Identification and Analysis of the Expression of microRNAs during the Low-Temperature Dormancy Release of Tulipa thianschanica Seeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection and Pre-Treatment of Seeds
2.2. Total RNA Extraction and Establishment of the miRNA Library
2.3. Identification and Functional Annotation of the Target Genes of miRNAs
2.4. Analysis of the Differential Expression of miRNA
2.5. Verification of the Target Genes of miRNAs by qRT-PCR
2.6. Statistical Analysis
3. Results and Analysis
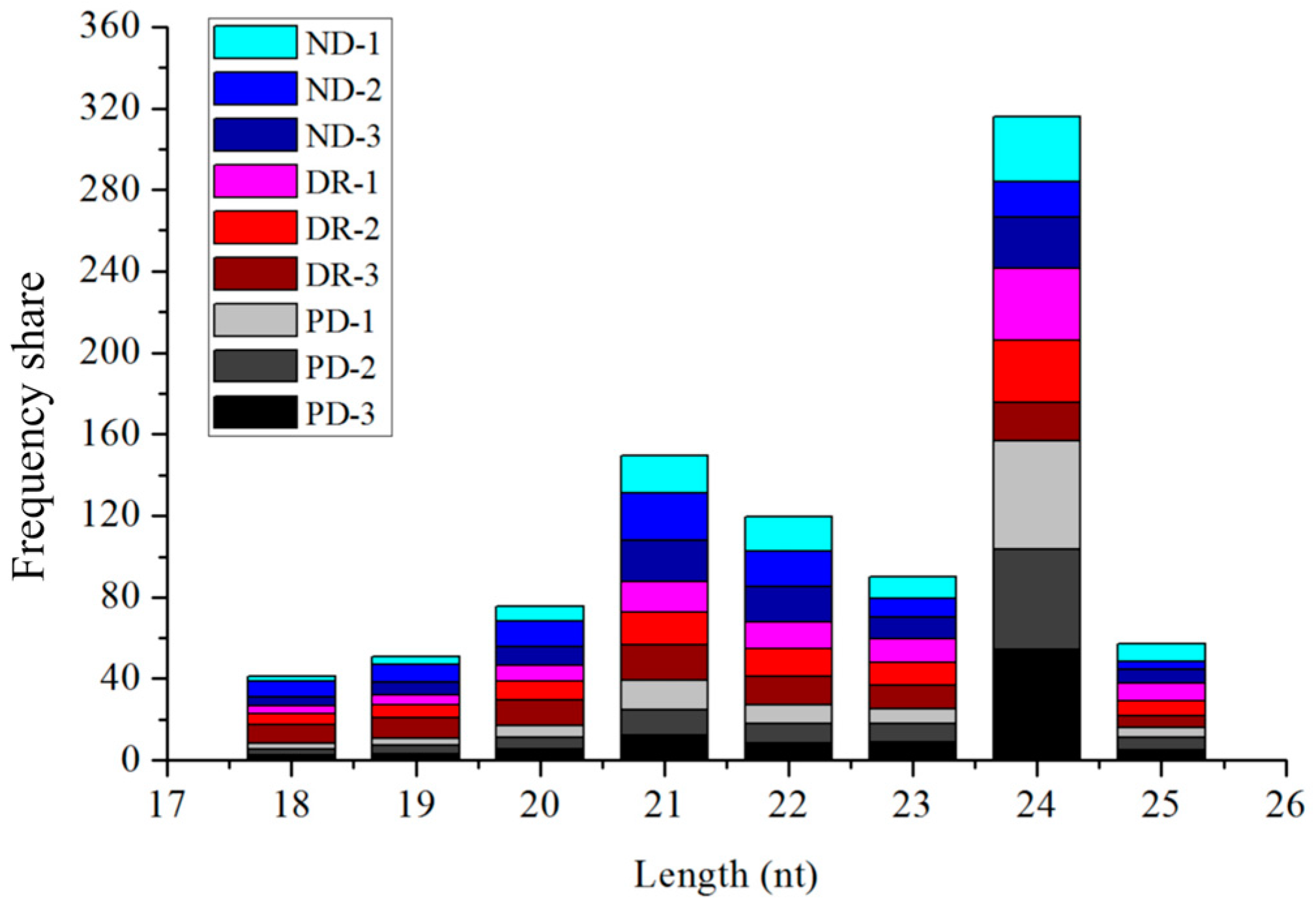
3.1. miRNA Sequencing of Seeds at Different Stages of Low-Temperature Stratification
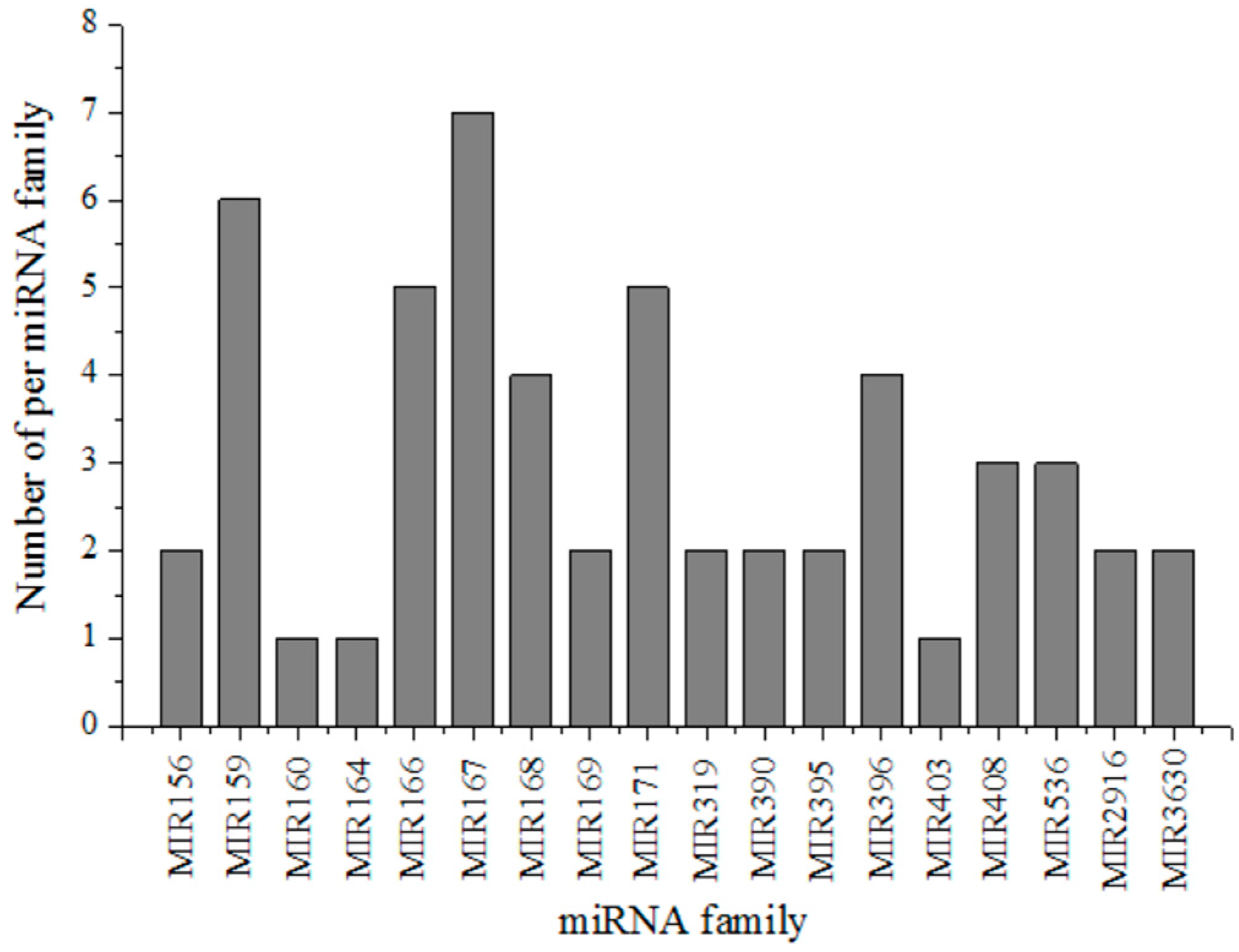
3.2. Identification of the Conserved miRNAs
3.3. Identification of Novel miRNAs
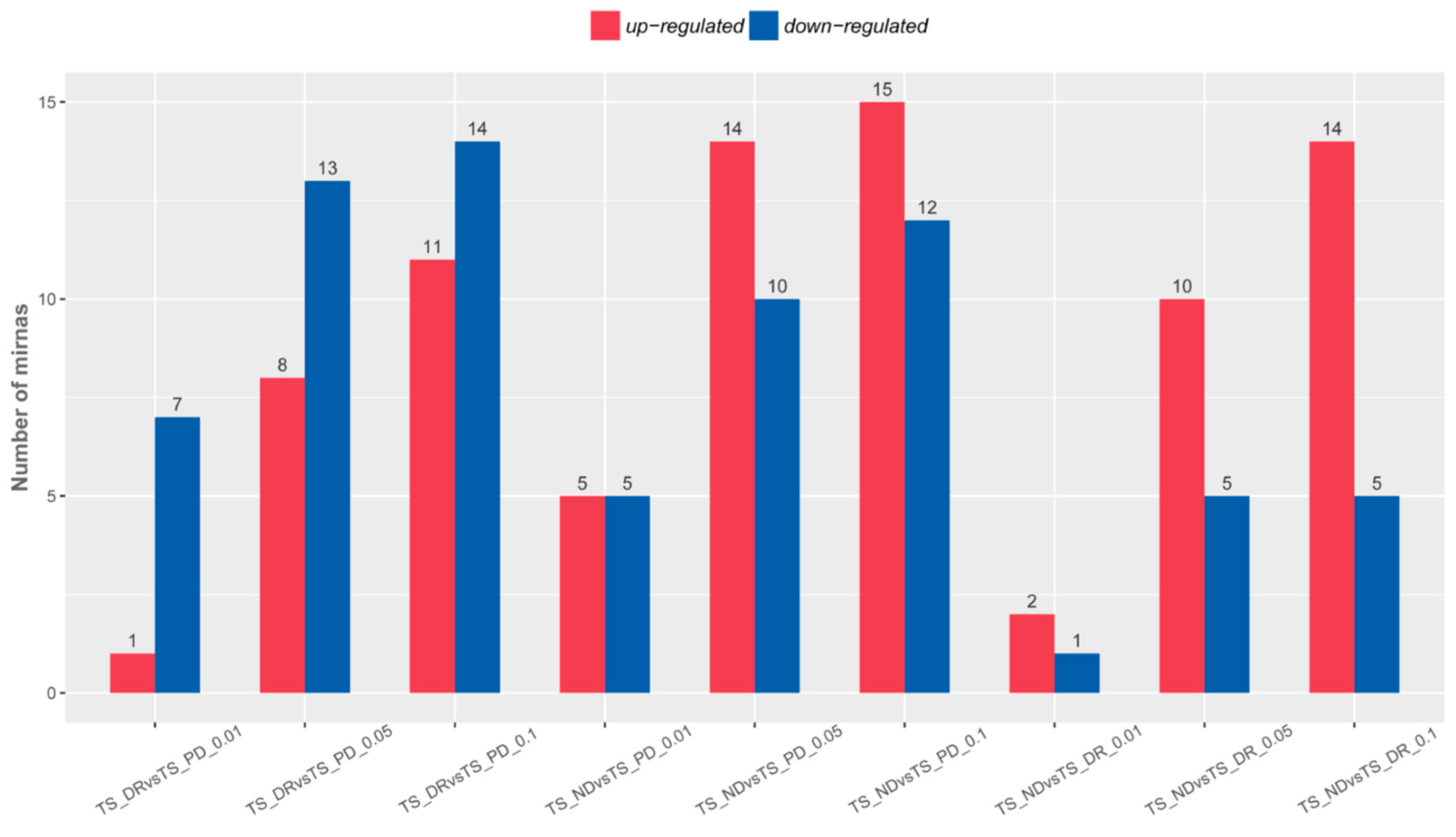
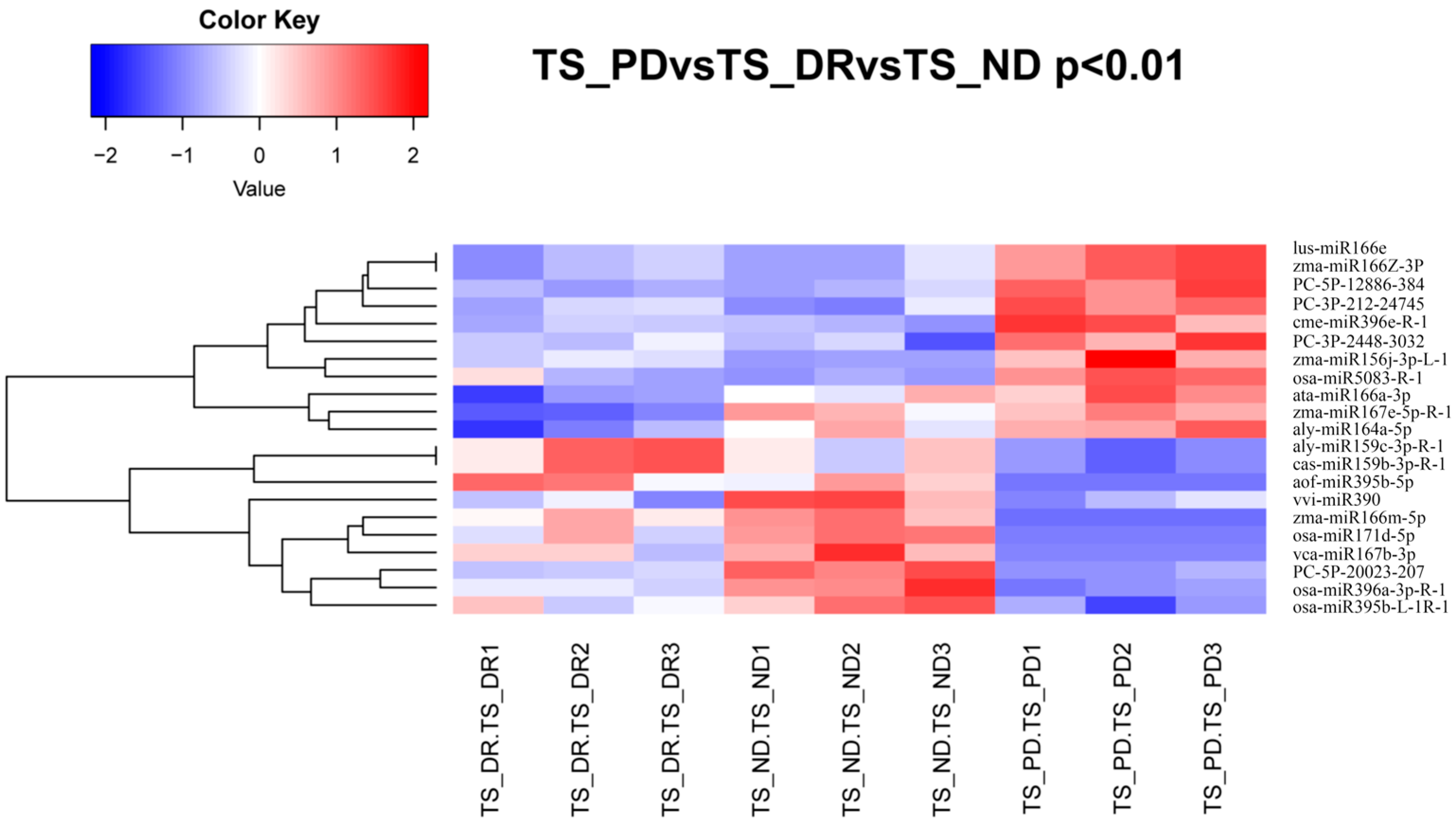
3.4. Analysis of the Differentially Expressed miRNAs during the Release of Seed Dormancy
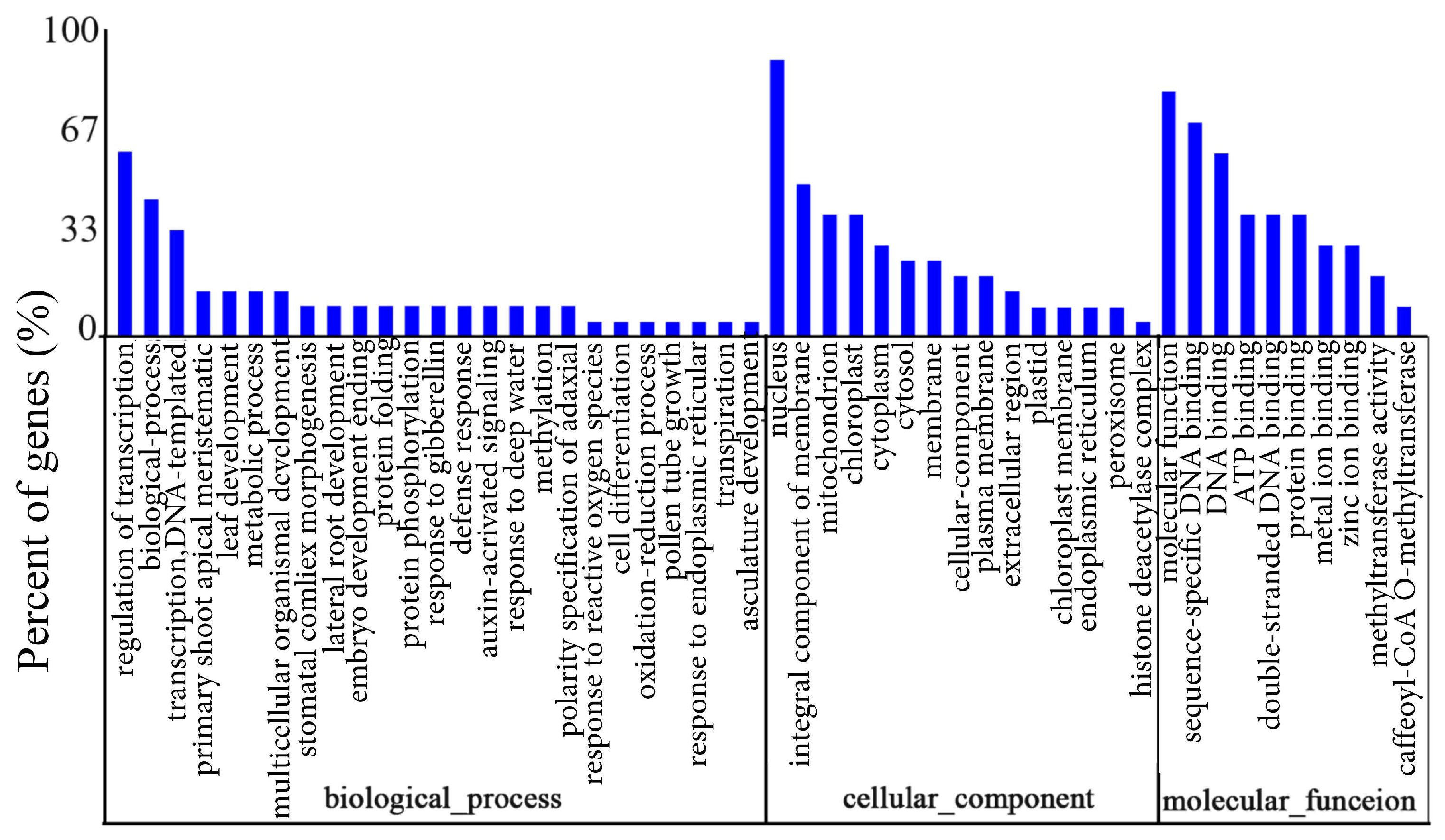
3.5. Prediction and Functional Annotation of the Target Genes of the miRNAs
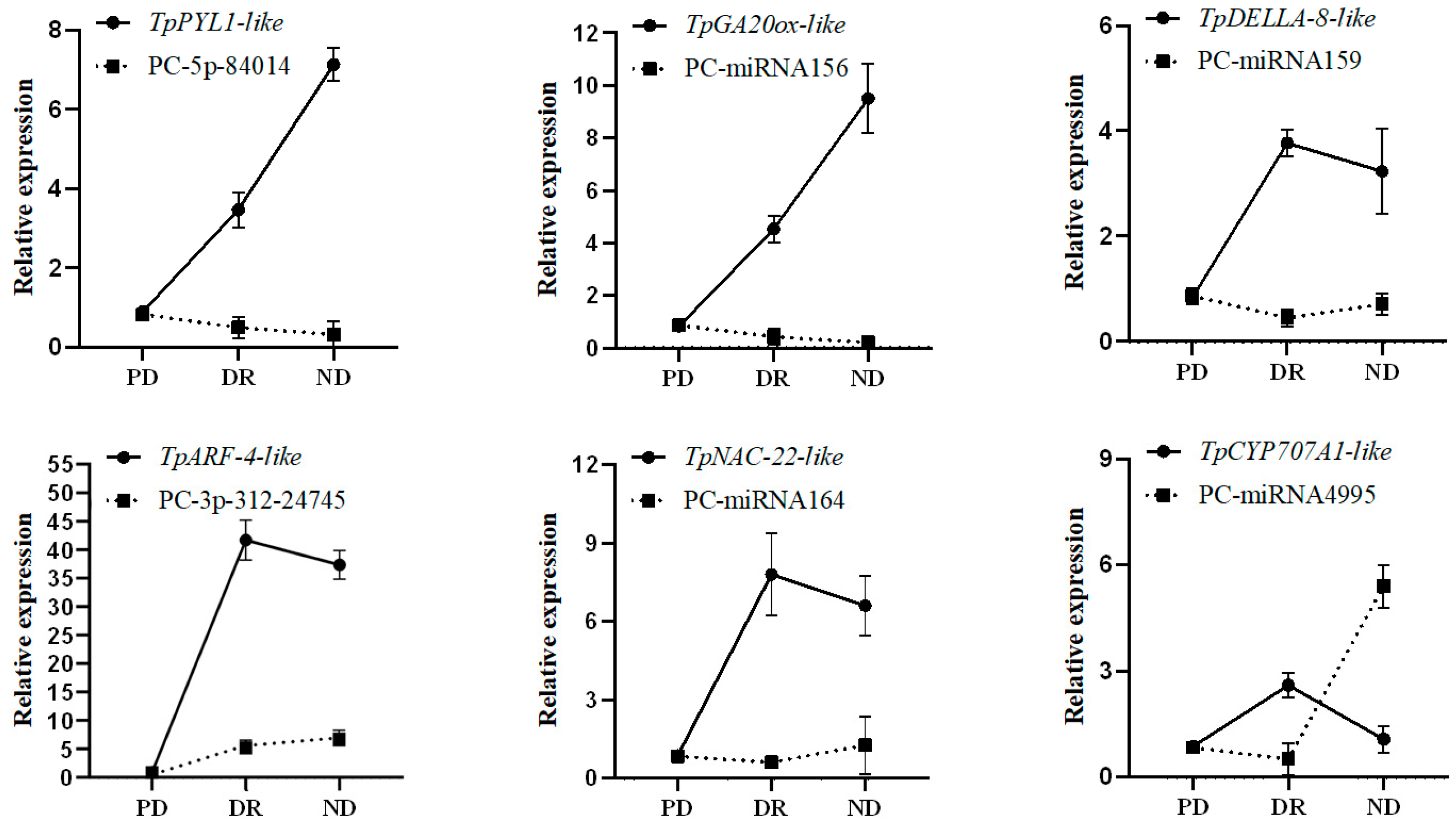
3.6. Analysis of the Expression of miRNAs Related to Dormancy and Their Target Genes
4. Discussion
4.1. miRNA Dynamic Patterns of Expression during the Dormancy Release of T. thianschanica Seeds
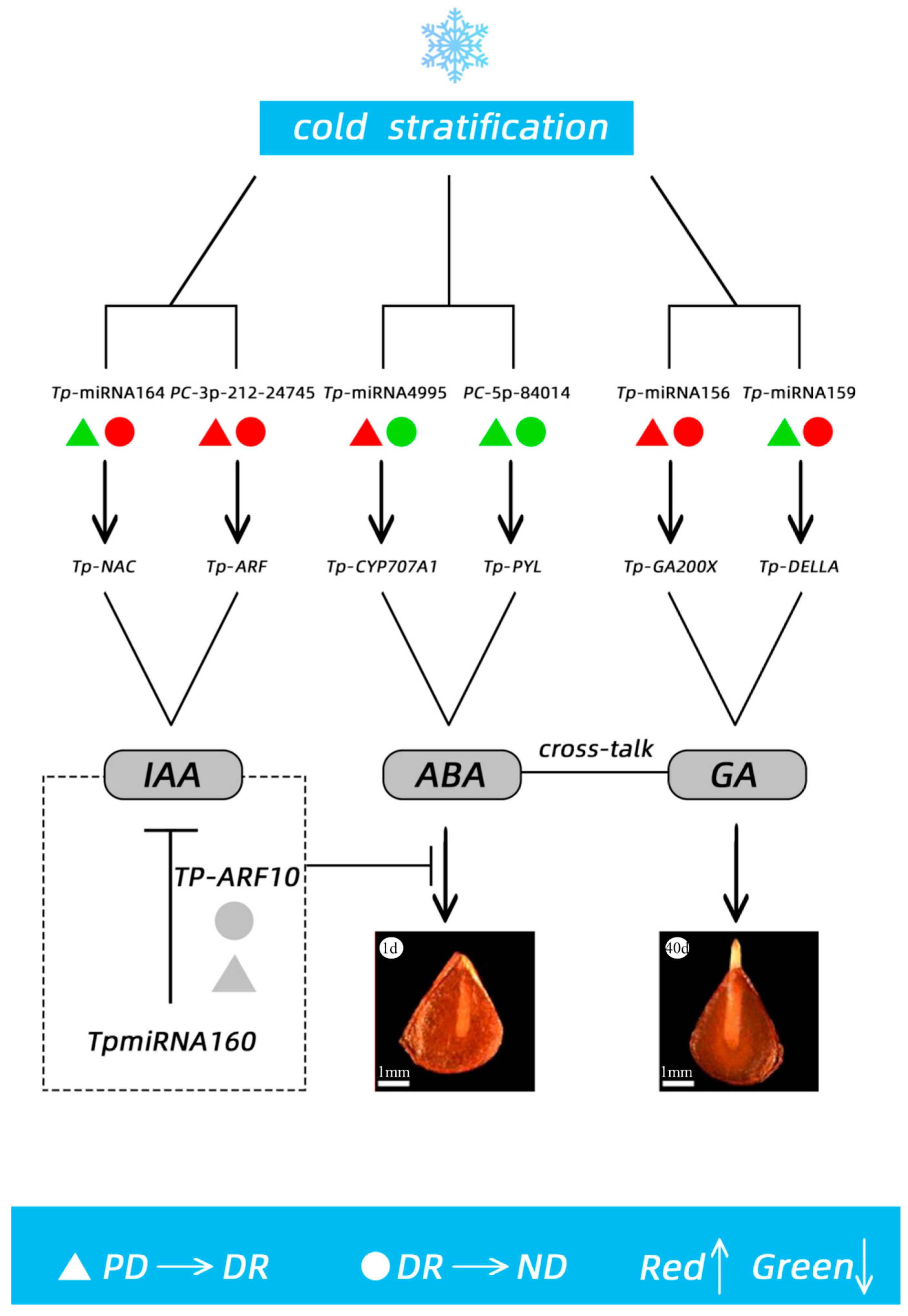
4.2. Regulatory Mechanism of the miRNA of T. thianschanica Seeds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qu, L.W.; Xue, L.; Xing, G.M.; Zhang, Y.Q.; Chen, J.J.; Zhang, W.; Lei, J.J. Karyotype analysis of eight wild Tulipa species native to China and the interspecific hybridization with tulip cultivars. Euphytica 2018, 214, 2–12. [Google Scholar] [CrossRef]
- Xing, G.M.; Qu, L.W.; Zhang, Y.Q.; Xue, L.; Su, J.W.; Lei, J.J. Collection and evaluation of wild tulip (Tulipa spp.) resources in China. Genet. Resour. Crop Evol. 2017, 64, 641–652. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, J.; Xue, L.; Dai, H.P.; Lei, J.J. Seed Morphology and Germination of Native Tulipa Species. Agriculture 2023, 13, 466. [Google Scholar] [CrossRef]
- Chen, X.M. MicroRNA biogenesis and function in plants. FEBS Lett. 2005, 579, 5923–5931. [Google Scholar] [CrossRef] [PubMed]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of miRNA genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef]
- Bai, B.; Peviani, A.; van der Horst, S.; Gamm, M.; Snel, B.; Bentsink, L.; Hanson, J. Extensive translational regulation during seed germination revealed by polysomal profiling. New Phytol. 2017, 214, 233–244. [Google Scholar] [CrossRef]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, 31–36. [Google Scholar] [CrossRef]
- Ramya, R.; Herv, V.; Jerry, T.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar]
- Huang, D.Q.; Koh, C.; Feurtado, J.A.; Tsang, E.W.T.; Cutler, A.J. MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genom. 2013, 14, 140. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, H.J.; Jung, H.J.; Maruyama, K.; Suzuki, N.; Kang, H. Over expression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta 2010, 232, 1447–1454. [Google Scholar] [CrossRef]
- Martin, R.C.; Liu, P.P.; Goloviznina, N.A.; Nonogaki, H. MicroRNA, seeds, and Darwin: Diverse function of miRNA in seed biology and plant responses to stress. J. Exp. Bot. 2010, 61, 2229–2234. [Google Scholar] [CrossRef]
- Li, J.Y.; Reichel, M.; Li, Y.J.; Millar, A.A. The functional scope of plant microRNA-mediated silencing. Trends Plant Sci. 2014, 19, 750–756. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of mi156 and mi172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; El-Kassaby, Y.A. Regulatory crosstalk between microRNAs and hormone signalling cascades controls the variation on seed dormancy phenotype at Arabidopsis thaliana seed set. Plant Cell Rep. 2017, 36, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.F.; Jiao, F.C.; Xu, H.C.; Jiao, Q.Q.; Zhang, X.; Zhang, Y.; Du, S.Y.; Xi, M.H.; Wang, A.; Chen, J.T.; et al. The role of RNA-binding protein, microRNA and alternative splicing in seed germination: A field need to be discovered. BMC Plant Biol. 2021, 21, 194. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef] [PubMed]
- Weitbrecht, K.; Muller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Howell, M.D. miRNAs in the biogenesis of transacting siRNAs in higher plants. Semin. Cell Dev. Biol. 2010, 21, 798–804. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Jan, C.; Rajagopalan, R.; Bartel, D.P. A two-hit trigger for siRNA biogenesis in plants. Cell 2006, 127, 565–577. [Google Scholar] [CrossRef]
- Griffiths, J.; Murase, K.; Rieu, I.; Zentella, R.; Zhang, Z.L.; Powers, S.J.; Gong, F.; Phillips, A.L.; Hedden, P.; Sun, T.P. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2006, 18, 3399–3414. [Google Scholar] [CrossRef]
- Wang, J.W.; Wang, L.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, Y.; Liu, Z.; Kong, D.; Duan, M.; Luo, L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 2010, 61, 4157–4168. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Z.; Deng, M.; Liu, R.; Niu, S.; Fan, G. Identification and functional analysis of microRNAs and their targets in Platanus acerifolia under lead (Pb) stress. Int. J. Mol. Sci. 2015, 16, 7098–7111. [Google Scholar] [CrossRef] [PubMed]
- Saminathan, T.; Bodunrin, A.; Singh, N.V.; Devarajan, R.; Nimmakayala, P.; Jeff, M.; Aradhya, M.; Reddy, U.K. Genome-wide identification of microRNAs in pomegranate (Punica granatum L.) by high throughput sequencing. BMC Plant Biol. 2016, 16, 122. [Google Scholar] [CrossRef] [PubMed]
- Mei, M.; Wei, J.; Ai, W.F.; Zhang, L.J.; Lu, X.J. Integrated RNA and miRNA sequencing analysis reveals a complex regulatory network of Magnolia sieboldii seed germination. Sci. Rep. 2021, 11, 10842. [Google Scholar] [CrossRef]
- DeBoer, K.; Melser, S.; Sperschneider, J.; Kamphuis, L.G.; Garg, G.; Gao, L.L.; Frick, K.; Singh, K.B. Identification and profiling of narrow-leafed lupin (Lupinus angustifolius) microRNAs during seed development. BMC Genom. 2019, 20, 135. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.W.; Zhao, J.G.; Zhao, J.Y.; Xu, H.M.; Wang, L.; Jin, B. Cytological and miRNA expression changes during the vascular cambial transition from the dormant stage to the active stage in Ginkgo biloba L. Trees 2016, 30, 2177–2188. [Google Scholar] [CrossRef]
- Kajal, M.; Singh, K. Small RNA profiling for identification of miRNAs involved in regulation of saponins biosynthesis in Chlorophytum borivilianum. BMC Plant Biol. 2017, 17, 265. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, C.H.; Liu, H.N.; Tao, J.; Zhang, K.L. Seed dormancy and germination requirements of Torilis scabra (Apiaceae). Agronomy 2023, 13, 1250. [Google Scholar] [CrossRef]
- Wan, Y.L.; Zhang, M.; Liu, A.Q.; Yuan, Q.P.; Liu, Y. Morphology and physiology response to stratification during seeds epicotyl dormancy breaking of Paeonia emodi Wall. Ex. Agronomy 2022, 12, 1745. [Google Scholar] [CrossRef]
- Han, Z.P.; Jin, Y.Q.; Wang, B.; Guo, Y.Y. Multi-omics revealed the molecular mechanism of Maize (Zea mays L.) seed germination regulated by GA3. Agronomy 2023, 13, 1929. [Google Scholar] [CrossRef]
- Zhou, Y.W.; Wang, W.; Yang, L.H.; Su, X.X. Identification and expression analysis of microRNAs in response to dormancy release during cold storage of Lilium pumilum Bulbs. J. Plant Growth Regul. 2021, 40, 388–404. [Google Scholar] [CrossRef]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.H.; Liu, Y.G.; Ye, N.H.; Liu, R.; Zhang, J.H. Involvement of the abscisic acid catabolic gene CYP707A2 in the glucose-induced delay in seed germination and post-germination growth of Arabidopsis. Physiol. Plant. 2011, 143, 375–384. [Google Scholar] [CrossRef]
- Matakiadis, T.; Alboresi, A.; Jikumaru, Y.; Tatematsu, K.; Pichon, O.; Renou, J.P.; Kamiya, Y.; Nambara, E.; Truong, H.N. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 2009, 149, 949–960. [Google Scholar] [CrossRef]
- Patrick, A.; Alan, H.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar]

| Sample | Total Reads (%) | 3ADT and Length Filter (%) | Junk Reads (%) | Rfam (%) | Repeats (%) | Valid Reads (%) |
|---|---|---|---|---|---|---|
| PD-1 | 17,035,681 (100) | 3,287,565 (19.30) | 53,503 (0.31) | 1,425,279 (8.37) | 8550 (0.05) | 12,266,027 (72.00) |
| PD-2 | 13,530,443 (100) | 3,063,073 (22.64) | 38,381 (0.28) | 1,346,965 (9.96) | 7442 (0.06) | 9,079,240 (67.10) |
| PD-3 | 15,579,474 (100) | 3,112,538 (19.98) | 49,475 (0.32) | 1,286,262 (8.26) | 8658 (0.06) | 11,127,570 (71.42) |
| DR-1 | 14,669,850 (100) | 5,914,655 (40.32) | 18,357 (0.13) | 2,270,576 (15.48) | 45,245 (0.31) | 6,456,516 (44.01) |
| DR-2 | 11,288,606 (100) | 3,695,920 (32.74) | 27,623 (0.24) | 1,439,851 (12.75) | 16,100 (0.14) | 6,120,230 (54.22) |
| DR-3 | 13,881,039 (100) | 4,534,585 (32.67) | 36,418 (0.26) | 1,700,172 (12.25) | 15,765 (0.11) | 7,604,768 (54.79) |
| ND-1 | 14,222,034 (100) | 4,408,012 (30.99) | 27,732 (0.19) | 1,718,145 (12.08) | 21,017 (0.15) | 8,060,083 (56.67) |
| ND-2 | 12,884,793 (100) | 4,592,940 (35.65) | 18,327 (0.14) | 1,563,487 (12.13) | 26,889 (0.21) | 6,701,697 (52.01) |
| ND-3 | 12,143,228 (100) | 4,863,545 (40.05) | 25,445 (0.21) | 1,131,736 (9.32) | 11,006 (0.09) | 6,118,020 (50.38) |
| Sample | rRNA | tRNA | snoRNA | snRNA | Rfam RNA |
|---|---|---|---|---|---|
| PD-1 | 813,906 (4.78%) | 519,783 (3.05%) | 11,783 (0.07%) | 33,001 (0.19%) | 46,806 (0.27%) |
| PD-2 | 765,727 (5.66%) | 476,440 (3.52%) | 12,430 (0.09%) | 46,725 (0.35%) | 45,643 (0.34%) |
| PD-3 | 683,463 (4.39%) | 504,865 (3.24%) | 12,422 (0.08%) | 40,735 (0.26%) | 44,777 (0.29%) |
| DR-1 | 1,267,257 (8.64%) | 865,419 (5.90%) | 18,345 (0.13%) | 29,577 (0.20%) | 89,978 (0.61%) |
| DR-2 | 887,866 (7.87%) | 468,740 (4.15%) | 6274 (0.06%) | 28,979 (0.26%) | 47,992 (0.43%) |
| DR-3 | 1,091,435 (7.86%) | 501,606 (3.61%) | 7150 (0.05%) | 39,114 (0.28%) | 60,867 (0.44%) |
| ND-1 | 879,050 (6.82%) | 317,343 (2.61%) | 4989 (0.04%) | 26,628 (0.22%) | 48,241 (0.40%) |
| ND-2 | 734,535 (6.05%) | 571,427 (4.43%) | 9242 (0.07%) | 43,361 (0.34%) | 60,407 (0.47%) |
| ND-3 | 1,061,289 (7.46%) | 541,407 (3.81%) | 8030 (0.06%) | 41,243 (0.29%) | 66,176 (0.47%) |
| miRNA | miRNA Sequence | Length (nt) | CG % | dG | Level of Expression |
|---|---|---|---|---|---|
| PC-5p-12886_384 | TGGACCAATCACAGCAAGAAT | 21 | 35.40 | −101.90 | high |
| PC-3p-2448_3032 | TATTCTTGCTGTGATTGGTCC | 21 | 35.40 | −101.90 | high |
| PC-5p-29011_123 | ACGGAACGCACCTCCGAACGCACC | 24 | 61.00 | −104.10 | high |
| PC-3p-190777_6 | TGCGTTTCGGAGGTGCGTTCC | 21 | 61.00 | −104.10 | low |
| PC-5p-26456_140 | ACCCCAAATCCCCCGAGAAGC | 21 | 54.00 | −48.10 | high |
| PC-3p-212_24745 | TTTTCGGGTGATTTGAGGTGG | 21 | 54.00 | −48.10 | high |
| PC-5p-20023_207 | GGAATGGTGTGATCGGTAAAT | 21 | 44.00 | −53.30 | high |
| PC-5p-84014_23 | GTTCCCTCCGGCACTTCACC | 20 | 54.40 | −51.70 | high |
| PC-5p-80803_25 | AAGCAAAGTGAACTAGAGC | 19 | 46.30 | −68.90 | high |
| PC-3p-73564_29 | TTGTATCTCACTTGTAGTCCT | 21 | 41.90 | −79.60 | high |
| PC-3p-104391_16 | CTCAATTGTAGACTTGACC | 19 | 37.80 | −28.70 | high |
| PC-3p-95380_19 | TTGTAGACCTAACCTTGTAGA | 21 | 43.60 | −101.30 | high |
| PC-5p-54971_47 | TGATAGTGATTGATGAAGCTC | 21 | 34.30 | −31.50 | high |
| PC-5p-46859_61 | ACCTCAAATCCCCCGAAAAGC | 21 | 55.10 | −66.30 | high |
| GO Term | GO ID | Annotated | Significant | % |
|---|---|---|---|---|
| Biological Process | ||||
| Primary shoot apical meristem specification | GO:0010072 | 5 | 3 | 60 |
| Regulation of transcription, DNA-templated | GO:0006355 | 15 | 12 | 80 |
| Response to deep water | GO:0030912 | 2 | 2 | 100 |
| Stomatal complex morphogenesis | GO:0010103 | 2 | 2 | 100 |
| Leaf development | GO:0048366 | 3 | 3 | 100 |
| Polarity specification of adaxial/abaxial axis | GO:0009944 | 4 | 2 | 50 |
| Lateral root development | GO:0048527 | 2 | 2 | 100 |
| Transcription, DNA-templated | GO:0006351 | 9 | 7 | 77.8 |
| Regulation of lignin biosynthetic process | GO:1901141 | 1 | 1 | 100 |
| Meristem initiation | GO:0010014 | 3 | 1 | 33.3 |
| Regulation of RNA biosynthetic process | GO:2001141 | 1 | 1 | 100 |
| Determination of bilateral symmetry | GO:0009855 | 3 | 1 | 33.3 |
| Cellular response to freezing | GO:0071497 | 1 | 1 | 100 |
| Integument development | GO:0080060 | 3 | 1 | 33.3 |
| Response to gibberellin | GO:0009739 | 2 | 2 | 100 |
| Auxin-activated signaling pathway | GO:0009734 | 2 | 2 | 100 |
| Cellular component | ||||
| Histone deacetylase complex | GO:0000118 | 1 | 1 | 100 |
| Mitochondrion | GO:0005739 | 9 | 5 | 55.6 |
| Nuclear chromosome | GO:0000228 | 1 | 1 | 100 |
| Nucleus | GO:0005634 | 20 | 8 | 40 |
| Integral component of mitochondrial outer membrane | GO:0031307 | 1 | 1 | 100 |
| Peroxisome | GO:0005739 | 8 | 2 | 25 |
| Molecular function | ||||
| Double-stranded DNA binding | GO:0003690 | 4 | 4 | 100 |
| Sequence-specific DNA binding transcription factor activity | GO:0003700 | 9 | 7 | 77.8 |
| Sigma factor activity | GO:0016987 | 1 | 1 | 100 |
| Plastid sigma factor activity | GO:0001053 | 1 | 1 | 100 |
| Carboxypeptidase activity | GO:0004180 | 1 | 1 | 100 |
| Acetyl-CoA C-acetyltransferase activity | GO:0003985 | 1 | 1 | 100 |
| Chorismate mutase activity | GO:0004106 | 2 | 1 | 50 |
| L-ascorbate peroxidase activity | GO:0016688 | 2 | 1 | 50 |
| Methyltransferase activity | GO:0008168 | 2 | 2 | 100 |
| Calcium-transporting ATPase activity | GO:0005388 | 1 | 1 | 100 |
| Caffeoyl-CoA O-methyltransferase activity | GO:0042409 | 1 | 1 | 100 |
| DNA binding | GO:0003677 | 8 | 6 | 75 |
| Signal transducer activity | GO:0004871 | 2 | 1 | 50 |
| ATP binding | GO:0005524 | 4 | 2 | 50 |
| Oxidoreductase activity | GO:0016491 | 1 | 1 | 100 |
| miRNA Family | Target Genes | Annotation |
|---|---|---|
| zma-miR156j-3p_L-1_2ss14 | TRINITY_DN30618_c0 | Uncharacterized protein LOC108987072 |
| osa-miR156a | TRINITY_DN30823_c0 | Gibberellin 20 oxidase 1-D-like |
| cas-miR159b-3p_R+1 | TRINITY_DN42042_c1 | Chorismate mutase 3, chloroplastic |
| aly-miR159c-3p_R+1_1ss20CT | TRINITY_DN42042_c1 | Chorismate mutase 3, chloroplastic |
| aof- miR159f | TRINITY_DN46102_c0 | DELLA protein DWARF8-like |
| bdi-miR162 | TRINITY_DN41525_c2 | Plant UBX domain-containing protein 7 |
| aly-miR164a-5p | TRINITY_DN32775_c0 | NAC domain-containing protein 21/22 |
| TRINITY_DN31804_c0 | NAC domain-containing protein 21/22-like | |
| TRINITY_DN35987_c1 | NAC domain-containing protein 100-like | |
| zma-miR166m-5p | TRINITY_DN42896_c0 | Uncharacterized protein LOC103700952 |
| TRINITY_DN42749_c0 | Probable calcium-binding protein CML21 | |
| lus-miR166e | TRINITY_DN45178_c0 | Homeobox-leucine zipper protein HOX32 |
| TRINITY_DN38326_c0 | NDR1/HIN1-like protein 12 | |
| ata-miR166a-3p | TRINITY_DN45178_c0 | Homeobox-leucine zipper protein HOX32 |
| vca-miR167b-3p | TRINITY_DN27282_c0 | Non-specific lipid-transfer protein-like |
| vca-miR168a-5p | TRINITY_DN45301_c1 | Hypothetical protein POPTR_0012s03410g |
| osa-miR171d-5p | TRINITY_DN33637_c1 | -- |
| vvi-miR390 | TRINITY_DN43406_c0 | Serine/threonine-protein kinase BAM3 |
| TRINITY_DN39224_c0 | LRR receptor-like serine/threonine-protein kinase | |
| TRINITY_DN46340_c0 | - | |
| TRINITY_DN37038_c0 | LRR receptor-like serine/threonine-protein kinase | |
| osa-miR395b_L-1R+1 | TRINITY_DN43730_c0 | ATP sulfurylase 1, chloroplastic-like |
| TRINITY_DN16935_c0 | - | |
| TRINITY_DN40931_c0 | Transcriptional corepressor LEUNIG isoform X4 | |
| aof-MIR395b-p5_2ss11AG20 | TRINITY_DN46806_c1 | Calcium-transporting ATPase 8 |
| cme-miR396e_R+1 | TRINITY_DN36128_c0 | Growth-regulating factor 4-like |
| TRINITY_DN34323_c1 | Growth-regulating factor 1 | |
| TRINITY_DN45792_c0 | Growth-regulating factor 10-like isoform X2 | |
| TRINITY_DN44678_c0 | CSC1-like protein ERD4 | |
| TRINITY_DN34323_c1 | Growth-regulating factor 1-like | |
| zma-miR166l-3p | TRINITY_DN45178_c0 | Homeobox-leucine zipper protein HOX32 |
| TRINITY_DN38326_c0 | NDR1/HIN1-like protein 12 | |
| gma-miR4995 | TRINITY_DN37500_c0 | Abscisic acid 8’-hydroxylase 1-like |
| osa-miR5083_R-1 | TRINITY_DN41267_c1 | Uncharacterized protein LOC105045431 isoform |
| TRINITY_DN39322_c1 | -- | |
| TRINITY_DN44382_c0 | -- | |
| TRINITY_DN42545_c0 | Uncharacterized protein LOC103973876 isoform | |
| Novel miRNA | ||
| PC-3p-212_24745 | TRINITY_DN36128_c0 | Growth-regulating factor 4-like |
| PC-3p-2448_3032 | TRINITY_DN41146_c2 | -- |
| TRINITY_DN37385_c0 | Uncharacterized protein LOC103721212 | |
| TRINITY_DN22436_c0 | -- | |
| TRINITY_DN31474_c0 | Transcription factor vib-1 | |
| PC-5p-20023_207 | TRINITY_DN34607_c0 | Uncharacterized protein LOC103979330 isoform |
| PC-5p-12886_384 | TRINITY_DN41146_c2 | -- |
| PC-5p-29011_123 | TRINITY_DN39092_c1 | Ethylene-responsive transcription factor 4 |
| PC-5p-84014_23 | TRINITY_DN39500_c3 | Abscisic acid receptor PYR1-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Wang, F.; Chen, Y.; Niu, X.; Li, C.; Yang, X.; Li, S. Identification and Analysis of the Expression of microRNAs during the Low-Temperature Dormancy Release of Tulipa thianschanica Seeds. Agronomy 2023, 13, 3067. https://doi.org/10.3390/agronomy13123067
Zhang W, Wang F, Chen Y, Niu X, Li C, Yang X, Li S. Identification and Analysis of the Expression of microRNAs during the Low-Temperature Dormancy Release of Tulipa thianschanica Seeds. Agronomy. 2023; 13(12):3067. https://doi.org/10.3390/agronomy13123067
Chicago/Turabian StyleZhang, Wei, Feihan Wang, Yuwei Chen, Xiaorun Niu, Chaoyang Li, Xiu Yang, and Sen Li. 2023. "Identification and Analysis of the Expression of microRNAs during the Low-Temperature Dormancy Release of Tulipa thianschanica Seeds" Agronomy 13, no. 12: 3067. https://doi.org/10.3390/agronomy13123067
APA StyleZhang, W., Wang, F., Chen, Y., Niu, X., Li, C., Yang, X., & Li, S. (2023). Identification and Analysis of the Expression of microRNAs during the Low-Temperature Dormancy Release of Tulipa thianschanica Seeds. Agronomy, 13(12), 3067. https://doi.org/10.3390/agronomy13123067





