γ-aminobutyric acid (GABA), should be promoted [197,198,199].
In the pursuit of improving rice quality, the objectives may vary depending on the demands of different regional markets and consumers, but the fundamental strategy for achieving this goal is relatively uniform (Figure 3), that is, in summary, fully utilizing cloned rice quality-related QTLs/genes and employing various breeding methods, pyramiding multiple elite alleles to enhance head rice recovery and ECQ while maintaining high yield. On this foundation, enhancing the appearance quality and aroma of rice grains becomes feasible, resulting in an output characterized by uniform shape, high translucency, and an appealing fragrance.
Figure 3.
Breeding rice variety with superior grain quality.
Author Contributions
Writing—original draft preparation, D.G. and F.H.; Writing—review, editing, and financial support, G.Y.; Collection of information and data, X.Z., Y.C., R.L., J.Y., M.Z. and W.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by earmarked funding for the project from the President’s Fund Project of Liaoning Academy of Agricultural Sciences (2023QN2408), the Liaoning Provincial Doctoral Research Start-up Fund in 2022 (2022-BS-050), the Liaoning Academy of Agricultural Sciences Collaborative Innovation Plan 2022 “Revelation and Commanding” project (2022XTCX0501003), the China Agriculture Research System (CARS-01-55), and the Applied Basic Research Project of Liaoning Province (2022JH2/101300283).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rao, Y.; Li, Y.; Qian, Q. Recent progress on molecular breeding of rice in China. Plant Cell Rep. 2014, 33, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.-Q.; Jiang, W.; Ham, T.-H.; Chu, S.-H.; Lestari, P.; Lee, J.-H.; Kim, M.-K.; Xu, F.-R.; Han, L.; Dai, L.-Y. Comparison of grain quality traits between japonica rice cultivars from Korea and Yunnan Province of China. J. Crop Sci. Biotechnol. 2008, 11, 135–140. [Google Scholar]
- Chen, Y.; Wang, M.; Ouwerkerk, P.B. Molecular and environmental factors determining grain quality in rice. Food Energy Secur. 2012, 1, 111–132. [Google Scholar] [CrossRef]
- Bao, J. Genes and QTLs for rice grain quality improvement. In Rice; InTech: London, UK, 2014; pp. 239–278. [Google Scholar]
- Custodio, M.C.; Cuevas, R.P.; Ynion, J.; Laborte, A.G.; Velasco, M.L.; Demont, M. Rice quality: How is it defined by consumers, industry, food scientists, and geneticists? Trends Food Sci. Technol. 2019, 92, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xia, D.; He, Y. Rice grain quality—Traditional traits for high quality rice and health-plus substances. Mol. Breed. 2020, 40, 1. [Google Scholar] [CrossRef]
- Cheng, J.; Lin, X.; Long, Y.; Zeng, Q.; Zhao, K.; Hu, P.; Peng, J. Rice grain quality: Where we are and where to go? Adv. Agron. 2022, 172, 211–252. [Google Scholar]
- Li, P.; Chen, Y.-H.; Lu, J.; Zhang, C.-Q.; Liu, Q.-Q.; Li, Q.-F. Genes and their molecular functions determining seed structure, components, and quality of rice. Rice 2022, 15, 18. [Google Scholar] [CrossRef]
- Zhou, H.; Yun, P.; He, Y. Rice appearance quality. In Rice; Elsevier: Amsterdam, The Netherlands, 2019; pp. 371–383. [Google Scholar]
- Fitzgerald, M.A.; McCouch, S.R.; Hall, R.D. Not just a grain of rice: The quest for quality. Trends Plant Sci. 2009, 14, 133–139. [Google Scholar] [CrossRef]
- Lu, S.; Luh, B.S. Properties of the rice caryopsis. In Rice: Volume I. Production/Volume II. Utilization; Springer: Boston, MA, USA, 1991; pp. 389–419. [Google Scholar] [CrossRef]
- Huang, R.; Jiang, L.; Zheng, J.; Wang, T.; Wang, H.; Huang, Y.; Hong, Z. Genetic bases of rice grain shape: So many genes, so little known. Trends Plant Sci. 2013, 18, 218–226. [Google Scholar] [CrossRef]
- Zuo, J.; Li, J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 2014, 48, 99–118. [Google Scholar] [CrossRef]
- Li, N.; Li, Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 2016, 33, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, R.; Li, Y. Molecular Networks of Seed Size Control in Plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zhang, C.; Li, Q.; Liu, Q. Genetic control of grain appearance quality in rice. Biotechnol. Adv. 2022, 60, 108014. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Ding, C.; Qian, Q. Molecular bases of rice grain size and quality for optimized productivity. Sci. Bull. 2023, 68, 314–350. [Google Scholar]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Takano-Kai, N.; Jiang, H.; Kubo, T.; Sweeney, M.; Matsumoto, T.; Kanamori, H.; Padhukasahasram, B.; Bustamante, C.; Yoshimura, A.; Doi, K.; et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 2009, 182, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef]
- Trusov, Y.; Chakravorty, D.; Botella, J.R. Diversity of heterotrimeric G-protein gamma subunits in plants. BMC Res. Notes 2012, 5, 608. [Google Scholar] [CrossRef]
- Sun, S.; Wang, L.; Mao, H.; Shao, L.; Li, X.; Xiao, J.; Ouyang, Y.; Zhang, Q. A G-protein pathway determines grain size in rice. Nat. Commun. 2018, 9, 851. [Google Scholar] [CrossRef]
- Qi, P.; Lin, Y.S.; Song, X.J.; Shen, J.B.; Huang, W.; Shan, J.X.; Zhu, M.Z.; Jiang, L.; Gao, J.P.; Lin, H.X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012, 22, 1666–1680. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, X.; Lan, H.; Huang, J.; Wang, J.; Zhang, H. The additive effects of GS3 and qGL3 on rice grain length regulation revealed by genetic and transcriptome comparisons. BMC Plant Biol. 2015, 15, 156. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Zhou, H.; Liu, R.; Dan, W.; Li, P.; Wu, B.; Chen, J.; Wang, L.; Gao, G.; Zhang, Q.; et al. GL3.3, a Novel QTL Encoding a GSK3/SHAGGY-like Kinase, Epistatically Interacts with GS3 to Produce Extra-long Grains in Rice. Mol. Plant 2018, 11, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef]
- Duan, P.; Xu, J.; Zeng, D.; Zhang, B.; Geng, M.; Zhang, G.; Huang, K.; Huang, L.; Xu, R.; Ge, S.; et al. Natural Variation in the Promoter of GSE5 Contributes to Grain Size Diversity in Rice. Mol. Plant 2017, 10, 685–694. [Google Scholar] [CrossRef]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef]
- Zhang, X.F.; Yang, C.Y.; Lin, H.X.; Wang, J.W.; Xue, H.W. Rice SPL12 coevolved with GW5 to determine grain shape. Sci. Bull. 2021, 66, 2353–2357. [Google Scholar] [CrossRef]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef]
- Xu, C.; Liu, Y.; Li, Y.; Xu, X.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. Differential expression of GS5 regulates grain size in rice. J. Exp. Bot. 2015, 66, 2611–2623. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J.; et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Miao, J.; Gu, H.; Peng, X.; Leburu, M.; Yuan, F.; Gu, H.; Gao, Y.; Tao, Y.; Zhu, J.; et al. Natural Variations in SLG7 Regulate Grain Shape in Rice. Genetics 2015, 201, 1591–1599. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.S.; Li, Q.F.; Zhang, C.Q.; Zhang, C.; Yang, Q.Q.; Pan, L.X.; Ren, X.Y.; Lu, J.; Gu, M.H.; Liu, Q.Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef]
- Zhao, D.-s.; Liu, J.-y.; Ding, A.-q.; Zhang, T.; Ren, X.-y.; Zhang, L.; Li, Q.-f.; Fan, X.-l.; Zhang, C.-q.; Liu, Q.-q. Improving grain appearance of erect-panicle japonica rice cultivars by introgression of the null gs9 allele. J. Integr. Agric. 2021, 20, 2032–2042. [Google Scholar] [CrossRef]
- Lu, L.; Shao, D.; Qiu, X.; Sun, L.; Yan, W.; Zhou, X.; Yang, L.; He, Y.; Yu, S.; Xing, Y. Natural variation and artificial selection in four genes determine grain shape in rice. New. Phytol. 2013, 200, 1269–1280. [Google Scholar] [CrossRef]
- Fan, C.; Yu, S.; Wang, C.; Xing, Y. A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 2009, 118, 465–472. [Google Scholar] [CrossRef]
- Wang, C.; Chen, S.; Yu, S. Functional markers developed from multiple loci in GS3 for fine marker-assisted selection of grain length in rice. Theor. Appl. Genet. 2011, 122, 905–913. [Google Scholar] [CrossRef]
- Nan, J.; Feng, X.; Wang, C.; Zhang, X.; Wang, R.; Liu, J.; Yuan, Q.; Jiang, G.; Lin, S. Improving rice grain length through updating the GS3 locus of an elite variety Kongyu 131. Rice 2018, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Han, R.; Wu, K.; Zhang, J.; Ye, Y.; Wang, S.; Chen, J.; Pan, Y.; Li, Q.; Xu, X.; et al. G-protein betagamma subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 2018, 9, 852. [Google Scholar] [CrossRef]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhong, H.; Jiang, X.; Zhang, J.; Huang, R.; Liao, F.; Deng, Y.; Liu, Q.; Huang, Y.; Wang, H.; et al. Identification and Pleiotropic Effect Analysis of GSE5 on Rice Chalkiness and Grain Shape. Front. Plant Sci. 2021, 12, 814928. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Yun, P.; Luo, L.; Yan, B.; Peng, B.; Xie, W.; Wang, G.; Li, X.; et al. Chalk5 encodes a vacuolar H(+)-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 2014, 46, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Yun, P.; Zhou, H.; Xia, D.; Gu, Y.; Li, P.; Yao, J.; Zhou, Z.; Chen, J.; Liu, R.; et al. Natural variation in WHITE-CORE RATE 1 regulates redox homeostasis in rice endosperm to affect grain quality. Plant Cell 2022, 34, 1912–1932. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y.; Zhu, Z.; Chen, T.; Zhao, L.; Lin, J.; Zhou, L. Development of a new japonica rice variety Nan-jing 46 with good eating quality by marker assisted selection. Rice Genom. Genet. 2010, 1. [Google Scholar] [CrossRef]
- Zhou, P.H.; Tan, Y.F.; He, Y.Q.; Xu, C.G.; Zhang, Q. Simultaneous improvement for four quality traits of Zhenshan 97, an elite parent of hybrid rice, by molecular marker-assisted selection. Theor. Appl. Genet. 2003, 106, 326–331. [Google Scholar] [CrossRef]
- Liu, Q.Q.; Li, Q.F.; Cai, X.L.; Wang, H.M.; Tang, S.Z.; Yu, H.X.; Wang, Z.Y.; Gu, M.H. Molecular marker-assisted selection for improved cooking and eating quality of two elite parents of hybrid rice. Crop Sci. 2006, 46, 2354–2360. [Google Scholar] [CrossRef]
- Ni, D.; Zhang, S.; Chen, S.; Xu, Y.; Li, L.; Li, H.; Wang, Z.; Cai, X.; Li, Z.; Yang, J. Improving cooking and eating quality of Xieyou57, an elite indica hybrid rice, by marker-assisted selection of the Wx locus. Euphytica 2011, 179, 355–362. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Y.; Chen, S.; Liu, X.; Zhu, J.; Zhou, L.; Lu, Y.; Li, Q.; Fan, X.; Tang, S.; et al. A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant Biol. 2021, 63, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Lu, Y.; Shao, Y.; Zhang, G.; Xiao, P.; Shen, S.; Corke, H.; Bao, J. Molecular marker assisted selection for improvement of the eating, cooking and sensory quality of rice (Oryza sativa L.). J. Cereal Sci. 2010, 51, 159–164. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, M.; Li, R.; Shen, L.; Wang, W.; Liu, M.; Zhu, Q.; Hu, Z.; He, Q.; Xue, Y. Identification of quantitative trait loci responsible for rice grain protein content using chromosome segment substitution lines and fine mapping of qPC-1 in rice (Oryza sativa L.). Mol. Breed. 2015, 35, 1–9. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, Z.; Xu, C.; Guo, M.; Li, Y.; Zhang, Y.; Zhong, C.; Sun, S.; Yan, C. Genetic improvement of panicle-erectness japonica rice toward both yield and eating and cooking quality. Mol. Breed. 2020, 40, 1–12. [Google Scholar] [CrossRef]
- Wu, K.; Xu, X.; Zhong, N.; Huang, H.; Yu, J.; Ye, Y.; Wu, Y.; Fu, X. The rational design of multiple molecular module-based assemblies for simultaneously improving rice yield and grain quality. J. Genet. Genom. 2018, 45, 337–341. [Google Scholar] [CrossRef]
- Yang, T.; Gu, H.; Yang, W.; Liu, B.; Liang, S.; Zhao, J. Artificially Selected Grain Shape Gene Combinations in Guangdong Simiao Varieties of Rice (Oryza sativa L.). Rice 2023, 16, 3. [Google Scholar] [CrossRef]
- Lisle, A.; Martin, M.; Fitzgerald, M. Chalky and translucent rice grains differ in starch composition and structure and cooking properties. Cereal Chem. 2000, 77, 627–632. [Google Scholar] [CrossRef]
- Gong, J.; Miao, J.; Zhao, Y.; Zhao, Q.; Feng, Q.; Zhan, Q.; Cheng, B.; Xia, J.; Huang, X.; Yang, S.; et al. Dissecting the Genetic Basis of Grain Shape and Chalkiness Traits in Hybrid Rice Using Multiple Collaborative Populations. Mol. Plant 2017, 10, 1353–1356. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Y.; Fang, Y.; Zeng, L.; Xu, J.; Yu, H.; Shi, Z.; Pan, J.; Zhang, D.; Kang, S.; et al. A Rare Allele of GS2 Enhances Grain Size and Grain Yield in Rice. Mol. Plant 2015, 8, 1455–1465. [Google Scholar] [CrossRef]
- Ayaad, M.; Han, Z.; Zheng, K.; Hu, G.; Abo-Yousef, M.; Sobeih, S.E.S.; Xing, Y. Bin-based genome-wide association studies reveal superior alleles for improvement of appearance quality using a 4-way MAGIC population in rice. J. Adv. Res. 2021, 28, 183–194. [Google Scholar] [CrossRef]
- Tian, Z.; Qian, Q.; Liu, Q.; Yan, M.; Liu, X.; Yan, C.; Liu, G.; Gao, Z.; Tang, S.; Zeng, D.; et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 2009, 106, 21760–21765. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Sun, S.S. Profiling the expression of genes controlling rice grain quality. Plant Mol. Biol. 2005, 59, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gilbert, R.G. Starch molecular structure: The basis for an improved understanding of cooked rice texture. Carbohydr. Polym. 2018, 195, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Q.; Liu, W.J.; Xu, Y.; He, Y.Q.; Luo, L.J.; Xing, Y.Z.; Xu, C.G.; Zhang, Q. Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain. Theor. Appl. Genet. 2007, 115, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, Y.; Chen, Z.; Chen, F.; Pan, L.; Lu, Y.; Li, Q.; Fan, X.; Sun, Z.; Liu, Q. Characteristics of Grain Physicochemical Properties and the Starch Structure in Rice Carrying a Mutated ALK/SSIIa Gene. J. Agric. Food Chem. 2020, 68, 13950–13959. [Google Scholar] [CrossRef]
- Jeon, J.S.; Ryoo, N.; Hahn, T.R.; Walia, H.; Nakamura, Y. Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 2010, 48, 383–392. [Google Scholar] [CrossRef]
- Pfister, B.; Zeeman, S.C. Formation of starch in plant cells. Cell. Mol. Life Sci. 2016, 73, 2781–2807. [Google Scholar] [CrossRef]
- Bao, J. Rice starch. In Rice; Elsevier: Amsterdam, The Netherlands, 2019; pp. 55–108. [Google Scholar]
- Huang, L.; Tan, H.; Zhang, C.; Li, Q.; Liu, Q. Starch biosynthesis in cereal endosperms: An updated review over the last decade. Plant Commun. 2021, 2, 100237. [Google Scholar] [CrossRef]
- Zeeman, S.C.; Kossmann, J.; Smith, A.M. Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 2010, 61, 209–234. [Google Scholar] [CrossRef]
- Ball, S.G.; van de Wal, M.H.; Visser, R.G. Progress in understanding the biosynthesis of amylose. Trends Plant Sci. 1998, 3, 462–467. [Google Scholar] [CrossRef]
- Gu, M.-H.; Liu, Q.-Q.; Yan, C.-J.; Tang, S.-Z. Grain quality of hybrid rice: Genetic variation and molecular improvement. Accel. Hybrid. Rice Dev. Los. Banos (Philipp.) Int. Rice Res. Inst. 2010, 345–356. [Google Scholar]
- Wang, Z.Y.; Wu, Z.L.; Xing, Y.Y.; Zheng, F.G.; Guo, X.L.; Zhang, W.G.; Hong, M.M. Nucleotide sequence of rice waxy gene. Nucleic Acids Res. 1990, 18, 5898. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Zheng, F.Q.; Shen, G.Z.; Gao, J.P.; Snustad, D.P.; Li, M.G.; Zhang, J.L.; Hong, M.M. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995, 7, 613–622. [Google Scholar] [CrossRef]
- Cai, X.L.; Wang, Z.Y.; Xing, Y.Y.; Zhang, J.L.; Hong, M.M. Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 1998, 14, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Suzuki, Y.; Sakai, M.; Imbe, T. Molecular characterization of Wx-mq, a novel mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.). Breed. Sci. 2002, 52, 131–135. [Google Scholar] [CrossRef]
- Wanchana, S.; Toojinda, T.; Tragoonrung, S.; Vanavichit, A. Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci. 2003, 165, 1193–1199. [Google Scholar] [CrossRef]
- Bao, J.S.; Corke, H.; Sun, M. Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor. Appl. Genet. 2006, 113, 1171–1183. [Google Scholar] [CrossRef]
- Mikami, I.; Uwatoko, N.; Ikeda, Y.; Yamaguchi, J.; Hirano, H.Y.; Suzuki, Y.; Sano, Y. Allelic diversification at the wx locus in landraces of Asian rice. Theor. Appl. Genet. 2008, 116, 979–989. [Google Scholar] [CrossRef]
- Liu, L.; Ma, X.; Liu, S.; Zhu, C.; Jiang, L.; Wang, Y.; Shen, Y.; Ren, Y.; Dong, H.; Chen, L.; et al. Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 2009, 71, 609–626. [Google Scholar] [CrossRef]
- Yang, J.; Wang, J.; Fan, F.J.; Zhu, J.Y.; Chen, T.; Wang, C.L.; Zheng, T.Q.; Zhang, J.; Zhong, W.G.; Xu, J.L. Development of AS-PCR marker based on a key mutation confirmed by resequencing of Wx-mp in Milky P rincess and its application in japonica soft rice (Oryza sativa L.) breeding. Plant Breed. 2013, 132, 595–603. [Google Scholar] [CrossRef]
- Zhang, C.; Zhu, J.; Chen, S.; Fan, X.; Li, Q.; Lu, Y.; Wang, M.; Yu, H.; Yi, C.; Tang, S.; et al. Wx(lv), the Ancestral Allele of Rice Waxy Gene. Mol. Plant 2019, 12, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xia, D.; Zhao, D.; Li, Y.; Li, P.; Wu, B.; Gao, G.; Zhang, Q.; Wang, G.; Xiao, J.; et al. The origin of Wx(la) provides new insights into the improvement of grain quality in rice. J. Integr. Plant Biol. 2021, 63, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chen, S.; Yang, G.; Zha, W.; Cai, H.; Li, S.; Chen, Z.; Liu, K.; Xu, H.; You, A. A perfect functional marker for the gene of intermediate amylose content Wx-in in rice (Oryza sativa L.). Crop Breed. Appl. Biotechnol. 2018, 18, 103–109. [Google Scholar] [CrossRef]
- Tan, Y.F.; Li, J.X.; Yu, S.B.; Xing, Y.Z.; Xu, C.G.; Zhang, Q. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor. Appl. Genet. 1999, 99, 642–648. [Google Scholar] [CrossRef]
- Tian, R.; Jiang, G.-H.; Shen, L.-H.; Wang, L.-Q.; He, Y.-Q. Mapping quantitative trait loci underlying the cooking and eating quality of rice using a DH population. Mol. Breed. 2005, 15, 117–124. [Google Scholar] [CrossRef]
- Su, Y.; Rao, Y.; Hu, S.; Yang, Y.; Gao, Z.; Zhang, G.; Liu, J.; Hu, J.; Yan, M.; Dong, G.; et al. Map-based cloning proves qGC-6, a major QTL for gel consistency of japonica/indica cross, responds by Waxy in rice (Oryza sativa L.). Theor. Appl. Genet. 2011, 123, 859–867. [Google Scholar] [CrossRef]
- Gao, Z.; Zeng, D.; Cui, X.; Zhou, Y.; Yan, M.; Huang, D.; Li, J.; Qian, Q. Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci. China C. Life Sci. 2003, 46, 661–668. [Google Scholar] [CrossRef]
- Nakamura, Y.; Francisco, P.B., Jr.; Hosaka, Y.; Sato, A.; Sawada, T.; Kubo, A.; Fujita, N. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 2005, 58, 213–227. [Google Scholar] [CrossRef]
- Gao, Z.; Zeng, D.; Cheng, F.; Tian, Z.; Guo, L.; Su, Y.; Yan, M.; Jiang, H.; Dong, G.; Huang, Y.; et al. ALK, the key gene for gelatinization temperature, is a modifier gene for gel consistency in rice. J. Integr. Plant Biol. 2011, 53, 756–765. [Google Scholar] [CrossRef]
- Waters, D.L.; Henry, R.J.; Reinke, R.F.; Fitzgerald, M.A. Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol. J. 2006, 4, 115–122. [Google Scholar] [CrossRef]
- Miura, S.; Crofts, N.; Saito, Y.; Hosaka, Y.; Oitome, N.F.; Watanabe, T.; Kumamaru, T.; Fujita, N. Starch Synthase IIa-Deficient Mutant Rice Line Produces Endosperm Starch With Lower Gelatinization Temperature Than Japonica Rice Cultivars. Front. Plant Sci. 2018, 9, 645. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, Y.; Feng, L.; Hao, W.; Li, C.; Yang, Y.; Fan, X.; Li, Q.; Zhang, C.; Liu, Q. Genetic Dissection and Functional Differentiation of ALK(a) and ALK(b), Two Natural Alleles of the ALK/SSIIa Gene, Responding to Low Gelatinization Temperature in Rice. Rice 2020, 13, 39. [Google Scholar] [CrossRef]
- Xiang, X.; Kang, C.; Xu, S.; Yang, B. Combined effects of Wx and SSIIa haplotypes on rice starch physicochemical properties. J. Sci. Food Agric. 2017, 97, 1229–1234. [Google Scholar] [CrossRef]
- Bradbury, L.M.; Fitzgerald, T.L.; Henry, R.J.; Jin, Q.; Waters, D.L. The gene for fragrance in rice. Plant Biotechnol. J. 2005, 3, 363–370. [Google Scholar] [CrossRef]
- Chen, S.; Wu, J.; Yang, Y.; Shi, W.; Xu, M. The fgr gene responsible for rice fragrance was restricted within 69kb. Plant Sci. 2006, 171, 505–514. [Google Scholar] [CrossRef]
- Chen, S.; Yang, Y.; Shi, W.; Ji, Q.; He, F.; Zhang, Z.; Cheng, Z.; Liu, X.; Xu, M. Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell 2008, 20, 1850–1861. [Google Scholar] [CrossRef]
- Shi, W.; Yang, Y.; Chen, S.; Xu, M. Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol. Breed. 2008, 22, 185–192. [Google Scholar] [CrossRef]
- Kovach, M.J.; Calingacion, M.N.; Fitzgerald, M.A.; McCouch, S.R. The origin and evolution of fragrance in rice (Oryza sativa L.). Proc. Natl. Acad. Sci. USA 2009, 106, 14444–14449. [Google Scholar] [CrossRef]
- Shao, G.; Tang, A.; Tang, S.; Luo, J.; Jiao, G.; Wu, J.; Hu, P. A new deletion mutation of fragrant gene and the development of three molecular markers for fragrance in rice. Plant Breed. 2011, 130, 172–176. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, G.; Xu, X.; Li, J. Discovery of a new fragrance allele and development of functional markers for identifying diverse fragrant genotypes in rice. Mol. Breed. 2014, 33, 701–708. [Google Scholar] [CrossRef]
- Bindusree, G.; Natarajan, P.; Kalva, S.; Madasamy, P. Whole genome sequencing of Oryza sativa L. cv. Seeragasamba identifies a new fragrance allele in rice. PLoS ONE 2017, 12, e0188920. [Google Scholar] [CrossRef]
- Shao, G.; Tang, S.; Chen, M.; Wei, X.; He, J.; Luo, J.; Jiao, G.; Hu, Y.; Xie, L.; Hu, P. Haplotype variation at Badh2, the gene determining fragrance in rice. Genomics 2013, 101, 157–162. [Google Scholar] [CrossRef]
- Myint, K.M.; Arikit, S.; Wanchana, S.; Yoshihashi, T.; Choowongkomon, K.; Vanavichit, A. A PCR-based marker for a locus conferring the aroma in Myanmar rice (Oryza sativa L.). Theor. Appl. Genet. 2012, 125, 887–896. [Google Scholar] [CrossRef]
- Chuba, M.; Sakurada, H.; Yuki, K.; Sano, T.; Chuba, R.; Sato, K.; Yokoo, N.; Honma, T.; Sato, S.; Miyano, H. Breeding of a new rice cultivar with low amylose content “Yukinomai” (Yamagata84). Bull. Agric. Res. Yamagata Prefect. 2006, 38, 1–23. [Google Scholar]
- Ando, I.; Sato, H.; Aoki, N.; Suzuki, Y.; Hirabayashi, H.; Kuroki, M.; Shimizu, H.; Ando, T.; Takeuchi, Y. Genetic analysis of the low-amylose characteristics of rice cultivars Oborozuki and Hokkai-PL9. Breed. Sci. 2010, 60, 187–194. [Google Scholar] [CrossRef]
- Fujino, K.; Hirayama, Y.; Kaji, R. Marker-assisted selection in rice breeding programs in Hokkaido. Breed. Sci. 2019, 69, 383–392. [Google Scholar] [CrossRef]
- Shao, Y.; Peng, Y.; Mao, B.; Lv, Q.; Yuan, D.; Liu, X.; Zhao, B. Allelic variations of the Wx locus in cultivated rice and their use in the development of hybrid rice in China. PLoS ONE 2020, 15, e0232279. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Zhang, C.; Tiozon, R.N., Jr.; Liu, Q. Post-genomics revolution in the design of premium quality rice in a high-yielding background to meet consumer demands in the 21st century. Plant Commun. 2022, 3, 100271. [Google Scholar] [CrossRef]
- Golestan Hashemi, F.S.; Rafii, M.Y.; Razi Ismail, M.; Mohamed, M.T.; Rahim, H.A.; Latif, M.A.; Aslani, F. Opportunities of marker-assisted selection for rice fragrance through marker-trait association analysis of microsatellites and gene-based markers. Plant Biol. 2015, 17, 953–961. [Google Scholar] [CrossRef]
- Addison, C.K.; Angira, B.; Kongchum, M.; Harrell, D.L.; Baisakh, N.; Linscombe, S.D.; Famoso, A.N. Characterization of Haplotype Diversity in the BADH2 Aroma Gene and Development of a KASP SNP Assay for Predicting Aroma in U.S. Rice. Rice 2020, 13, 47. [Google Scholar] [CrossRef]
- Ashokkumar, S.; Jaganathan, D.; Ramanathan, V.; Rahman, H.; Palaniswamy, R.; Kambale, R.; Muthurajan, R. Creation of novel alleles of fragrance gene OsBADH2 in rice through CRISPR/Cas9 mediated gene editing. PLoS ONE 2020, 15, e0237018. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Abdelrahman, M.; Li, J.; Wang, F.; Ji, Z.; Qi, H.; Wang, C.; Zhao, K. CRISPR/Cas9 induces exon skipping that facilitates development of fragrant rice. Plant Biotechnol. J. 2021, 19, 642–644. [Google Scholar] [CrossRef] [PubMed]
- Hui, S.; Li, H.; Mawia, A.M.; Zhou, L.; Cai, J.; Ahmad, S.; Lai, C.; Wang, J.; Jiao, G.; Xie, L.; et al. Production of aromatic three-line hybrid rice using novel alleles of BADH2. Plant Biotechnol. J. 2022, 20, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Shafiq, S.; Tang, X. CRISPR-Cas9-mediated editing of BADH2 gene triggered fragrance revolution in rice. Physiol. Plant 2023, 175, e13871. [Google Scholar] [CrossRef] [PubMed]
- Sakthivel, K.; Sundaram, R.M.; Shobha Rani, N.; Balachandran, S.M.; Neeraja, C.N. Genetic and molecular basis of fragrance in rice. Biotechnol. Adv. 2009, 27, 468–473. [Google Scholar] [CrossRef]
- Huang, L.; Gu, Z.; Chen, Z.; Yu, J.; Chu, R.; Tan, H.; Zhao, D.; Fan, X.; Zhang, C.; Li, Q.; et al. Improving rice eating and cooking quality by coordinated expression of the major starch synthesis-related genes, SSII and Wx, in endosperm. Plant Mol. Biol. 2021, 106, 419–432. [Google Scholar] [CrossRef]
- Zeng, D.; Tian, Z.; Rao, Y.; Dong, G.; Yang, Y.; Huang, L.; Leng, Y.; Xu, J.; Sun, C.; Zhang, G.; et al. Rational design of high-yield and superior-quality rice. Nat. Plants 2017, 3, 17031. [Google Scholar] [CrossRef]
- Zhao, M.; Lin, Y.; Chen, H. Improving nutritional quality of rice for human health. Theor. Appl. Genet. 2020, 133, 1397–1413. [Google Scholar] [CrossRef]
- Lin, R.; Luo, Y.; Liu, D.; Huang, C. Determination and analysis on principal qualitative characters of rice germplasm. In Rice germplasm resources in China; Agricultural Science and Technology Publisher of China: Beijing, China, 1993; pp. 83–93. [Google Scholar]
- Zhou, L.; Liu, Q.; Zhang, C.; Xu, Y.; Tang, S.; Gu, M. Variation and distribution of seed storage protein content and composition among different rice varieties. Acta Agron. Sin. 2009, 35, 884–891. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, M.; Sun, S.; Zou, Y.; Yin, S.; Liu, Y.; Tang, S.; Gu, M.; Yang, Z.; Yan, C. Natural variation of OsGluA2 is involved in grain protein content regulation in rice. Nat. Commun. 2019, 10, 1949. [Google Scholar] [CrossRef]
- He, W.; Wang, L.; Lin, Q.; Yu, F. Rice seed storage proteins: Biosynthetic pathways and the effects of environmental factors. J. Integr. Plant Biol. 2021, 63, 1999–2019. [Google Scholar] [CrossRef] [PubMed]
- Xinkang, L.; Chunmin, G.; Lin, W.; Liting, J.; Xiangjin, F.; Qinlu, L.; Zhengyu, H.; Chun, L. Rice Storage Proteins: Focus on Composition, Distribution, Genetic Improvement and Effects on Rice Quality. Rice Sci. 2023, 30, 207–221. [Google Scholar] [CrossRef]
- Li, H.; Yang, J.; Yan, S.; Lei, N.; Wang, J.; Sun, B. Molecular causes for the increased stickiness of cooked non-glutinous rice by enzymatic hydrolysis of the grain surface protein. Carbohydr. Polym. 2019, 216, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jang, S.G.; Lar, S.M.; Lee, A.R.; Cao, F.Y.; Seo, J.; Kwon, S.W. Genome-Wide Identification and Genetic Variations of the Starch Synthase Gene Family in Rice. Plants 2021, 10, 1154. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Sun, C.; Wang, R.; Wang, R.; Wang, X.; Zhang, Y.; Zhu, J. The Key Metabolites in Rice Quality Formation of Conventional japonica Varieties. Curr. Issues Mol. Biol. 2023, 45, 990–1001. [Google Scholar] [CrossRef]
- Tan, Y.; Sun, M.; Xing, Y.; Hua, J.; Sun, X.; Zhang, Q.; Corke, H. Mapping quantitative trait loci for milling quality, protein content and color characteristics of rice using a recombinant inbred line population derived from an elite rice hybrid. Theor. Appl. Genet. 2001, 103, 1037–1045. [Google Scholar] [CrossRef]
- Wang, L.; Zhong, M.; Li, X.; Yuan, D.; Xu, Y.; Liu, H.; He, Y.; Luo, L.; Zhang, Q. The QTL controlling amino acid content in grains of rice (Oryza sativa) are co-localized with the regions involved in the amino acid metabolism pathway. Mol. Breed. 2008, 21, 127–137. [Google Scholar] [CrossRef]
- Lou, J.; Chen, L.; Yue, G.; Lou, Q.; Mei, H.; Xiong, L.; Luo, L. QTL mapping of grain quality traits in rice. J. Cereal Sci. 2009, 50, 145–151. [Google Scholar] [CrossRef]
- Ye, G.; Liang, S.; Wan, J. QTL mapping of protein content in rice using single chromosome segment substitution lines. Theor. Appl. Genet. 2010, 121, 741–750. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, W.; Chen, X.; Ma, J.; Chen, W.; Zhao, Z.; Zhai, H.; Wan, J. Dynamic QTL analysis of rice protein content and protein index using recombinant inbred lines. J. Plant Biol. 2011, 54, 321–328. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, W.; Liu, S.; Chen, L.; Liu, X.; Chen, X.; Ma, J.; Chen, W.; Zhao, Z.; Jiang, L. Genetic relationship between grain chalkiness, protein content, and paste viscosity properties in a backcross inbred population of rice. J. Cereal Sci. 2012, 56, 153–160. [Google Scholar] [CrossRef]
- Cheng, L.; Xu, Q.; Zheng, T.; Ye, G.; Luo, C.; Xu, J.; Li, Z. Identification of stably expressed quantitative trait loci for grain yield and protein content using recombinant inbred line and reciprocal introgression line populations in rice. Crop Sci. 2013, 53, 1437–1446. [Google Scholar] [CrossRef]
- Kashiwagi, T.; Munakata, J. Identification and characteristics of quantitative trait locus for grain protein content, TGP12, in rice (Oryza sativa L.). Euphytica 2018, 214, 165. [Google Scholar] [CrossRef]
- Chattopadhyay, K.; Behera, L.; Bagchi, T.B.; Sardar, S.S.; Moharana, N.; Patra, N.R.; Chakraborti, M.; Das, A.; Marndi, B.C.; Sarkar, A.; et al. Detection of stable QTLs for grain protein content in rice (Oryza sativa L.) employing high throughput phenotyping and genotyping platforms. Sci. Rep. 2019, 9, 3196. [Google Scholar] [CrossRef]
- Peng, B.; Kong, H.; Li, Y.; Wang, L.; Zhong, M.; Sun, L.; Gao, G.; Zhang, Q.; Luo, L.; Wang, G.; et al. OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 2014, 5, 4847. [Google Scholar] [CrossRef]
- Hamaker, B.R.; Griffin, V.K. Effect of disulfide bond-containing protein on rice starch gelatinization and pasting. Cereal Chem. 1993, 70, 377–380. [Google Scholar]
- Martin, M.; Fitzgerald, M. Proteins in rice grains influence cooking properties! J. Cereal Sci. 2002, 36, 285–294. [Google Scholar] [CrossRef]
- Yoshida, H.; Tanigawa, T.; Yoshida, N.; Kuriyama, I.; Tomiyama, Y.; Mizushina, Y. Lipid components, fatty acid distributions of triacylglycerols and phospholipids in rice brans. Food Chem. 2011, 129, 479–484. [Google Scholar] [CrossRef]
- Tong, C.; Liu, L.; Waters, D.L.; Huang, Y.; Bao, J. The contribution of lysophospholipids to pasting and thermal properties of nonwaxy rice starch. Carbohydr. Polym. 2015, 133, 187–193. [Google Scholar] [CrossRef]
- Concepcion, J.C.T.; Calingacion, M.; Garson, M.J.; Fitzgerald, M.A. Lipidomics reveals associations between rice quality traits. Metabolomics 2020, 16, 54. [Google Scholar] [CrossRef]
- Concepcion, J.C.T.; Ouk, S.; Riedel, A.; Calingacion, M.; Zhao, D.; Ouk, M.; Garson, M.J.; Fitzgerald, M.A. Quality evaluation, fatty acid analysis and untargeted profiling of volatiles in Cambodian rice. Food Chem. 2018, 240, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xia, D.; Li, P.; Ao, Y.; Xu, X.; Wan, S.; Li, Y.; Wu, B.; Shi, H.; Wang, K.; et al. Genetic architecture and key genes controlling the diversity of oil composition in rice grains. Mol. Plant 2021, 14, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, Z.; Yang, C.; Liu, Z.; Shen, S.; Zhan, C.; Lyu, Y.; Zhang, F.; Li, K.; Shi, Y.; et al. The NET locus determines the food taste, cooking and nutrition quality of rice. Sci. Bull. 2022, 67, 2045–2049. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Rosental, L.; Xu, Y.; Xu, D.; Orf, I.; Wang, W.; Hu, Z.; Su, S.; Bai, S.; Ashraf, M.; et al. Genetic architecture of seed glycerolipids in Asian cultivated rice. Plant Cell. Environ. 2023, 46, 1278–1294. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, N.; Butardo, V.M., Jr.; Misra, G.; Cuevas, R.P.; Anacleto, R.; Kavi Kishor, P.B. Designing climate-resilient rice with ideal grain quality suited for high-temperature stress. J. Exp. Bot. 2015, 66, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, B.; Chen, L.; Li, P.; Cao, C. The different influences of drought stress at the flowering stage on rice physiological traits, grain yield, and quality. Sci. Rep. 2019, 9, 3742. [Google Scholar] [CrossRef]
- Huang, M.; Cao, J.; Liu, Y.; Zhang, M.; Hu, L.; Xiao, Z.; Chen, J.; Cao, F. Low-temperature stress during the flowering period alters the source–sink relationship and grain quality in field-grown late-season rice. J. Agron. Crop Sci. 2021, 207, 833–839. [Google Scholar] [CrossRef]
- Zheng, C.; Liu, C.; Liu, L.; Tan, Y.; Sheng, X.; Yu, D.; Sun, Z.; Sun, X.; Chen, J.; Yuan, D. Effect of salinity stress on rice yield and grain quality: A meta-analysis. Eur. J. Agron. 2023, 144, 126765. [Google Scholar] [CrossRef]
- Lin, C.J.; Li, C.Y.; Lin, S.K.; Yang, F.H.; Huang, J.J.; Liu, Y.H.; Lur, H.S. Influence of high temperature during grain filling on the accumulation of storage proteins and grain quality in rice (Oryza sativa L.). J. Agric. Food Chem. 2010, 58, 10545–10552. [Google Scholar] [CrossRef]
- Counce, P.; Bryant, R.; Bergman, C.; Bautista, R.; Wang, Y.J.; Siebenmorgen, T.; Moldenhauer, K.; Meullenet, J.F. Rice milling quality, grain dimensions, and starch branching as affected by high night temperatures. Cereal Chem. 2005, 82, 645–648. [Google Scholar] [CrossRef]
- Aboubacar, A.; Moldenhauer, K.A.; McClung, A.M.; Beighley, D.H.; Hamaker, B.R. Effect of growth location in the United States on amylose content, amylopectin fine structure, and thermal properties of starches of long grain rice cultivars. Cereal Chem. 2006, 83, 93–98. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, L.; Zhu, Z.; Lu, H.; Zhou, X.; Qian, Y.; Li, Q.; Lu, Y.; Gu, M.; Liu, Q. Characterization of Grain Quality and Starch Fine Structure of Two Japonica Rice (Oryza Sativa) Cultivars with Good Sensory Properties at Different Temperatures during the Filling Stage. J. Agric. Food Chem. 2016, 64, 4048–4057. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, H.; Hirose, T.; Kuroda, M.; Yamaguchi, T. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 2007, 144, 258–277. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Dian, W.; Wu, P. Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry 2003, 63, 53–59. [Google Scholar] [CrossRef]
- Umemoto, T.; Terashima, K. Research note: Activity of granule-bound starch synthase is an important determinant of amylose content in rice endosperm. Funct. Plant Biol. 2002, 29, 1121–1124. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.K.; Chang, M.C.; Tsai, Y.G.; Lur, H.S. Proteomic analysis of the expression of proteins related to rice quality during caryopsis development and the effect of high temperature on expression. Proteomics 2005, 5, 2140–2156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Duan, L.; Dai, J.S.; Zhang, C.Q.; Li, J.; Gu, M.H.; Liu, Q.Q.; Zhu, Y. Major QTLs reduce the deleterious effects of high temperature on rice amylose content by increasing splicing efficiency of Wx pre-mRNA. Theor. Appl. Genet. 2014, 127, 273–282. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, H.; Feng, M.; Zhu, Y. Suppression of OsMADS7 in rice endosperm stabilizes amylose content under high temperature stress. Plant Biotechnol. J. 2018, 16, 18–26. [Google Scholar] [CrossRef]
- Fan, X.; Li, Y.; Lu, Y.; Zhang, C.; Li, E.; Li, Q.; Tao, K.; Yu, W.; Wang, J.; Chen, Z.; et al. The interaction between amylose and amylopectin synthesis in rice endosperm grown at high temperature. Food Chem. 2019, 301, 125258. [Google Scholar] [CrossRef]
- Zhao, Q.; Ye, Y.; Han, Z.; Zhou, L.; Guan, X.; Pan, G.; Asad, M.A.; Cheng, F. SSIIIa-RNAi suppression associated changes in rice grain quality and starch biosynthesis metabolism in response to high temperature. Plant Sci. 2020, 294, 110443. [Google Scholar] [CrossRef]
- Tashiro, T.; Wardlaw, I. The effect of high temperature on kernel dimensions and the type and occurrence of kernel damage in rice. Aust. J. Agric. Res. 1991, 42, 485–496. [Google Scholar] [CrossRef]
- Tsukaguchi, T.; Iida, Y. Effects of assimilate supply and high temperature during grain-filling period on the occurrence of various types of chalky kernels in rice plants (Oryza sativa L.). Plant Prod. Sci. 2008, 11, 203–210. [Google Scholar] [CrossRef]
- Hakata, M.; Kuroda, M.; Miyashita, T.; Yamaguchi, T.; Kojima, M.; Sakakibara, H.; Mitsui, T.; Yamakawa, H. Suppression of alpha-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol. J. 2012, 10, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Nakata, M.; Fukamatsu, Y.; Miyashita, T.; Hakata, M.; Kimura, R.; Nakata, Y.; Kuroda, M.; Yamaguchi, T.; Yamakawa, H. High Temperature-Induced Expression of Rice alpha-Amylases in Developing Endosperm Produces Chalky Grains. Front. Plant Sci. 2017, 8, 2089. [Google Scholar] [CrossRef] [PubMed]
- Suriyasak, C.; Harano, K.; Tanamachi, K.; Matsuo, K.; Tamada, A.; Iwaya-Inoue, M.; Ishibashi, Y. Reactive oxygen species induced by heat stress during grain filling of rice (Oryza sativa L.) are involved in occurrence of grain chalkiness. J. Plant Physiol. 2017, 216, 52–57. [Google Scholar] [CrossRef]
- Xu, H.; Li, X.; Zhang, H.; Wang, L.; Zhu, Z.; Gao, J.; Li, C.; Zhu, Y. High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice. Plant Cell Env. 2020, 43, 1879–1896. [Google Scholar] [CrossRef]
- Ren, Y.; Huang, Z.; Jiang, H.; Wang, Z.; Wu, F.; Xiong, Y.; Yao, J. A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J. Exp. Bot. 2021, 72, 2947–2964. [Google Scholar] [CrossRef]
- Nevame, A.Y.M.; Emon, R.M.; Malek, M.A.; Hasan, M.M.; Alam, M.A.; Muharam, F.M.; Aslani, F.; Rafii, M.Y.; Ismail, M.R. Relationship between High Temperature and Formation of Chalkiness and Their Effects on Quality of Rice. Biomed. Res. Int. 2018, 2018, 1653721. [Google Scholar] [CrossRef]
- Yang, W.; Liang, J.; Hao, Q.; Luan, X.; Tan, Q.; Lin, S.; Zhu, H.; Liu, G.; Liu, Z.; Bu, S.; et al. Fine mapping of two grain chalkiness QTLs sensitive to high temperature in rice. Rice 2021, 14, 33. [Google Scholar] [CrossRef]
- Murata, K.; Iyama, Y.; Yamaguchi, T.; Ozaki, H.; Kidani, Y.; Ebitani, T. Identification of a novel gene (Apq1) from the indica rice cultivar ‘Habataki’ that improves the quality of grains produced under high temperature stress. Breed. Sci. 2014, 64, 273–281. [Google Scholar] [CrossRef]
- Park, J.R.; Kim, E.G.; Jang, Y.H.; Kim, K.M. Screening and identification of genes affecting grain quality and spikelet fertility during high-temperature treatment in grain filling stage of rice. BMC Plant Biol. 2021, 21, 263. [Google Scholar] [CrossRef] [PubMed]
- Leesawatwong, M.; Jamjod, S.; Kuo, J.; Dell, B.; Rerkasem, B. Nitrogen fertilizer increases seed protein and milling quality of rice. Cereal Chem. 2005, 82, 588–593. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, Y.; Jia, B.; Wang, Y.; Wang, Y.; Xu, Q.; Li, R.; Wang, S.; Dou, F. Effects of cultivar, nitrogen rate, and planting density on rice-grain quality. Agronomy 2018, 8, 246. [Google Scholar] [CrossRef]
- Zhu, D.-w.; Zhang, H.-c.; Guo, B.-w.; Ke, X.; Dai, Q.-g.; Wei, H.-y.; Hui, G.; Hu, Y.-j.; Cui, P.-y.; Huo, Z.-y. Effects of nitrogen level on yield and quality of japonica soft super rice. J. Integr. Agric. 2017, 16, 1018–1027. [Google Scholar] [CrossRef]
- Gu, J.; Chen, J.; Chen, L.; Wang, Z.; Zhang, H.; Yang, J. Grain quality changes and responses to nitrogen fertilizer of japonica rice cultivars released in the Yangtze River Basin from the 1950s to 2000s. Crop J. 2015, 3, 285–297. [Google Scholar] [CrossRef]
- Liang, H.; Gao, S.; Ma, J.; Zhang, T.; Wang, T.; Zhang, S.; Wu, Z. Effect of nitrogen application rates on the nitrogen utilization, yield and quality of rice. Food Nutr. Sci. 2021, 12, 13–27. [Google Scholar] [CrossRef]
- Cao, X.; Sun, H.; Wang, C.; Ren, X.; Liu, H.; Zhang, Z. Effects of late-stage nitrogen fertilizer application on the starch structure and cooking quality of rice. J. Sci. Food Agric. 2018, 98, 2332–2340. [Google Scholar] [CrossRef]
- Liang, H.; Tao, D.; Zhang, Q.; Zhang, S.; Wang, J.; Liu, L.; Wu, Z.; Sun, W. Nitrogen fertilizer application rate impacts eating and cooking quality of rice after storage. PLoS ONE 2021, 16, e0253189. [Google Scholar] [CrossRef]
- Ali, I.; Iqbal, A.; Ullah, S.; Muhammad, I.; Yuan, P.; Zhao, Q.; Yang, M.; Zhang, H.; Huang, M.; Liang, H.; et al. Effects of Biochar Amendment and Nitrogen Fertilizer on RVA Profile and Rice Grain Quality Attributes. Foods 2022, 11, 625. [Google Scholar] [CrossRef]
- Xia, D.; Wang, Y.; Shi, Q.; Wu, B.; Yu, X.; Zhang, C.; Li, Y.; Fu, P.; Li, M.; Zhang, Q.; et al. Effects of Wx Genotype, Nitrogen Fertilization, and Temperature on Rice Grain Quality. Front. Plant Sci. 2022, 13, 901541. [Google Scholar] [CrossRef]
- Gong, D.; Xu, X.; Wu, L.; Dai, G.; Zheng, W.; Xu, Z. Effect of biochar on rice starch properties and starch-related gene expression and enzyme activities. Sci. Rep. 2020, 10, 16917. [Google Scholar] [CrossRef]
- Chen, L.; Guo, L.; Deng, X.; Pan, X.; Liao, P.; Xiong, Q.; Gao, H.; Wei, H.; Dai, Q.; Zeng, Y. Effects of biochar on rice yield, grain quality and starch viscosity attributes. J. Sci. Food Agric. 2023. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, M.; Reuling, G.; Karssen, C.M. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant 1984, 61, 377–383. [Google Scholar] [CrossRef]
- Fitzgerald, M.A.; Sackville Hamilton, N.R.; Calingacion, M.N.; Verhoeven, H.A.; Butardo, V.M. Is there a second fragrance gene in rice? Plant Biotechnol. J. 2008, 6, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Liu, Y.; Gong, C.; Chen, B.; Wang, T. Waxy is an important factor for grain fissure resistance and head rice yield as revealed by a genome-wide association study. J. Exp. Bot. 2022, 73, 6942–6954. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Pan, C.; Li, Y.; Wu, Y.; Cai, Y.; Lu, Y.; Wang, R.; Yu, L.; Shi, W.; Kang, H.; et al. Genomic insight into balancing high yield, good quality, and blast resistance of japonica rice. Genome Biol. 2021, 22, 283. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Ye, X.; Al-Babili, S.; Kloti, A.; Zhang, J.; Lucca, P.; Beyer, P.; Potrykus, I. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 2000, 287, 303–305. [Google Scholar] [CrossRef]
- Wirth, J.; Poletti, S.; Aeschlimann, B.; Yakandawala, N.; Drosse, B.; Osorio, S.; Tohge, T.; Fernie, A.R.; Gunther, D.; Gruissem, W.; et al. Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol. J. 2009, 7, 631–644. [Google Scholar] [CrossRef]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef] [PubMed]
- Kusaba, M.; Miyahara, K.; Iida, S.; Fukuoka, H.; Takano, T.; Sassa, H.; Nishimura, M.; Nishio, T. Low glutelin content1: A dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 2003, 15, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, L.; Liu, G.; Meng, X.; Jing, Y.; Shu, X.; Kong, X.; Sun, J.; Yu, H.; Smith, S.M.; et al. Critical roles of soluble starch synthase SSIIIa and granule-bound starch synthase Waxy in synthesizing resistant starch in rice. Proc. Natl. Acad. Sci. USA 2016, 113, 12844–12849. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.C.; Xie, M.X.; Wang, Y.C.; Li, J.Y. Molecular Mechanisms Underlying gamma-Aminobutyric Acid (GABA) Accumulation in Giant Embryo Rice Seeds. J. Agric. Food Chem. 2017, 65, 4883–4889. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).