Abstract
Drought stress is one of the main environmental challenges that dramatically reduce global rice production within several agricultural ecosystems. Breeding drought-tolerant rice genotypes is an important sustainable strategy to overcome this constraint. In this work, drought tolerance levels were assessed according to biochemical, anatomical, and molecular aspects, which led to selecting three promising crosses (Sakha 107 × Sakha super 300, Sakha 107 × M206, and Sakha 107 × Sakha 108) that were compared with their parents as controls. The antioxidant capabilities of the chosen potential crosses, such as the ascorbate peroxidase activity (APX), superoxide dismutase activity (SOD), catalase activity (CAT), and total phenolics, were significantly higher compared with their parents under drought stress. Moreover, the promising selected crosses could accumulate greater proline and chlorophyll contents. The potential superiority of the three selected rice crosses was anatomically represented throughout cross-sections of roots, stems, and leaves, which recorded higher values of cross-section diameter, epidermal thickness, cortex thickness, mesophyll thickness, and bundle sheath thickness as well as a broader range of xylem vessel diameters than their parents under a water deficit. The observed superiority of the antioxidant activities in the overall drought-tolerance mechanisms and anatomical characteristics reflected their protective role in the adaptation process under water stress. Molecular analyses using inter-simple sequence repeat (ISSR) markers suggested two promising crosses (Sakha 107 × Sakha super 300 and Sakha 107 × M206) to be the most suitable crosses for saving water. They had the highest similarity values and were grouped in a distinct cluster. The relative gene expression of OsACS2, OsCML31, OsCYP94C2a, and OsSRO1c was significantly elevated in the two selected drought-tolerant rice genotypes (Sakha 107 × Sakha super 300 and Sakha 107 × M206).
1. Introduction
Rice (Oryza sativa L.) is the primary source of caloric intake for more than half of the world population [1,2]. Drought, salinity, heavy metals, cold, and high temperatures are many forms of abiotic stress that rice experiences during the growth stage [3]. Drought has mainly reduced rice yields by 25.4% over the previous two decades [4], making it one of the most severe stress factors that prevent rice grain yields to reach optimal production. Rice is a crop that is very sensitive to drought stress and notoriously consumes a large amount of irrigation water [5]. Rice’s vulnerability to drought stress has hindered efforts to minimize water use related to this semi-aquatic crop [6]. Thus, the greatest method to boost rice output is to create new drought-resistant rice genotypes with high yield potential [7].
According to the USDA’s Foreign Agricultural Services (FAS) in Cairo, Egypt’s rice output is expected to exceed 4.0 million metric tons (MMT) in 2020/2021, with cultivated land covering 451,164 hectares [8]. The amount of irrigation water accessible from the Nile River is not only limited; it is also annually declining. In addition, a shortage of water reduced the yields of 15.2% of the rice fields [9]. As a result, breeding for drought-tolerant rice lines is critical for Egypt’s rice cultivation.
Drought resistance is a multifaceted feature that is regulated by four main physiological mechanisms: escape, avoidance, tolerance, and recovery [10]. The characteristics of drought tolerance might be morphological, physiological, or yield-related. Drought-tolerant genotypes may be easily identified by testing for many drought-tolerance-related traits simultaneously [11]. One of the main goals of drought tolerance is to breed a new genotype with the best yield potential [12,13].
Rice has a relatively efficient antioxidant defense system to counter ROS-induced oxidative damage [14]. There is a greater increase in antioxidant enzyme and metabolite concentrations in tolerant cultivars than in susceptible cultivars [15]. Abscisic acid (ABA), which is transported from roots to the canopy, causes water shortages in the leaf organs [16]. Rice cultivars with varying levels of drought tolerance should possess better antioxidant defense systems and proline contents.
Water deficiency can alter anatomical traits at the level of damage and trigger a successful adaptive response [17]. Drought-exposed leaf, stem, and root tissues exhibit variations in their responses [18]. The likely contributions of leaf, stem, and root structures to rice genotypes’ drought resistance may be addressed succinctly by analyzing variations in tissue structures from distinct genotypes under normal and drought circumstances [19].
Molecular methods indirectly aid in determining complicated features without the requirement for extensive and labor-intensive phenotypic assessments by facilitating the identification of genomic regions connected to traits of interest [20]. ISSR markers are PCR-based markers that are useful in various phases of the breeding process and are very informative and cost-effective in finding genetic linkages between different rice genotypes [21,22]. ISSR markers have been employed to investigate cereal genetic diversity and evolutionary relationships [23] as well as gene tagging in molecular assisted selection [24] and DNA fingerprinting [25].
Given the above, the biochemical and anatomical reactions in the new promising rice crosses were used to investigate the degree of tolerance under drought stress. Furthermore, ISSR markers and gene expression were recruited to evaluate genetic diversity and identify new promising crosses with distinct responses to drought.
2. Materials and Methods
Rice is grown annually in Egypt during the summer season, which ranges from April until the end of October, and is distributed in the Nile Delta areas, which resemble Mediterranean climates, especially the Kafr-El-Sheikh Governorate (31.3° N 30.93° E).
The average temperatures were 28.30 °C; 28.30 °C; 31.32 °C; and 32.33 °C during the rice growth periods from May to August for 2020 and 2022, respectively.
The research was carried out at the experimental farm of the Rice Research and Training Center (RRTC) of the Sakha Agricultural Station at Kafr El-Skeikh and the Agricultural Genetic Engineering Research Institute (AGERI) of Egypt’s Agricultural Research Center (ARC) during the period from the 2020 to 2022 seasons. Line by tester system analyses were used in the 2020 season to identify superior crosses based on various vegetative and yield characteristics during the 2021 season. Moreover, a gene expression analysis was conducted during the 2022 season.
2.1. Plant Materials
Giza 177, Sakha 105, Sakha 106, Sakha 107, Sakha 108, GZ10101-5, Hispagran, Sakha super 300, and M206 were chosen for our current research to represent a broad range of genetic variation and various responses to drought. That year’s growing season yielded a total of 18 crossbreeding combinations of the genotypes described above. During the rice-growing season of 2020, the parental lines were crossed, and during the 2021 season the parents and their F1 crosses were assessed in normal and drought circumstances. These genotypes were tested for their morphological, physiological, grain yield, and other characteristics. Three novel drought-tolerant potential crosses and their parental genotypes were chosen for the research based on the outcomes of these studies. Listed below are the genotypes’ codes, names, pedigrees, and drought tolerance levels (Table 1). The Agricultural Research Center, Egypt, provided the genotypes.

Table 1.
Codes, names, drought tolerance degrees, and pedigrees of rice genotypes that were used in this study.
2.2. Drought Stress Induction
A cyclic irrigation system was imposed two weeks after transplanting every 4 and 12 d in the cases of normal irrigation and the drought condition, respectively. After 21 d, the water supply was terminated after complete flooding with 5 cm of standing water.
2.3. Biochemical Analysis
Rice genotype leaves were investigated under normal and drought conditions for biochemical alterations to better understand how drought affects plants’ biochemical processes. A pre-chilled mortar and pestle were used to grind the leaves for 20 min at 4 °C in an extraction buffer comprising a 100 mmol/L potassium phosphate buffer (pH 7.0), 0.5% Triton X-100, and 1% polyvinylpyrrolidone (PVP). Enzyme tests were performed using the centrifuged supernatant as a sample. The technique described by Nakano et al. in 1987 [26] was used to assess the APX activity. The assessment of SOD activity was performed using a previously described technique [27]. The CAT activity was measured using a standard technique [28]. The Total Soluble Phenols (TSPs) assay was conducted using a previously described method [29]. Proline content determination was performed using a related assay [30]. Finally, the total chlorophyll (Chl.) concentration was determined as mg/g fresh weight per 1 g of fresh leaves overnight at 5 °C using 5 mL of dimethylformamide, where chlorophyll a, chlorophyll b, and the carotenoid concentration were separately spectrophotometrically determined at wavelengths of 663, 645, and 470 nm [31].
2.4. Anatomical Investigation
Drought stress caused certain anatomical alterations in the structure of the rice genotypes’ leaves, stems, and roots, which were anatomically investigated. Samples were obtained from the tip of the second leaf, 0.5 cm after the second stem node, and 0.5 cm from the tip of the chosen genotypes. A series of butyl alcohols were used to kill, fix, wash, and dehydrate the specimens (56–58 °C). Sections with a thickness of 15 µ were produced using a model 820 rotary microtome, dyed with a safranin–light green combination, glued on slides, and mounted on Canada balsam [32]. Photographic micrographs were taken after microscopic examinations of the slides. Scientists used a Zeiss Axiostar plus Research Microscope that could be upgraded to a professional digital image processing system to conduct the morphometric analyses (Carl Zeiss Axiovision Product Suite DVD 30). Measurements of various cells and tissues were taken with an ocular micrometer, where the exact values were calculated with a factor derived by comparing the ocular with stage micrometers.
2.5. Molecular Analysis
These genotypes were genetically selected for their ability to cope with drought. A previously described procedure [33] was used to harvest genomic DNA from 5–6 fresh seedling leaves for each genotype. Using a spectrophotometer and ethidium bromide staining of a 0.5% agarose gel, the quality and amount of DNA were determined. A sterile TE buffer was used to dilute the stock DNA accessions to working concentrations of 10 mg/mL, which were then used in the PCR analysis. For the research, ‘IDT, Integrated DNA technologies’ produced 10 ISSR-PCR (inter-simple sequence repeat–polymerase chain reaction) primers (Table 2). It was necessary to perform the amplification reactions using the previously provided instructions [34]. For the electrophoresis of ISSR products, agarose gels containing 1.4% (w/w) agarose were immersed in 1x TBE buffer and then stained with a solution of ethidium bromide for 20 min. Genomic DNA was analyzed using Biorad’s Gel Documentation System employing a UV trans-illuminator (USA). We conducted the PCR procedures three times for each primer to ensure the repeatability and reliability of the ISSR markers. A Bio-Rad Ladder (100 pb) was run in the gel as a standard size marker to assess ISSR fragment sizes. The amplified bands were given a score of 1 if they were present and a score of 0 if they were absent. A dice coefficient was used to obtain the genetic similarity coefficient (GS) among the genotypes analyzed [35]. The cluster analysis made use of a similarity matrix. Taxonomies were created using a cluster analysis that helped to organize the observed data. As a first stage, the distances between each accession were specified by the distance metric used for that accession (dice coefficient). However, the average distance between all pairs of accessions in two separate clusters was used to compute the distance between two clusters after numerous accessions had been connected. This technique is known as the Unweight Pair Group Method with Arithmetic Mean (UPGMA) [35].

Table 2.
The ISSR primer sequences utilized to identify variation in novel drought-tolerant crosses and their parents.
2.6. Gene Expression Analysis
The relative expression of the OsACS2, OsCML31, OsCYP94C2a, and OsSRO1c genes in response to drought stress (osmotic treatments) was investigated in the three promising crosses (Sakha 107 × M206, Sakha 107 × Sakha super 300, and Sakha 108 × M206), along with a drought-sensitive check cross (Giza 177 × Hispagran). To analyze the expression pattern of these genes, the kernels of all selected rice genotypes were manually de-husked and then surface-disinfected [36] during the 2022 season. The seeds were soaked in 70% EtOH for 1 min and then washed two times with dH2O. Subsequently, the seeds were treated with a 1% solution of NaClO (sodium hypochlorite) plus a few drops of tween 20% for 30 min with gentle agitation. The seeds were then washed 5 times with sterilized H2O under a hood and allowed to air dry for 5 min on sterile fertile paper. The seeds were then sown on sterile manually made cotton beads settled in sterile glass baby food jars. The seeds were hydrated with sterilized H2O and allowed to grow until the age of 10 d. At this time, the third leaf should have been well extended. At this stage, the osmotic stress of a −0.5 MPa osmotic potential was triggered by removing the water from the jars and replacing it with 205 mM sorbitol. The control plants were supplied with new dH2O. Both the control and treated plants were incubated at room temperature (23 ± 1 °C) for 3 d until the osmotic stress symptoms started to be noted. The third leaves of at least 3–5 plants were dissected and immediately transitioned into Liquid Nitrogen (LN) and then stored at −80 °C for subsequent molecular analysis.
2.6.1. Total RNA Isolation and First-Strand cDNA Synthesis
Total RNA was isolated from control and treated rice leaves (approximately 100 mg for each sample) using Direct-zolTM RNA miniprep (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions. The quality of the isolated RNA was spectrophotometrically examined using a Nanodrop system (Thermo Fisher Scientific, Waltham, MA, USA) and visually examined using 1.5% agarose gel electrophoresis stained with ethidium bromide to a final concentration of 0.5 µg/mL. The samples were separated using a Mupid-exu electrophoresis (Mupid, Tokyo, Japan) cell unit by applying an electric current of 7 V/cm; applicably, 100 V was applied for 10 min for all casted agarose gels. Consequently, agarose gel images were obtained using a Gel Documentation System (Biorad, Hercules, CA, USA). The first-strand cDNA was synthesized using 400 ng of total RNA for all samples to normalize the expression level using a COSMO cDNA synthesis kit (Willowfort, Birmingham, UK). The synthesized cDNA was diluted at a ratio of 1 to 5 using DNase/RRNase-free water and was then used for subsequent PCR tests to measure the relative expression of several drought stress marker genes [37,38].
2.6.2. Quantifying Relative Gene Expression
The relative gene expression was quantified via a Gel express method using a conventional PCR assay [39]. The PCR test for each sample constituted (I) 10 µL of PCR master mix including Taq DNA polymerase, buffer, MgCl2, and dNTPs mix (amaR OnePCR, Genedirex, Taiwan); (II) 0.5 µL of each forward and reverse primer (10 µM each); (III) 5 µL of diluted cDNA; and (IV) 4 µL of sterilized dH2O. The thermal cycler program (T100 Thermal Cycler, Biorad, USA) was as follows: 94 °C for 3 min as an initial denaturation; 29 cycles of 94 °C for 1 min, 58 °C for 30 s, and 72 °C for 1 min; then incubation at 72 °C for 5 min as a final extension step. The reaction was stopped by incubating the samples for at least 20 min at 4 °C. Four target genes were investigated in this work in addition to the reference gene. They are listed in Table 3. As an essential part of the Gel express method, PCR amplicon products were visualized in 1.5% agarose gels. All PCR samples were typically electrophoresed under the same conditions. The agarose gels were stained with 0.5 µg/mL ethidium bromide (final concentration), the loaded PCR product volume for each sample was ideally 5 µL, and the applied current was 100V for 10 min. Image production was achieved using the Gel Doc System (Biorad, Hercules, CA, USA), meaning that each gel image was ideally produced under the same conditions for the zoom value, image enhancement, agarose gel size, and number of wells.

Table 3.
The sequences of forward and reverse primers for target and reference genes.
2.7. Statistical Analyses
The selected promising crosses and their parents were subjected to a separated randomized complete block design (RCBD) with three replications under cyclic normal irrigation (every 4 d) or water deficit (every 12 d). An analysis of variance (ANOVA) was used to test for significant differences among different drought levels (PTreatments), rice genotypes (PGenotypes), and their interaction (PTreatments × genotypes). Tukey’s honestly significant difference (HSD) test was used for post hoc analysis (PTreatments × genotypes < 0.05). On the other hand, all other trials were performed in a completely randomized design [40]. An ANOVA followed by an HSD test was used to test the significant differences among different rice genotypes (p < 0.05).
3. Results
3.1. Effect of Drought Stress on the Biochemical Characterization of Leaves of New Promising Rice (Oryza sativa L.) Genotypes and Their Parents under Normal and Drought Conditions
The activities of APX (Figure 1A), SOD (Figure 1B), CAT (Figure 1C), and TSP (Figure 1D) were significantly increased to various extents in the leaves of the promising rice genotypes under drought stress compared to the normal irrigation. The highest activity was recorded with the crosses of Sakha 107 × Sakha super 300 and Sakha 107 × M206. The results also noted that the proline accumulation varied across all genotypes. The proline quantity of the dryness-subjected plants’ leaves dramatically grew (Figure 1E). A significant enhancement in proline accumulation was noted with Sakha 107 × Sakha super 300 and Sakha 107 × M206 compared to the remaining crosses and parents. The Chl concentration was decreased in all genotypes under drought stress compared to the normal conditions. However, in the Sakha 107 × Sakha super 300 and Sakha 107 × M206 crosses, the Chl concentration was increased significantly compared with the other genotypes under drought conditions (Figure 1F).
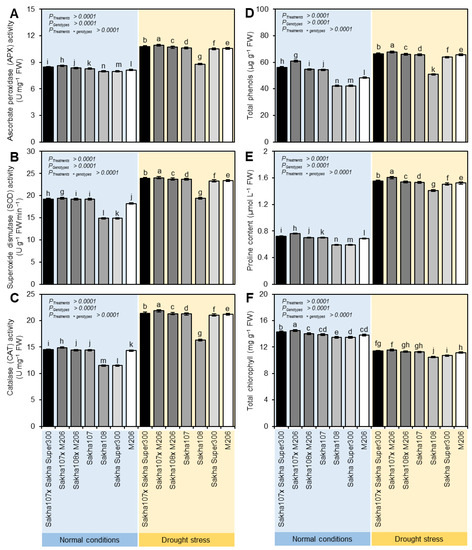
Figure 1.
Biochemical characterization of leaves of new promising rice (Oryza sativa L.) genotypes and their parents under normal and drought conditions. (A) Ascorbate peroxidase (APX) activity, (B) superoxide dismutase (SOD) activity, (C) catalase (CAT) activity, (D) total phenols, (E) proline content, and (F) total chlorophyll. Data presented are the means ± SDs of three biological replicates. Significance letters were generated based on the p-value of the interaction between treatments (as the main plots) and genotypes (as subplots) that were mentioned (p treatment × genotypes). Different letters indicate statistically significant differences among treatments according to Tukey’s honestly significant difference (HSD) test (p Treatment × genotypes ≤ 0.05), whereas the same letters indicate no statistically significant differences among them.
3.2. Effect of Drought Stress on Root Anatomical Features of New Promising Rice (Oryza sativa L.) Genotypes and Their Parents under Drought Conditions 60 d after Sowing
Figure 2 elucidates the differences in root anatomical parameters between the promising crosses and their parental lines when grown during a water shortage. The crosses Sakha 107 × Sakha super 300 and Sakha 107 × M206 showed larger root diameters (Figure 2A) compared to their parents. Anatomical parameters such as the thickness of the epidermis (μm) (Figure 2B), the thickness of the cortex (μm) (Figure 2C), and the vascular cylinder diameter (μ) (Figure 2D) were increased in the promising crosses compared to their parents. However, the xylem vessel numbers (Figure 2F) were markedly increased in the parents Sakha 107 and M206, and the xylem vessel diameters (μ) (Figure 2E) in the promising crosses were greatly increased compared to those in their parents.

Figure 2.
Root anatomical features of new promising rice (Oryza sativa L.) genotypes and their parents under drought conditions 60 d after sowing. (A) Root diameter, (B) epidermis thickness, (C) cortex thickness, (D) vascular cylinder diameter, (E) xylem vessel diameter, and (F) number of xylem vessels. Data presented are the means ± SDs of three biological replicates. Different letters indicate statistically significant differences among treatments according to Tukey’s honestly significant difference (HSD) test (p ≤ 0.05), whereas the same letters indicate no statistically significant differences among them.
3.3. Effect of Drought Stress on Stem Anatomical Features of New Promising Rice (Oryza sativa L.) Genotypes and Their Parents under Drought Conditions 60 d after Sowing
Figure 3 reflects the underlying results of stem anatomical parameter measurement during drought stress. The stem cross-sectional (Figure 3A) (µm) and xylem vessel diameters (Figure 3G) (µm) increased in the crosses Sakha 107 × M206 and Sakha 107 × Sakha super 300 compared to their parents. In addition, the epidermis thickness (Figure 3B) (µm) and cortex thickness (Figure 3C) (µm) were significantly increased in the same crosses compared to their parents. The vascular bundle number (Figure 3E) (µm) of the crosses’ stems was unchanged from that of their parents. However, a significant increase in the vascular bundle diameter was observed in the promising crosses’ stems.

Figure 3.
Stem anatomical features of new promising rice (Oryza sativa L.) genotypes and their parents under drought conditions 60 d after sowing. (A) Stem thickness, (B) epidermis thickness, (C) cortex thickness, (D) vascular bundle diameter, (E) number of vascular bundles, (F) xylem diameter, and (G) pith diameter. Data presented are the means ± SDs of three biological replicates. Different letters indicate statistically significant differences among treatments according to Tukey’s honestly significant difference (HSD) test (p ≤ 0.05), whereas the same letters indicate no statistically significant differences among them.
3.4. Effect of Drought Stress on Leaves’ Anatomical Features in New Promising Rice (Oryza sativa L.) Genotypes and Their Parents under Drought Conditions 60 d after Sowing
Figure 4 reveals the structural changes in leaves of crosses and parents under drought stress. The leaf thickness (Figure 4A) (µm) and mesophyll thickness (Figure 4B) (µm) significantly increased in the crosses Sakha 107 × M206 and Sakha 107 × Sakha super 300 compared to their parents. In terms of vascular components, the two mentioned crosses showed considerable increases in the xylem and vascular bundle diameters (Figure 4F) (µm) compared to their parents. We discovered a considerable increase in the mid-vein leaf thickness (Figure 4D) (µm) of crosses for the mid-vein parameters. Furthermore, analogously with their parents, the bundle sheath thickness (Figure 4C) (µm) was dramatically increased in the leaf mid-vein of the promising crosses.

Figure 4.
Leaf anatomical features of new promising rice (Oryza sativa L.) genotypes and their parents under drought conditions 60 d after sowing. (A) Leaf thickness, (B) mesophyll thickness, (C) bundle sheath thickness, (D) mid-vein thickness, (E) vascular bundle thickness, and (F) xylem vessel diameter. Data presented are the means ± SDs of three biological replicates. Different letters indicate statistically significant differences among treatments according to Tukey’s honestly significant difference (HSD) test (p ≤ 0.05), whereas the same letters indicate no statistically significant differences among them.
3.5. Molecular Analyses of Rice Genotypes
Genetic relationships among the genotypes (parents and crosses) with different responses to drought, namely, Sakha 107 × Sakha super 300, Sakha 107 × M206, Sakha 108 × M206, Sakha 107, Sakha 108, Sakha super 300, and M206, were determined via ISSR analysis. Ten ISSR primers were used to measure the similarity degree among the selected genotypes (Table 4 and Figure 5). The 10 primers yielded a total of 82 alleles, including 44 polymorphic and 38 monomorphic markers. Primer 3 was found to have a high polymorphism rate of 92.30%. Primers 6, 7, 9, and 10 were, inversely, found to have a low polymorphism rate of 0.00%. Polymorphism rates ranged from 42.85 to 77.77% for the other primers. In the seven chosen genotypes, the overall proportion of polymorphic markers for all primers was 53.33%, indicating modest genetic diversity. The ISSR-5 primer amplified the most ISSR loci (14 bands); however, the ISSR-3 primer had the greatest polymorphism ratio of 92.30% (Table 4).

Table 4.
The base sequence, total number of alleles, polymorphic bands, monomorphic bands, and percentage of polymorphism revealed by each of 10 primers in chosen rice genotypes that were wholly calculated.
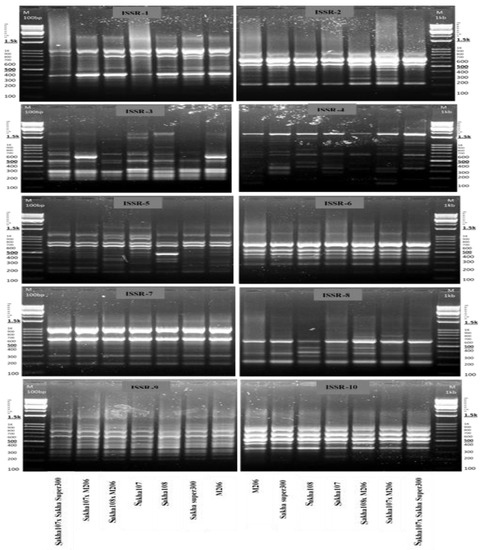
Figure 5.
Amplification profiles of 10 ISSR markers in 7 new promising rice (Oryza sativa L.) genotypes and their parents under drought conditions 60 d after sowing.
The genotypes were divided into two major groups by a distance tree demonstrating the genetic distance using a cluster analysis (Figure 6). The first major cluster had just one genotype: Sakha 108. Meanwhile, there were two sub-clusters in the second major cluster of genotypes. The first sub-cluster consisted of one genotype: Sakha 108 × M206. The second sub-cluster was divided into two groups: Sakha 107 × Sakha Super 300, Sakha 107 × M206, and Sakha 107 grouped together in the first group, and the two genotypes Sakha Super 300 and M206 grouped together in the second group. Table 5 tabulates the pair-wise genetic similarity estimates for the seven genotypes utilized herein. The similarity coefficient varied between 88.2 and 78.1%. The crosses Sakha 107 × Sakha Super 300 and Sakha 107 × M206 had the most similarities (88.2%). The cross Sakha 107 × Sakha Super 300 and its parent Sakha 107 had the second greatest similarity (88.1%), while the cross Sakha 107 × M206 and Sakha 107 had the third highest similarity (88.0%). By considering the large distances among other genotypes, these results were also substantiated by the fact that these genotypes have different origins with a high genetic variation level.
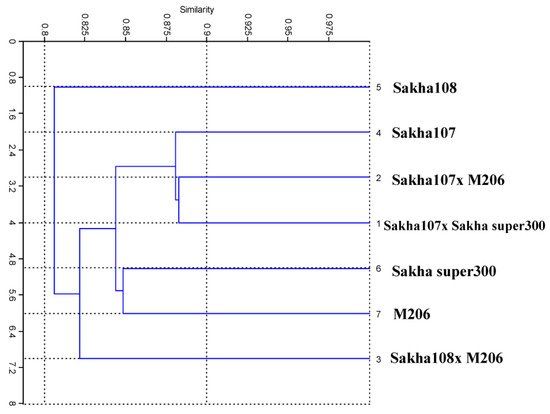
Figure 6.
UPGMA tree declaring the genetic relationships among 7 new promising rice (Oryza sativa L.) genotypes and their parents based on ISSR markers.

Table 5.
Genetic similarity of 10 ISSR markers among seven rice genotypes.
3.6. Gene Expression Changes in the Rice Genotypes due to Drought Stress
The relative expression of osACS2 (encodes 1-aminocyclopropane-1-carboxylic acid synthase, a key ethylene biosynthesis enzyme) was investigated in four rice genotypes (Giza 177 × Hispagran, Sakha 108 × M206, Sakha 107 × M206, and Sakha 107 × Sakha Super 300) under control and drought stress conditions. It was found that OsACS2 was highly expressed in response to drought stress compared to the control condition, with the Sakha 107 × M206 and Sakha 107 × Sakha Super 300 genotypes showing 3.4- and 2.7-fold increases, respectively (Figure 7A,C). Furthermore, both rice genotypes showed a higher level of expression of OsACS2, averaging a 7.25-fold increase compared to the genotypes Giza 177 × Hispagran and Sakha 108 × M206. The OsCML31 gene encodes the calcium signaling protein calmodulin-related sensor protein. As portrayed in Figure 7B,D, the two rice genotypes Sakha 107 × M206 and Sakha 107 ×Sakha super 300 significantly elevated their expression of CML31 compared to control conditions. The stressed genotype Sakha 107 × Sakha super 300 elevated OsCML31 expression to approximately 21 times higher than in the control plants, while the genotype Sakha 107 × M206, under stress conditions, could increase OsCML31 expression by nearly 130-fold compared with the control condition. Both rice genotypes could express the OsCML31 gene more than the other two genotypes (Giza 177 × Hispagran and Sakha 108 × M206), with an average increase of 6.2-fold.

Figure 7.
Quantifying gene expression profile of OsACS2 (encodes 1-aminocyclopropane-1-carboxylic acid synthase, a key ethylene biosynthesis enzyme) and OsCML31 (encodes Calmodulin-related calcium sensor protein) in response to osmotic stress in 4 new genetically different Egyptian rice lines. (A,B) Agarose gel electrophoresis shows the migration of PCR products representing OsACS2 and OsCML31 genes under control and salinity stress, respectively. (C,D) represent the quantification of relative OsACS2 and OsCML31 gene expression depending on image analysis of (A) and (B), respectively. M indicates 100 bp DNA ladder. The lowercase letters in (C,D) indicate statistically significant differences at p < 0.05.
The expression of OsCYP94C2a (Cytochrome P450 of the subfamily CYP94 subclade C member 2a) was also quantified. Like the previous findings of OsACS2 and OsCML31, both the Sakha 107 × M206 (2.2-fold) and Sakha 107 × Sakha super 300 (5.1-fold) genotypes increased the expression of OsCYP94C2a in response to drought stress compared to the control condition (Figure 8A,C). In addition, the expression of OsCYP94C2a in the Sakha 107 × M206 and Sakha 107 × Sakha super 300 genotypes was the highest under drought stress, with a 5.7-fold average increase compared to Giza 177 × Hispagran and Sakha 108 × M206 (Figure 8A,C). The expression of OsSRO1c (encodes Similar to Radical-induced cell death One protein) was also quantified and was found to be significantly up-regulated in all investigated genotypes in response to drought stress (Figure 8B,D). Nevertheless, the expression of OsSRO1c was drastically higher in drought-stressed Sakha 107 × M206 and Sakha 107 × Sakha super 300 plants compared to the Giza 177 × Hispagran and Sakha 108 × M206 genotypes relative to the corresponding control ones. Furthermore, the former two genotypes expressed the gene OsSRO1c significantly more than the last two genotypes, with an average increase of 4-fold.

Figure 8.
Quantifying gene expression profile of OsCYP94C2a (Cytochrome P450 of the subfamily CYP94 subclade C member 2a) and OsSRO1c (encodes Similar to Radical-induced cell death One protein) in response to osmotic stress in 4 new genetically different Egyptian rice lines. In (A) and (B), agarose gel electrophoresis shows the migration of PCR products representing OsCYP94C2a and OsSRO1c genes under control and salinity stress, respectively. (C,D) represent the quantification of relative OsACS2 and OsCML31 gene expression depending on image analysis of (A) and (B), respectively. M indicates 100 bp DNA ladder. The lowercase letters in (C,D) indicate statistically significant differences at p < 0.05.
The mean performances of three chosen superior crosses for some yields and their components compared with their parental lines are presented in Table 6. The line Sakha 108 and tester M206, as the parental genotypes, as well as their cross combinations Sakha 108 × M206 and Sakha 107 × M206 had superior values for panicles/plant numbers under both normal and water stress conditions.

Table 6.
Mean performances for some yields and their components of three chosen superior crosses and their parents under normal and drought stress conditions in 2022 growing season.
With respect to the 1000-grain weight, the parent M206 recorded the highest mean value (29.63 g) under the normal irrigation system, whilst the Sakha 107 rice cultivar reached the peak value (28.64 g) under the water stress condition. However, the peak mean value for the 1000-grain weight was recorded with the cross of Sakha 108 × M206 under the normal irrigation system (33.35 g), while the highest 1000-grain weight value was recorded with the cross of Sakha 107 × M206 (31.59 g) under water stress conditions. Regarding the grain yield/plant, both Sakha 108 and M206 significantly maximized the mean and par values (47.96 and 47.64 g/plant), followed by Sakha super 300 (45.06 g/plant) and Sakha 107 (44.56 g/plant), under the normal irrigation system. Meanwhile, Sakha 107 showed remarkable superiority (41.54 g/plant), followed by the rest of the parental cultivars, which significantly recorded the par values 39.84, 39.54, and 39.35 g/plant with the M206, Sakha super 300, and Sakha 108 rice cultivars under water stress conditions, respectively. On the other hand, the chosen cross Sakha 108 × M206 recorded the peak mean value for grain yield/plant (55.63 g/plant), followed by the second chosen cross Sakha 107 × M206 (55.21 g/plant), under the normal irrigation system. Under the drought stress condition, the opposite arrangement of the chosen crosses occurred and the cross Sakha 107 × M206 recorded the highest mean grain yield value (48.06 g/plant), followed by the two remaining selected crosses (46.91 and 46.84 g/plant, respectively).
4. Discussion
Rice, which requires a lot of water for growth and development [41], has a significant challenge during a water shortage. As a result, one of the most significant environmental variables restricting plant growth and development is drought [42]. The lack of knowledge of the genetics and inheritance of drought tolerance characteristics, alongside a full misunderstanding of physiological drought tolerance features and drought processes, is a huge setback in drought tolerance breeding. Selecting secondary features that contribute to drought tolerance in breeding programs may also boost the production and roots in water-limited conditions. To find the true potential of drought-resistant genotypes, we combined the findings of phenotypic, physiological, and grain yield research with genomic data and genetic diversity studies.
When plants are subjected to drought, their generation of reactive oxygen species (ROS) rises, resulting in lipid peroxidation, protein denaturation, DNA mutation, and cellular oxidative damage [43]. Induced antioxidant enzyme activities in plants are a natural way for plants to combat oxidative damage in a hostile environment [44]. Herein, we measured antioxidant enzyme activities. In a drought setting, APX, SOD, and CAT exhibited a greater degree of induction. The level of TSP was similarly increased in the promising crosses. Antioxidant enzymes and phenols are critical for scavenging H2O2 toxicity. The joint action of CAT and SOD turns the lethal superoxide radical (O2) and hydrogen H2O2 into the water and molecular oxygen under unfavorable conditions such as drought stress (O2) [45,46].
Additionally, the drought stress dramatically increased the content of proline in rice crosses’ leaves. These findings indicated that plants’ production of these osmotic modifications is a widespread response to drought. Osmotic adjustment via buildup of a cellular solute, such as proline, has been proposed as one of the probable ways to overcome osmotic stress produced by a decrease in cellular water [47]. Proline is a non-protein amino acid that develops in most tissues when they are exposed to water stress and is quickly reduced when a drought is terminated [48]. Proline is important for maintaining cell membrane integrity, the stability of enzymes, and proteins [49].
The observed changes in Chl agreed well with a previous study [50], which revealed that the synthesis of photosynthetic pigments being lost or reduced under drought stress is a common avenue and is directly associated with plant biomass and yield reduction. The two crosses Sakha 107 × Sakha super 300 and Sakha 107 × M206 had the greatest Chl among all genotypes when drought stress occurred. According to these results, drought stress reduced the chlorophyll variability in rice leaves in possibly tolerant genotypes [51].
Anatomical root, stem, and leaf alterations are required to explain the differences between parents and crosses under water stress, and therefore, to comprehend the processes utilized by promising crosses to cope with drought. The obtained results indicated that the root and xylem vessel diameters significantly increased in promising crosses. These findings fit well with those of Al-Khalifah et al. (2006) [52], who illustrated a link between the xylem channel diameter and water conductivity maintenance. Drought-tolerant genotypes may grow bigger xylem vessels and roots with greater diameters only on rare occasions to enhance water intake when it becomes available. The thickness of the epidermis (μ), cortex (μ), and vascular cylinder (μ) were all enhanced compared with their parents. Recently, the root cortex thickness has been shown to be decreased in sensitive rice cultivars exposed to water deprivation [53]. In the promising crosses subjected to water shortage, the cross-sectional stem area, pith, and xylem vessel diameter all increased. The epidermis, cortex thickness, and number of vascular bundles were also significantly increased in the crosses compared to the parents. Most leaf anatomical structures are strongly impacted by water deprivation in plants [54]. Since leaves are the principal organs of internal water removal, drought-tolerant rice genotypes perform leaf anatomical modifications to conserve water. Similarly, it was discovered that the mid-vein thickness of leaves, mesophyll, and bundle sheaths for the promising crosses significantly increased compared with the parents’ values. Overall, from the anatomical point of view, stems were more resistant to water shortage than roots and leaves. However, the rice cultivars with greater stem areas and higher stem xylem diameters maintained high leaf water potential under water restriction [55]. There was a positive link between the total stem and leaf xylem areas of drought-stressed rice [56]. All these qualities gave the morphological foundation for drought tolerance in promising rice genotypes via the capacity to modify their root, stem, and leaf structures. Consequently, they may live and develop in water-stressed circumstances.
For drought improvement, several studies have relied on phenotypic selection. As a result, developments in DNA molecular markers and their capacity to identify genomic areas linked with significant features would be more beneficial (e.g., drought tolerance). ISSR markers are useful because they offer a rapid, accurate, and comprehensive method that may also be utilized to develop genetic and genomic fingerprinting [57]. ISSR markers were employed herein to measure the genetic diversity and to identify the prospective crosses by comparing these parents at the molecular level.
The obtained results indicated that molecular polymorphisms were detected among the selected genotypes using 10 ISSR markers. Six of the ten tested primers showed polymorphisms for genotypes. A total of 44 amplified polymorphic fragments were detected, and the highest numbers of alleles were detected with the primers ISSR 6, 10, 9, 18, 3, and 5, with 12, 10, 9, 7, 3, and 3 alleles, respectively. These polymorphic bands were used to determine the drought tolerance in rice genotypes. The clustering pattern and similarity index indicated that there was a close relationship between Sakha 107 × Sakha super 300 and Sakha 107 × M206, mostly because they were developed from the tolerant parent (Sakha 107). On this premise, the crosses Sakha 107 × Sakha super 300 and Sakha 107 × M206 are proposed as the most acceptable crosses for drought tolerance, which have the greatest similarity value and clustered in a unique cluster.
These findings also suggest that ISSR markers may be a superior tool for studying drought tolerance. A relationship between GA repeats and rice variety abiotic stress resistance (salinity, drought, and flood) was previously claimed [58]. The results indicate that ISSR markers linked to GA 8 YG may be used to identify genes/new alleles associated with the three abiotic stresses in rice germplasm and can be used to differentiate three groups of stress-tolerant genotypes. ISSR markers based on AG and GA repeats were also employed to differentiate geographically different Oryza nivara accessions [59]. As previously reported for rice and tomato [58,59], the primer (GATA) might assist in recognizing all kinds and is acceptable for fingerprinting.
Drought tolerance is a multiplex plant trait with broad integrated molecular responses, which mainly comprise two main steps: (i) stress sensing/signaling and (ii) promoting molecular, physiological, and phonological adaptive changes [60]. The fine-tuning of hormone homeostasis is crucial for creating an adaptive and efficient link among different molecular responses to drought stress in a spatio-temporal manner [61]. Ethylene (C2H2) is a well-acknowledged gaseous plant stress hormone that contributes to plant development and the response to drought, flooding, low temperature, and salinity stress [62]. Under drought stress mimicked by osmotic stress, the expression of OsACS2 (encodes 1-aminocyclopropane-1-carboxylic acid synthase, a key ethylene biosynthesis enzyme) was significantly elevated in the two most drought-tolerant rice genotypes (Sakha 107 × M206 and Sakha 107 × Sakha super 300). Ethylene was reported to be associated with enhancing plant antioxidative machinery under stress. The two drought-tolerant rice genotypes obviously accumulated higher amounts of proline [63]. Furthermore, the ethylene-responsive factor OsWR1 was induced in transgenic rice lines and enhanced drought tolerance by increasing wax production in rice [64].
In this study, the contribution of calcium signaling in rice under drought stress was noted. Like OsACS2, the most drought-sensitive genotypes showed the highest level of OsCML31 gene expression. OsCML31 was reported to be significantly responsive under drought, salt, and alkalinity stress in rice [36]. Calcium ions are crucial second messengers in plants and are essential for efficient stress sensing and signal transduction, the process that eventually could trigger an adaptive response to drought stress in terms of early stomatal closure and H2O2 production [65,66]. This assumption could be supported by the significant up-regulation of OsSRO1c (related to stomatal closure) in the two drought-tolerant crosses Sakha 107 × M206 and Sakha 107 × Sakha super 300 in response to osmotic stress compared to other rice genotypes. OsROC1c is a rice homologue of SRO (similar to RCD one), which was identified as a direct target gene of SNAC1 (stress-responsive NAC 1), which is involved in the regulation of the stomatal aperture and the oxidative response [67,68].
Finally, the catabolic turnover of jasmonic acid was found to be associated with rice’s tolerance to abiotic stress [37]. OsCYP94C2a, which encodes the jasmonic-acid-degrading enzyme Cytochrome P450 of the subfamily CYP94 subclade C member 2a, was highly expressed in rice under osmotic and salt stress in the Egyptian rice genotype Sakha 101 [38]. In this study, OsCYP94C2a was strongly expressed in the most drought-sensitive rice genotypes. Jasmonic acid (JA) is a growth-inhibiting stress hormone associated with retarded root and shoot growth [68]. It is believed that the regulation of JA biosynthesis/turnover is essential in setting up the balance between growth and adaptation in plants under abiotic stress. Yang et al. (2012) [69] reported that jasmonic acid could interfere with the gibberellic acid signaling cascade and inhibit growth in favor of defense by re-allocating carbon energy towards adaptation or resistance mechanisms. It could be assumed that drought-tolerant genotypes could efficiently degrade jasmonic acid using the enzyme CYP94C2A more than other rice genotypes under drought stress [70].
It could be concluded that crossing genotypes with varying degrees of drought tolerance, as shown in this study, may have resulted in valuable transgressive segregates with improved drought resistance. The results have significant implications for rice breeding, particularly in terms of choosing drought-tolerant genotypes at the molecular level in the lab and facilitating drought breeding programs. Drought stress has become a severe threat to food security in the developing world as well as in Egypt. Although water is required during all growth periods of the rice plant, there are some critical growth stages when drought stress has a serious impact and creates massive reductions in the quantities of yields.
Rice’s responses to drought stress and its tolerance level can be measured by monitoring different anatomical, biochemical, and molecular changes during a drought period. The responses of plants to drought are complex, and different morphological and biochemical mechanisms are involved within plants during drought.
These mechanisms can occur via different avenues. One avenue was the genetic background of the parental lines that were used in the crosses to develop new combinations that were more tolerant to abiotic stress, especially water stress. According to the yield trails, the parents Sakha 108 and M206 registered higher numbers for panicles/plant, 1000-grain weight, and grain yield/plant under normal irrigation, while under drought stress the parents Sakha 107 and M206 recorded the highest values for all yield attributes. The cross of Sakha 108 × M206 occupied the first position for all studied yield trails under normal conditions. Similarly, under stress, the cross combinations Sakha 107 × M206 and Sakha 107 × Sakha super 300 ranked first and second for all yield attributes, respectively. With regards to the annual average temperature fluctuation during the life span of rice plants, it increased by 2 0, as the temperature was higher in 2022 than in the previous season, which resulted in earlier flowering and led to a greater yield. Secondly, the selected superior crosses showed remarkable evidence for their anatomical, biochemical, and molecular advanced differentiation and tended to be more tolerant to drought stress. Thirdly, drought is escaped by rapid development, which allows plants to finish their life span before severe water stress. Fourthly, drought is avoided by increasing water uptake and reducing the transpiration rate, stomatal conductance, and leaf area. Fifthly, drought tolerance is increased by maintaining tissue turgor during water stress via osmotic adjustment, allowing plants to maintain growth under water stress. The biosynthesis of the most famous amino acid (proline, which is related to the enzyme pyrroline-5-carboxylate synthetase) as an osmo-protectant indicates water stress.
5. Conclusions
From this study, it could be concluded that the crosses Sakha 107 × Sakha super 300 and Sakha 107 × M206 were tolerant of drought stress based on the positive responses of the antioxidant enzymes. Furthermore, the various structural factors may have aided drought tolerance, allowing the plant to alter its root, stem, and leaf structures, which were identified as highly significant outcomes for drought tolerance in the Egyptian rice genotypes. The genetic analysis using ISSR markers, gene expression, and polymorphic bands identified genetic markers that might be used to examine the speed of drought-tolerant genotypes that can be detected using a molecular laboratory. These crosses could be used in a breeding program to obtain a new promising hybrid that is more applicable during a water shortage.
Author Contributions
Conceptualization, M.I.A.-Y., A.B.E., A.A.E.-G., M.E.E.D., M.S.A.E., M.H., Y.N., A.E.-D.O. and M.S.A.E.; methodology, M.I.A.-Y., A.B.E., A.A.E.-G., M.E.E.D. and I.A.T.; software, M.I.A.-Y., A.B.E., A.A.E.-G., M.E.E.D. and W.H.E.-K.; validation, M.I.A.-Y., M.E., A.B.E., A.E.-D.O. and M.S.A.E.; formal analysis, A.B.E., I.A.T., M.H. and W.H.E.-K.; investigation, M.I.A.-Y., M.E., A.B.E., A.A.E.-G., M.S.A.E., Y.N. and A.E.-D.O.; resources, M.I.A.-Y., A.B.E., I.A.T. and W.H.E.-K.; data curation, A.B.E.; writing—original draft preparation, M.I.A.-Y. and A.B.E.; writing—review and editing, M.I.A.-Y., A.B.E. and A.E.-D.O.; visualization, M.I.A.-Y., A.B.E., M.H., Y.N. and A.E.-D.O.; supervision, M.I.A.-Y.; project administration, M.I.A.-Y. and A.B.E.; funding acquisition, M.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
This study did not report any data.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through the Large Groups Project under grant RGP2/164/44. All the authors are grateful for the support provided by the Faculty of Agriculture, Tanta University, Egypt, and the Rice Research Department, Field Crops Research Institute, Agricultural Research Center, Giza, Egypt.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhao, M.; Lin, Y.; Chen, H. Improving nutritional quality of rice for human health. Theor. Appl. Genet. 2020, 133, 1397–1413. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, N.K.; Ziska, L.H. Rice: Importance for global nutrition. J. Nutr. Sci. Vitaminol. 2019, 65, S2–S3. [Google Scholar] [CrossRef] [PubMed]
- Lafitte, H.R.; Ismail, A.; Bennett, J. Abiotic stress tolerance in rice for Asia: Progress and the future. In New Directions for a Diverse Planet, Proceedings of the 4th International Crop Science Congress, Brisbane, Australia, 26 September–1 October 2004; Fischer, T., Turner, N., Angus, J., McIntyre, L., Robertson, M., Borrell, A., Lloyd, D., Eds.; Crop Science Society of America: Madison, WI, USA. [Google Scholar]
- Zhang, J.; Zhang, S.; Cheng, M.; Jiang, H.; Zhang, X.; Peng, C.; Jin, J. Effect of drought on agronomic traits of rice and wheat: A meta-analysis. Int. J. Environ. Res. Public Health 2018, 15, 839. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Suenaga, K. Toward the genetic improvement of drought tolerance in crops. Jpn. Agric. Res. Q. JARQ 2017, 51, 1–10. [Google Scholar] [CrossRef][Green Version]
- Wassmann, R.; Jagadish SV, K.; Heuer, S.; Ismail, A.; Redona, E.; Serraj, R.; Sumfleth, K. Climate change affecting rice production: The physiological and agronomic basis for possible adaptation strategies. Adv. Agron. 2009, 101, 59–122. [Google Scholar]
- Rajiv, S.; Thivendran, P.; Deivanai, S. Genetic divergence of rice on some morphological and physiochemical responses to drought stress. Pertinaka J. 2010, 32, 315–328. [Google Scholar]
- FAS Foreign Agricultural Service. Grain and Feed Annual in Egypt. Report No: EG2020-0005; 2020. Available online: https://apps.fas.usda.gov/newgainapi/api/Report/DownloadReportByFileName?fileName=Grain%20and%20Feed%20Annual_Cairo_Egypt_EG2023-0003.pdf (accessed on 12 August 2023).
- Alzohairy, A.M.; Youssef, M.A.; Solliman, S.S. Molecular markers for new promising drought tolerant lines of rice under drought stress via RAPD-PCR and ISSR markers. J. Am. Sci. 2010, 6, 12. [Google Scholar]
- Nguyen, D.T.; Nguyen, T.K.L.; Pham, Q.C.; Nguyen, T.H.; Tran, Q.T.; Dao, X.H.; Nguyen, H.T. Mapping QTLs associated with root traits Related to drought resistance in Vietnamese upland rice. In Resilient Crops for Water Limited Environments: Proceedings of a Workshop Held at Cuernavaca Mexico; CIMMYT: Mexico City, Mexico, 2004; p. 234. [Google Scholar]
- Ghazy, M.I.; Salem, K.F.; Sallam, A. Utilization of genetic diversity and marker-trait to improve drought tolerance in rice (Oryza sativa L.). Mol. Biol. Rep. 2021, 48, 157–170. [Google Scholar] [CrossRef]
- Gewaily, E.E.; Hamad, H.S.; Mikhael, B.B.; Arafat, E.F. Performance of promising hybrid rice genotypes under different irrigation intervals. Menoufia J. Plant Prod. 2021, 6, 19–33. [Google Scholar] [CrossRef]
- Adhikari, M.; Adhikari, N.R.; Sharma, S.; Gairhe, J.; Bhandari, R.R.; Paudel, S. Evaluation of drought tolerant rice cultivars using drought tolerant indices under water stress and irrigated condition. Am. J. Clim. Change 2019, 8, 228. [Google Scholar] [CrossRef]
- Anjum, S.A.; Ashraf, U.; Tanveer, M.; Khan, I.; Hussain, S.; Shahzad, B.; Wang, L.C. Drought induced changes in growth, osmolyte accumulation and antioxidant metabolism of three maize hybrids. Front. Plant Sci. 2017, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Bosabalidis, A.M.; Kofidis, G. Comparative effects of drought stress on leaf anatomy of two olive cultivars. Plant Sci. 2002, 163, 375–379. [Google Scholar] [CrossRef]
- Olmos, E.; Sánchez-Blanco, M.J.; Ferrández, T.; Alarcon, J.J. Subcellular effects of drought stress in Rosmarinus officinalis. Plant Biol. 2007, 9, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Hazman, M.; Brown, K.M. Progressive drought alters architectural and anatomical traits of rice roots. Rice 2018, 11, 62. [Google Scholar] [CrossRef]
- Davierwala, A.P.; Chowdari, K.V.; Kumar, S.; Reddy AP, K.; Ranjekar, P.K.; Gupta, V.S. Use of three different marker systems to estimate genetic diversity of Indian elite rice varieties. Genetica 2000, 108, 269–284. [Google Scholar] [CrossRef]
- Pradeep Reddy, M.; Sarla, N.; Siddiq, E.A. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Sarla, N.; Bobba, S.; Siddiq, E.A. ISSR and SSR markers based on AG and GA repeats delineate geographically diverse Oryza nivara accessions and reveal rare alleles. Curr. Sci. 2003, 84, 683–690. [Google Scholar]
- Matos, M.; Pinto-Carnide, O.; Benito, C. Phylogenetic relationships among Portuguese rye based on isozyme, RAPD and ISSR markers. Hereditas 2001, 134, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.; Saini, N.; Jain, S.; Rana, P.; Singh, R.K.; Jain, R.K. Genetic analysis of a CSR10 (indica)× Taraori Basmati F3 population segregating for salt tolerance using ISSR markers. Euphytica 2003, 134, 231–238. [Google Scholar] [CrossRef]
- Carvalho, A.; Matos, M.; Lima-Brito, J.; Guedes-Pinto, H.; Benito, C. DNA fingerprint of F1 interspecific hybrids from the Triticeae tribe using ISSRs. Euphytica 2005, 143, 93–99. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Purification of ascorbate peroxidase in spinach chloroplasts; its inactivation in ascorbate-depleted medium and reactivation by monodehydroascorbate radical. Plant Cell Physiol. 1987, 28, 131–140. [Google Scholar]
- Gupta, A.S.; Heinen, J.L.; Holaday, A.S.; Burke, J.J.; Allen, R.D. Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA 1993, 90, 1629–1633. [Google Scholar] [CrossRef] [PubMed]
- Aebi, H. Catalase in vitro. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Zieslin, N.; Ben Zaken, R. Peroxidase activity and presence of phenolic substances in peduncles of rose flowers. Plant Physiol. Biochem. 1993, 31, 333–339. [Google Scholar]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Bhushan, D.; Pandey, A.; Choudhary, M.K.; Datta, A.; Chakraborty, S.; Chakraborty, N. Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol. Cell. Proteom. 2007, 6, 1868–1884. [Google Scholar] [CrossRef]
- Sutikno. Practical Guidience: Plant Microtechnics (BIO 30603); Faculty of Biology—Universitas Gadjah Mada: Yogyakarta, Indonesia, 2018. [Google Scholar]
- Couch, S.R.; Kochert, G.; Yu, Z.H.; Wang, Z.Y.; Khush, G.S.; Coffman, W.R.; Tanksley, S.D. Molecular mapping of rice chromosomes. Theor. Appl. Genet. 1988, 76, 815–829. [Google Scholar]
- Al-Turki, T.A.; Basahi, M.A. Assessment of ISSR based molecular genetic diversity of Hassawi rice in Saudi Arabia. Saudi J. Biol. Sci. 2015, 22, 591–599. [Google Scholar] [CrossRef]
- Sneath, P.H.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; W. H. Freeman: San Francisco, CA, USA, 1973. [Google Scholar]
- Hazman, M.; Hause, B.; Eiche, E.; Riemann, M.; Nick, P. Different Forms of Osmotic Stress Evoke Qualitatively Different Responses in Rice. J. Plant Physiol. 2016, 202, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Hazman, M.; Sühnel, M.; Schäfer, S.; Zumsteg, J.; Lesot, A.; Beltran, F.; Marquis, V.; Herrgott, L.; Miesch, L.; Riemann, M.; et al. Characterization of Jasmonoyl-Isoleucine (JA-Ile) Hormonal Catabolic Pathways in Rice upon Wounding and Salt Stress. Rice 2019, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Hazman, M.Y.; Mohamed, N.A.; Diab, N.E. Drought and salinity alter adaptive molecular response in two genetically unlike Egyptian rice cultivars. Egypt J. Exp. Biol. 2019, 15, 283–294. [Google Scholar] [CrossRef]
- Hazman, M. Gel express: A novel frugal method quantifies gene relative expression in conventional RT-PCR. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 11. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Tukey’s honestly significant difference (HSD) test. In Encyclopedia of Research Design; Sage: Thousand Oaks, CA, USA, 2010; Volume 3, pp. 1–5. [Google Scholar]
- Smirnoff, N. Tansley Review No. 52. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol. 1993, 125, 27–58. [Google Scholar] [CrossRef] [PubMed]
- Luna, C.M.; Pastori, G.M.; Driscoll, S.; Groten, K.; Bernard, S.; Foyer, C.H. Drought controls on H2O2 accumulation, catalase (CAT) activity and CAT gene expression in wheat. J. Exp. Bot. 2005, 56, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Chaitanya, K.V.; Sundar, D.; Masilamani, S.; Ramachandra Reddy, A. Variation in heat stress-induced antioxidant enzyme activities among three mulberry cultivars. Plant Growth Regul. 2002, 36, 175–180. [Google Scholar] [CrossRef]
- Noctor, G.; Veljovic-Jovanovic, S.; Foyer, C.H. Peroxide processing in photosynthesis: Antioxidant coupling and redox signalling. Philosophical Transactions of the Royal Society of London. Ser. B Biol. Sci. 2000, 355, 1465–1475. [Google Scholar] [CrossRef]
- Ibarra-caballero, J.O.R.G.E.; Villanueva-verduzco, C.; Molina-Galan, J.; Sanchez-De-Jimenez, E. Proline accumulation as a symptom of drought stress in maize: A tissue differentiation requirement. J. Exp. Bot. 1988, 39, 889–897. [Google Scholar] [CrossRef]
- Singh, D.K.; Sale, P.W.; Pallaghy, C.K.; Singh, V. Role of proline and leaf expansion rate in the recovery of stressed white clover leaves with increased phosphorus concentration. New Phytol. 2000, 146, 261–269. [Google Scholar] [CrossRef]
- Kishor, P.K.; Sangam, S.; Amrutha, R.N.; Laxmi, P.S.; Naidu, K.R.; Rao, K.S.; Sreenivasulu, N. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: Its implications in plant growth and abiotic stress tolerance. Curr. Sci. 2005, 88, 424–438. [Google Scholar]
- Pandey, V.; Shukla, A. Acclimation and tolerance strategies of rice under drought stress. Rice Sci. 2015, 22, 147–161. [Google Scholar] [CrossRef]
- Ghidan, W.; Khedr, R.A. Assessment of Some Agro-Physiological Traits and Genetic Markers in Rice (Oryza sativa L.) Under Normal and Water Stress Conditions. J. Plant Prod. 2021, 12, 73–86. [Google Scholar] [CrossRef]
- Makbul, S.; Güler, N.S.; Durmuş, N.; Güven, S. Changes in anatomical and physiological parameters of soybean under drought stress. Turk. J. Bot. 2011, 35, 369–377. [Google Scholar] [CrossRef]
- Pena-Valdivia, C.B.; Sanchez-Urdaneta, A.B. Effects of substrate water potential in root growth of Agave salmiana Otto ex Salm-Dyck seedlings. Biol. Res. 2009, 42, 239–248. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Al-Khalifah, N.S.; Khan, P.R.; Al-Abdulkader, A.M.; Nasroun, T. Impact of water stress on the sapwood anatomy and functional morphology of Calligonum comosum. IAWA J. 2006, 27, 299–312. [Google Scholar] [CrossRef]
- Sibounheuang, V.; Basnayake, J.; Fukai, S. Genotypic consistency in the expression of leaf water potential in rice (Oryza sativa L.). Field Crops Res. 2006, 97, 142–154. [Google Scholar] [CrossRef]
- Ouyang, W.; Yin, X.; Yang, J.; Struik, P.C. Do shoot anatomical characteristics allow rice to grow well under water deficit? J. Agron. Crop Sci. 2021, 208, 763–776. [Google Scholar] [CrossRef]
- Fernandez, M.; Figueiras, A.; Benito, C. The use of ISSR and RAPD markers for detecting DNA polymorphism, genotype identification and genetic diversity among barley cultivars with known origin. Theor. Appl. Genet. 2002, 104, 845–851. [Google Scholar] [CrossRef]
- Reddy, C.S.; Babu, A.P.; Swamy, B.P.; Kaladhar, K.; Sarla, N. ISSR markers based on GA and AG repeats reveal genetic relationship among rice varieties tolerant to drought, flood, or salinity. J. Zhejiang Univ. Sci. B 2009, 10, 133–141. [Google Scholar] [CrossRef]
- Davierwala, A.P.; Ramakrishna, W.; Chowdari, V.; Ranjekar, P.K.; Gupta, V.S. Potential of (GATA) n microsatellites from rice for inter-and intra-specific variability studies. BMC Evol. Biol. 2001, 1, 7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rao, R.; Corrado, G.; Bianchi, M.; Di Mauro, A. (GATA) 4 DNA fingerprinting identifies morphologically characterized ‘San Marzano’ tomato plants. Plant Breed. 2006, 125, 173–176. [Google Scholar] [CrossRef]
- Razi, K.; Muneer, S. Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit. Rev. Biotechnol. 2021, 41, 669–691. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gao, X.; Chen, S.; Li, D.; Chen, S.; Xie, M.; Yang, G. Genome-wide identification and expression analysis of ethylene responsive factor family transcription factors in Juglans regia. PeerJ 2021, 9, e12429. [Google Scholar] [CrossRef] [PubMed]
- Husain, T.; Fatima, A.; Suhel, M.; Singh, S.; Sharma, A.; Prasad, S.M.; Singh, V.P. A Brief Appraisal of Ethylene Signaling under Abiotic Stress in Plants. Plant Signal. Behav. 2020, 15, 1782051. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wan, L.; Zhang, L.; Zhang, Z.; Zhang, H.; Quan, R.; Huang, R. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 2012, 78, 275–288. [Google Scholar] [CrossRef]
- Han, Y.; Wang, Y.; Zhai, Y.; Wen, Z.; Liu, J.; Xi, C.; Han, S. OsOSCA1. 1 Mediates Hyperosmolality and Salt Stress Sensing in Oryza sativa. Biology 2022, 11, 678. [Google Scholar] [CrossRef]
- Xu, T.; Niu, J.; Jiang, Z. Sensing Mechanisms: Calcium Signaling Mediated Abiotic Stress in Plants. Front. Plant Sci. 2022, 13, 925863. [Google Scholar] [CrossRef]
- You, J.; Zong, W.; Li, X.; Ning, J.; Hu, H.; Li, X.; Xiong, L. The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 2013, 64, 569–583. [Google Scholar] [CrossRef]
- Hazman, M.; Fawzy, S.; Hamdy, A.; Khaled, A.; Mahmoud, A.; Khalid, E.; Ibrahim, H.M.; Gamal, M.; Elyazeed, N.A.; Saber, N.; et al. Enhancing rice resilience to drought by applying biochar–compost mixture in low-fertile sandy soil. Beni-Suef. Univ. J. Basic Appl. Sci. 2023, 12, 74. [Google Scholar] [CrossRef]
- To, H.T.M.; Nguyen, H.T.; Dang, N.T.M.; Nguyen, N.H.; Bui, T.X.; Lavarenne, J.; Champion, A. Unraveling the genetic elements involved in shoot and root growth regulation by jasmonate in rice using a genome-wide association study. Rice 2019, 12, 69. [Google Scholar] [CrossRef]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; He, S.Y. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef]
- Kurotani, K.I.; Yamanaka, K.; Toda, Y.; Ogawa, D.; Tanaka, M.; Kozawa, H.; Nakamura, H.; Hakata, M.; Ichikawa, H.; Hattori, T.; et al. Stress Tolerance Profiling of a Collection of Extant Salt-Tolerant Rice Varieties and Transgenic Plants Overexpressing Abiotic Stress Tolerance Genes. Plant Cell Physiol. 2015, 56, 1867–1876. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).