Isolation and Activity Analysis of Phytoene Synthase (ClPsy1) Gene Promoter of Canary-Yellow and Golden Flesh-Color Watermelon
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Promoter Isolation and Sequence Analysis
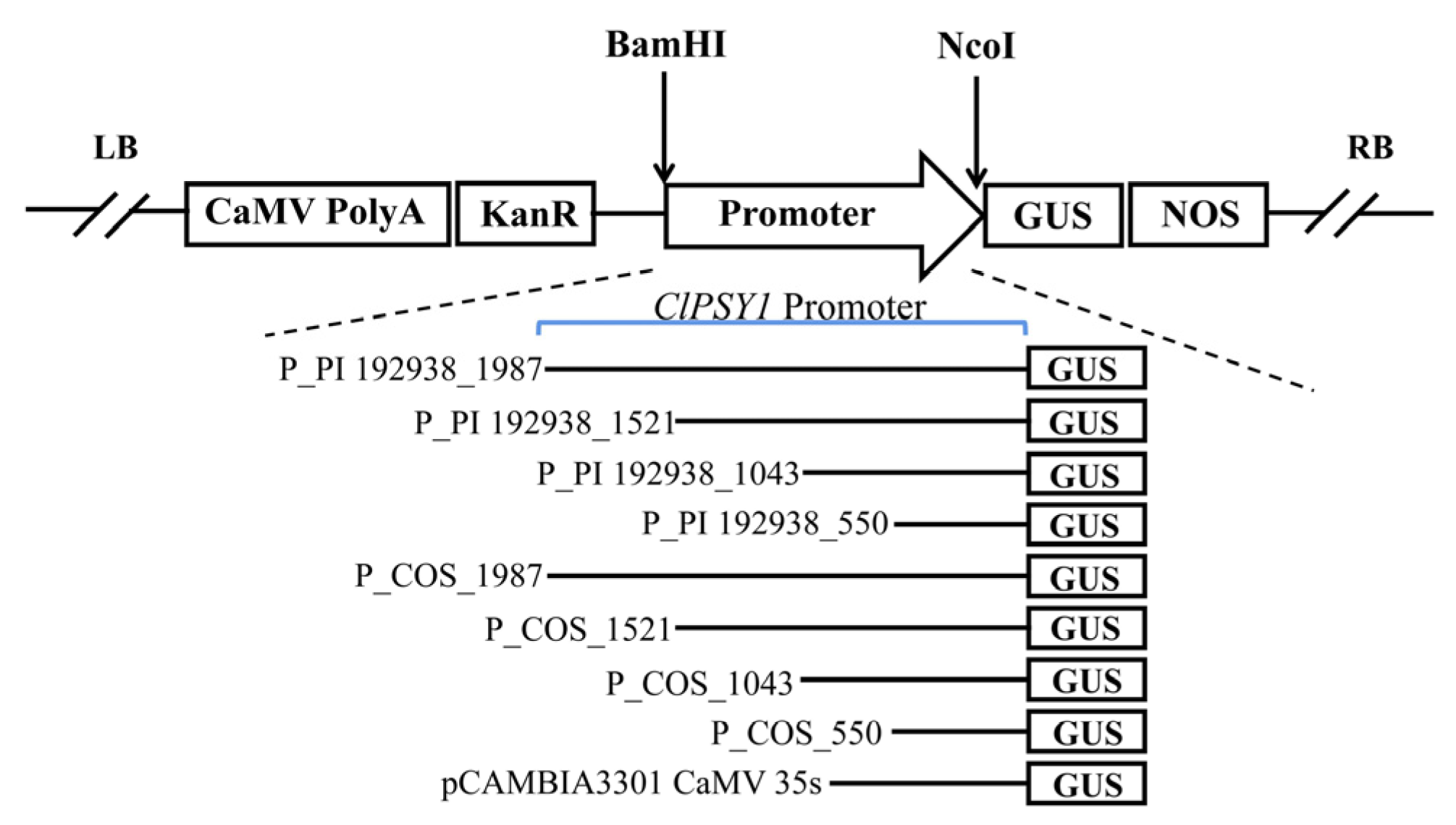
2.3. Promoter Deletion Fragments’ Vector Construction
2.4. Transient Transformation of Ripe Green Tomatoes
2.5. β-D-Glucuronidase Assays
2.6. Gene-Expression Pattern Analysis
2.7. Statistical Analysis
3. Results
3.1. ClPsy1 Promoter Sequence and cis-Acting Element Analysis
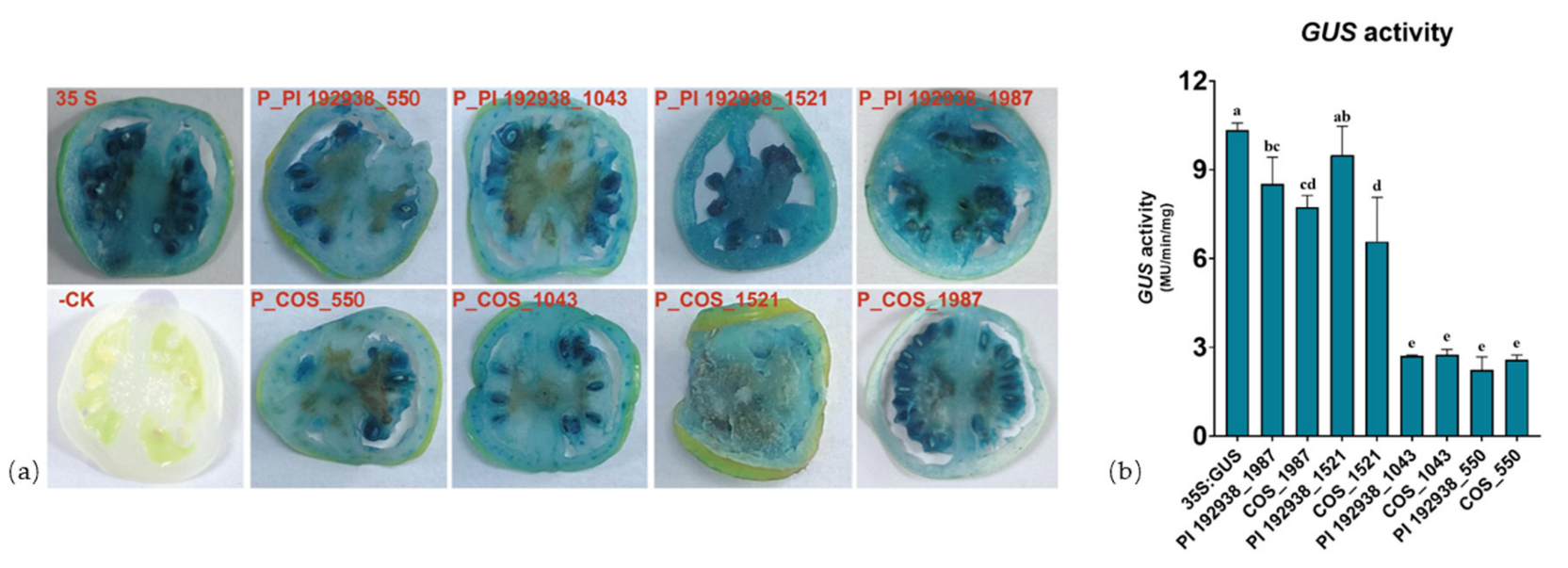
3.2. Analysis of ClPsy1 Promoter Activity in Transformed Tomato
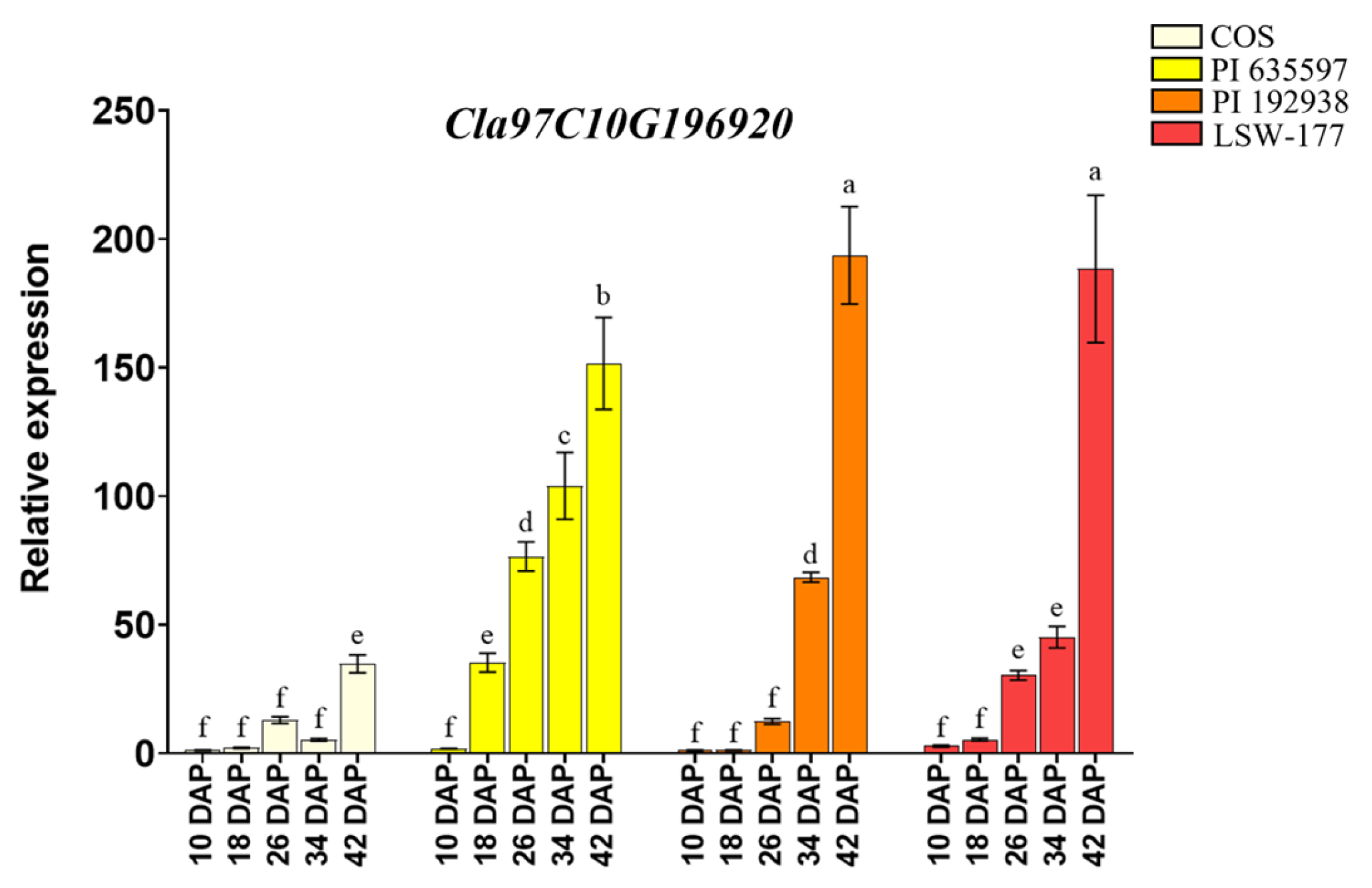
3.3. MYB TF Cla97C10G196920 Positively Regulates Carotenoid Metabolism in Watermelon Pulp
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yabuzaki, J. Carotenoids Database: Structures, chemical fingerprints and distribution among organisms. Database 2017, 2017, bax004. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shao, Z.; Zhang, M.; Wang, Q. Regulation of carotenoid metabolism in tomato. Mol. Plant 2015, 8, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Clagett-Dame, M.; Knutson, D. Vitamin A in reproduction and development. Nutrients 2011, 3, 385–428. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Rivers, J.; León, P.; McQuinn, R.P.; Pogson, B.J. Synthesis and Function of Apocarotenoid Signals in Plants. Trends Plant Sci. 2016, 21, 792–803. [Google Scholar] [CrossRef]
- Loreto, F.; Pollastri, S.; Fineschi, S.; Velikova, V. Volatile Isoprenoids and Their Importance for Protection against Environmental Constraints in the Mediterranean Area. Environ. Exp. Bot. 2014, 103, 99–106. Available online: https://www.sciencedirect.com/science/article/pii/S0098847213001263 (accessed on 15 November 2022). [CrossRef]
- Carotenoid Metabolism and Regulation in Horticultural Crops | Horticulture Research. Available online: https://www.nature.com/articles/hortres201536 (accessed on 15 November 2022).
- Fiedor, J.; Burda, K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef]
- Li, H.; Yang, X.; Shang, Y.; Zhang, Z.; Huang, S. biology and breeding in the genomics era. Sci. China Life Sci. 2023, 66, 226–250. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. Available online: https://www.sciencedirect.com/science/article/pii/S1360138510000294 (accessed on 15 November 2022). [CrossRef]
- Kaur, N.; Pandey, A.; Shivani; Kumar, P.; Pandey, P.; Kesarwani, A.K.; Mantri, S.S.; Awasthi, P.; Tiwari, S. Regulation of Banana Phytoene Synthase (MaPSY) Expression, Characterization and Their Modulation under Various Abiotic Stress Conditions. Front. Plant Sci. 2017, 8, 462. [Google Scholar] [CrossRef]
- Fantini, E.; Falcone, G.; Frusciante, S.; Giliberto, L.; Giuliano, G. Dissection of tomato lycopene biosynthesis through virus-induced gene silencing. Plant Physiol. 2013, 163, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vallabhaneni, R.; Yu, J.; Rocheford, T.; Wurtzel, E.T. The maize phytoene synthase gene family: Overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol. 2008, 147, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Welsch, R.; Maass, D.; Voegel, T.; Dellapenna, D.; Beyer, P. Transcription factor RAP2.2 and its interacting partner SINAT2: Stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007, 145, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Luo, D.; Lin, A.; Zhang, C.; Shan, L.; He, P.; Li, B.; Zhang, Q.; Hua, B.; Yuan, Z.; et al. A tomato B-box protein SlBBX20 modulates carotenoid biosynthesis by directly activating PHYTOENE SYNTHASE 1, and is targeted for 26S proteasome-mediated degradation. New Phytol. 2019, 221, 279–294. [Google Scholar] [CrossRef]
- Wu, M.; Xu, X.; Hu, X.; Liu, Y.; Cao, H.; Chan, H.; Gong, Z.; Yuan, Y.; Luo, Y.; Feng, B.; et al. SlMYB72 Regulates the Metabolism of Chlorophylls, Carotenoids, and Flavonoids in Tomato Fruit. Plant Physiol. 2020, 183, 854–868. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Rodríguez-Concepción, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef]
- Llorente, B.; D’Andrea, L.; Ruiz-Sola, M.A.; Botterweg, E.; Pulido, P.; Andilla, J.; Loza-Alvarez, P.; Rodriguez-Concepcion, M. Tomato fruit carotenoid biosynthesis is adjusted to actual ripening progression by a light-dependent mechanism. Plant J. 2016, 85, 107–119. [Google Scholar] [CrossRef]
- Vrebalov, J.; Pan, I.L.; Arroyo, A.J.M.; McQuinn, R.; Chung, M.; Poole, M.; Rose, J.; Seymour, G.; Grandillo, S.; Giovannoni, J.; et al. Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 2009, 21, 3041–3062. [Google Scholar] [CrossRef]
- Qin, G.; Wang, Y.; Cao, B.; Wang, W.; Tian, S. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 2012, 70, 243–255. [Google Scholar] [CrossRef]
- Bemer, M.; Karlova, R.; Ballester, A.R.; Tikunov, Y.M.; Bovy, A.G.; Wolters-Arts, M.; Rossetto, P.d.B.; Angenent, G.C.; de Maagd, R.A. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 2012, 24, 4437–4451. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Hu, Z.; Deng, L.; Wang, Y.; Zhu, M.; Zhang, J.; Chen, G. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 2013, 163, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Hu, Z.; Zhu, Z.; Dong, T.; Zhao, Z.; Cui, B.; Chen, G. Overexpression of a novel MADS-box gene SlFYFL delays senescence, fruit ripening and abscission in tomato. Sci. Rep. 2014, 4, 4367. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, Z.; Yao, Q.; Guo, X.; Nguyen, V.; Li, F.; Chen, G. A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 2018, 8, 3413. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The Citrus Transcription Factor CsMADS6 Modulates Carotenoid Metabolism by Directly Regulating Carotenogenic Genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zheng, X.; Zhu, K.; Xu, Q.; Deng, X. Isolation and Functional Characterization of a Lycopene β-cyclase Gene Promoter from Citrus. Front. Plant Sci. 2016, 7, 1367. [Google Scholar] [CrossRef]
- Cunningham, F.X.; Pogson, B.; Sun, Z.; McDonald, K.A.; DellaPenna, D.; Gantt, E. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 1996, 8, 1613–1626. [Google Scholar] [CrossRef]
- Gao, Y.; Fan, Z.-Q.; Zhang, Q.; Li, H.-L.; Liu, G.-S.; Jing, Y.; Zhang, Y.-P.; Zhu, B.-Z.; Zhu, H.-L.; Chen, J.-Y.; et al. A tomato NAC transcription factor, SlNAM1, positively regulates ethylene biosynthesis and the onset of tomato fruit ripening. Plant J. 2021, 108, 1317–1331. [Google Scholar] [CrossRef]
- Liu, S.; Gao, Z.; Wang, X.; Luan, F.; Dai, Z.; Yang, Z.; Zhang, Q. Nucleotide variation in the phytoene synthase (ClPsy1) gene contributes to golden flesh in watermelon (Citrullus lanatus L.). Appl. Genet. 2022, 135, 185–200. [Google Scholar] [CrossRef]
- Orzaez, D.; Mirabel, S.; Wieland, W.H.; Granell, A. Agroinjection of tomato fruits. A Tool for Rapid Functional Analysis of Transgenes Directly in Fruit. Plant Physiol. 2006, 140, 3–11. [Google Scholar] [CrossRef]
- Bustin, S.A.; Vandesompele, J.; Pfaffl, M. Standardization of qPCR and RT-qPCR. Gen. Eng. Biotechnol. News 2009, 29, 14. Available online: https://www.semanticscholar.org/paper/Standardization-of-qPCR-and-RT-qPCR-Bustin-Vandesompele/48364a69605bebda6783c474f8128f338ad43b30 (accessed on 10 February 2023).
- Fang, X.; Liu, S.; Gao, P.; Liu, H.; Wang, X.; Luan, F.; Zhang, Q.; Dai, Z. Expression of ClPAP and ClPSY1 in watermelon correlates with chromoplast differentiation, carotenoid accumulation, and flesh color formation. Sci. Hortic. 2020, 270, 109437. Available online: https://www.sciencedirect.com/science/article/pii/S030442382030265X (accessed on 15 November 2022). [CrossRef]
- Fang, X.; Gao, P.; Luan, F.; Liu, S. Identification and Characterization Roles of Phytoene Synthase (PSY) Genes in Watermelon Development. Genes 2022, 13, 1189. [Google Scholar] [CrossRef] [PubMed]
- Mapping of quantitative trait loci for lycopene content and fruit traits in Citrullus lanatus | SpringerLink. Available online: https://link.springer.com/article/10.1007/s10681-014-1308-9 (accessed on 13 February 2023).
- Liu, S.; Gao, P.; Zhu, Q.; Luan, F.; Davis, A.R.; Wang, X. Development of cleaved amplified polymorphic sequence markers and a CAPS-based genetic linkage map in watermelon (Citrullus lanatus [Thunb.] Matsum. Nakai) Constr. Using Whole-Genome Re-Seq. Data. Breed Sci. 2016, 66, 244–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Qiao, A.; Fang, X.; Sun, L.; Gao, P.; Davis, A.R.; Liu, S.; Luan, F. Fine Mapping of Lycopene Content and Flesh Color Related Gene and Development of Molecular Marker-Assisted Selection for Flesh Color in Watermelon (Citrullus lanatus). Front. Plant Sci. 2019, 10, 1240. [Google Scholar] [CrossRef]
- Liu, S.; Liu, M.; Cao, Y.; Xu, Y.; Liu, H.; Zhu, Q.; Zhang, X.; Luan, F. Identification of chromosome region and candidate genes for canary-yellow flesh (Cyf) locus in watermelon (Citrullus lanatus). Plant Sci. 2023, 329, 111594. [Google Scholar] [CrossRef]
- Branham, S.; Vexler, L.; Meir, A.; Tzuri, G.; Frieman, Z.; Levi, A.; Wechter, W.P.; Tadmor, Y.; Gur, A. Genetic mapping of a major codominant QTL associated with β-carotene accumulation in watermelon. Mol. Breed. 2017, 37, 146. Available online: https://link.springer.com/article/10.1007/s11032-017-0747-0 (accessed on 13 February 2023). [CrossRef]
- Cheng, J.; Wei, F.; Zhang, M.; Li, N.; Song, T.; Wang, Y.; Chen, D.; Xiang, J.; Zhang, X. Identification of a 193 bp promoter region of TaNRX1-D gene from common wheat that contributes to osmotic or ABA stress inducibility in transgenic Arabidopsis. Genes Genom. 2021, 43, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Berumen-Varela, G.; Ochoa-Jiménez, V.-A.; Burgara-Estrella, A.; Trillo-Hernández, E.-A.; Ojeda-Contreras, Á.-J.; Orozco-Avitia, A.; Rivera-Domínguez, M.; Troncoso-Rojas, R.; Báez-Sañudo, R.; Datsenka, T.; et al. Functional analysis of a tomato (Solanum lycopersicum L.) rhamnogalacturonan lyase promoter. J. Plant Physiol. 2018, 229, 175–184. [Google Scholar] [CrossRef]
- Kuang, J.-F.; Chen, J.-Y.; Liu, X.-C.; Han, Y.-C.; Xiao, Y.-Y.; Shan, W.; Tang, Y.; Wu, K.-Q.; He, J.-X.; Lu, W.-J. The transcriptional regulatory network mediated by banana (Musa acuminata) dehydration-responsive element binding (MaDREB) transcription factors in fruit ripening. New Phytol. 2017, 214, 762–781. [Google Scholar] [CrossRef]
- Sabir, I.A.; Manzoor, M.A.; Shah, I.H.; Liu, X.; Zahid, M.S.; Jiu, S.; Wang, J.; Abdullah, M.; Zhang, C. MYB transcription factor family in sweet cherry (Prunus avium L.): Genome-wide investigation, evolution, structure, characterization and expression patterns. BMC Plant Biol. 2022, 22, 2. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhang, L.; Liu, L.; Tang, X.-F.; Yang, W.-J.; Wu, Y.-M.; Huang, Y.-B.; Tang, Y.-X. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry 2009, 74, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 9, 359–378. Available online: https://onlinelibrary.wiley.com/doi/full/10.1111/tpj.14330 (accessed on 15 November 2022). [CrossRef]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef]
- Xi, W.; Feng, J.; Liu, Y.; Zhang, S.; Zhao, G. The R2R3-MYB transcription factor PaMYB10 is involved in anthocyanin biosynthesis in apricots and determines red blushed skin. BMC Plant Biol. 2019, 19, 287. [Google Scholar] [CrossRef]
- Li, J.; An, Y.; Wang, L. Transcriptomic Analysis of Ficus carica Peels with a Focus on the Key Genes for Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2020, 21, 1245. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Fang, X.; Liu, S.; Luan, F. Isolation and Activity Analysis of Phytoene Synthase (ClPsy1) Gene Promoter of Canary-Yellow and Golden Flesh-Color Watermelon. Agronomy 2023, 13, 1080. https://doi.org/10.3390/agronomy13041080
Cao Y, Fang X, Liu S, Luan F. Isolation and Activity Analysis of Phytoene Synthase (ClPsy1) Gene Promoter of Canary-Yellow and Golden Flesh-Color Watermelon. Agronomy. 2023; 13(4):1080. https://doi.org/10.3390/agronomy13041080
Chicago/Turabian StyleCao, Yue, Xufeng Fang, Shi Liu, and Feishi Luan. 2023. "Isolation and Activity Analysis of Phytoene Synthase (ClPsy1) Gene Promoter of Canary-Yellow and Golden Flesh-Color Watermelon" Agronomy 13, no. 4: 1080. https://doi.org/10.3390/agronomy13041080
APA StyleCao, Y., Fang, X., Liu, S., & Luan, F. (2023). Isolation and Activity Analysis of Phytoene Synthase (ClPsy1) Gene Promoter of Canary-Yellow and Golden Flesh-Color Watermelon. Agronomy, 13(4), 1080. https://doi.org/10.3390/agronomy13041080




