Estimation of Genetic Parameters and Identification of Leaf Blast-Resistant Rice RILs Using Cluster Analysis and MGIDI
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Design
2.3. Measurement of Phenotypic Traits Related to the Disease
2.4. Statistical Analysis
3. Results
3.1. Combined Analysis of Variance, Heritability, and Selection Accuracy
3.2. Grouping of Rice Genotypes
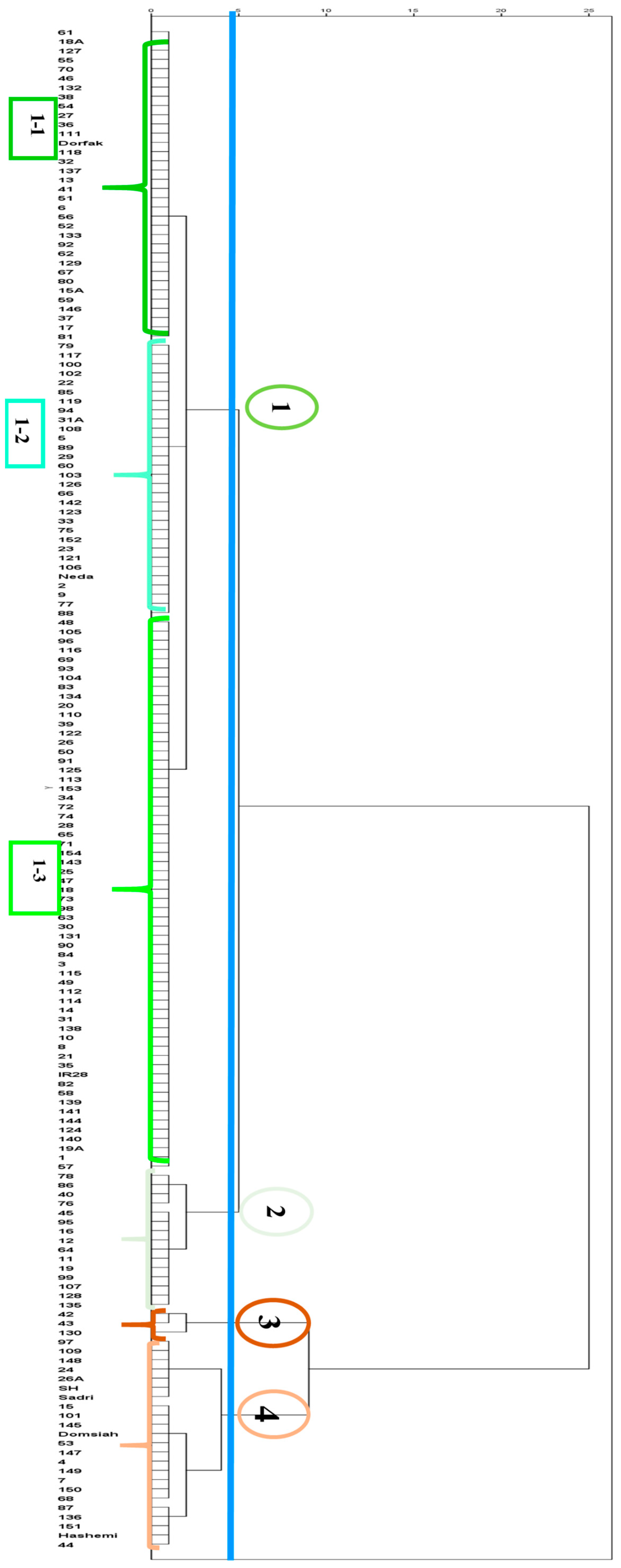
3.2.1. Cluster Analysis of the 2021 Dataset
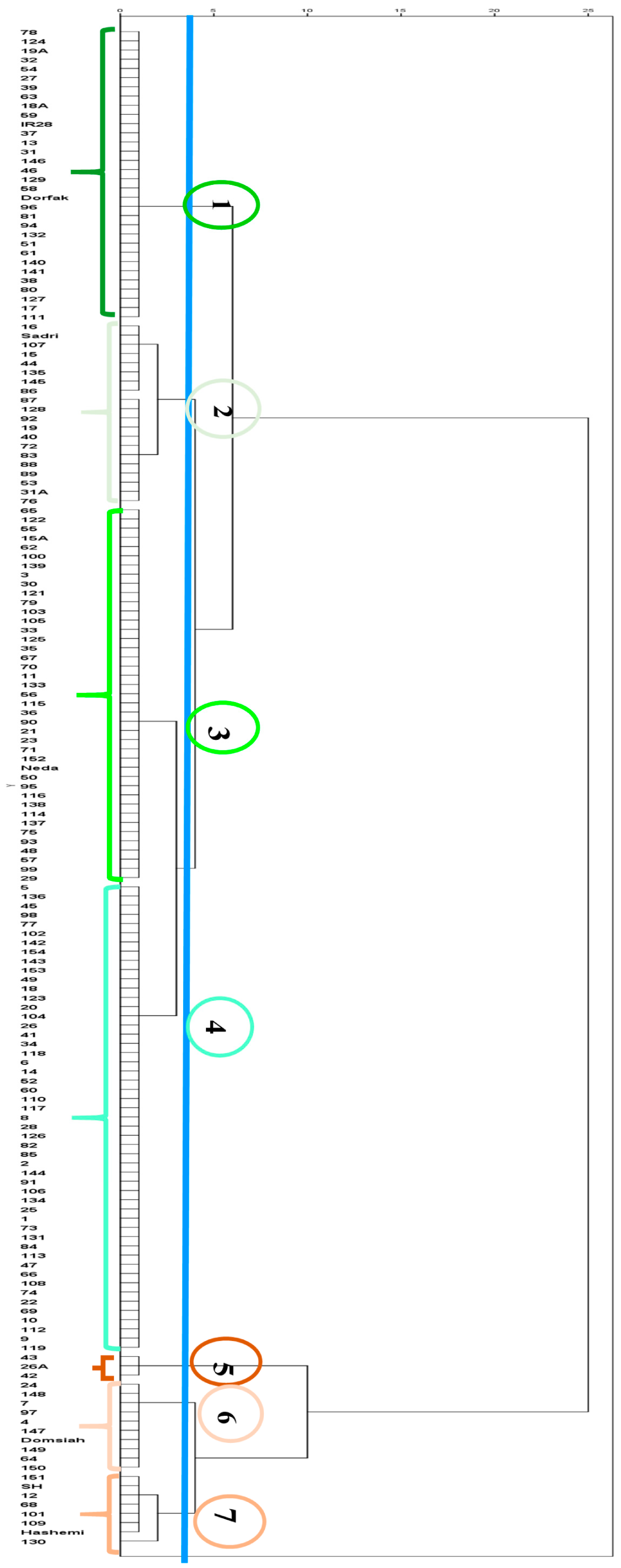
3.2.2. Cluster Analysis of the 2022 Dataset
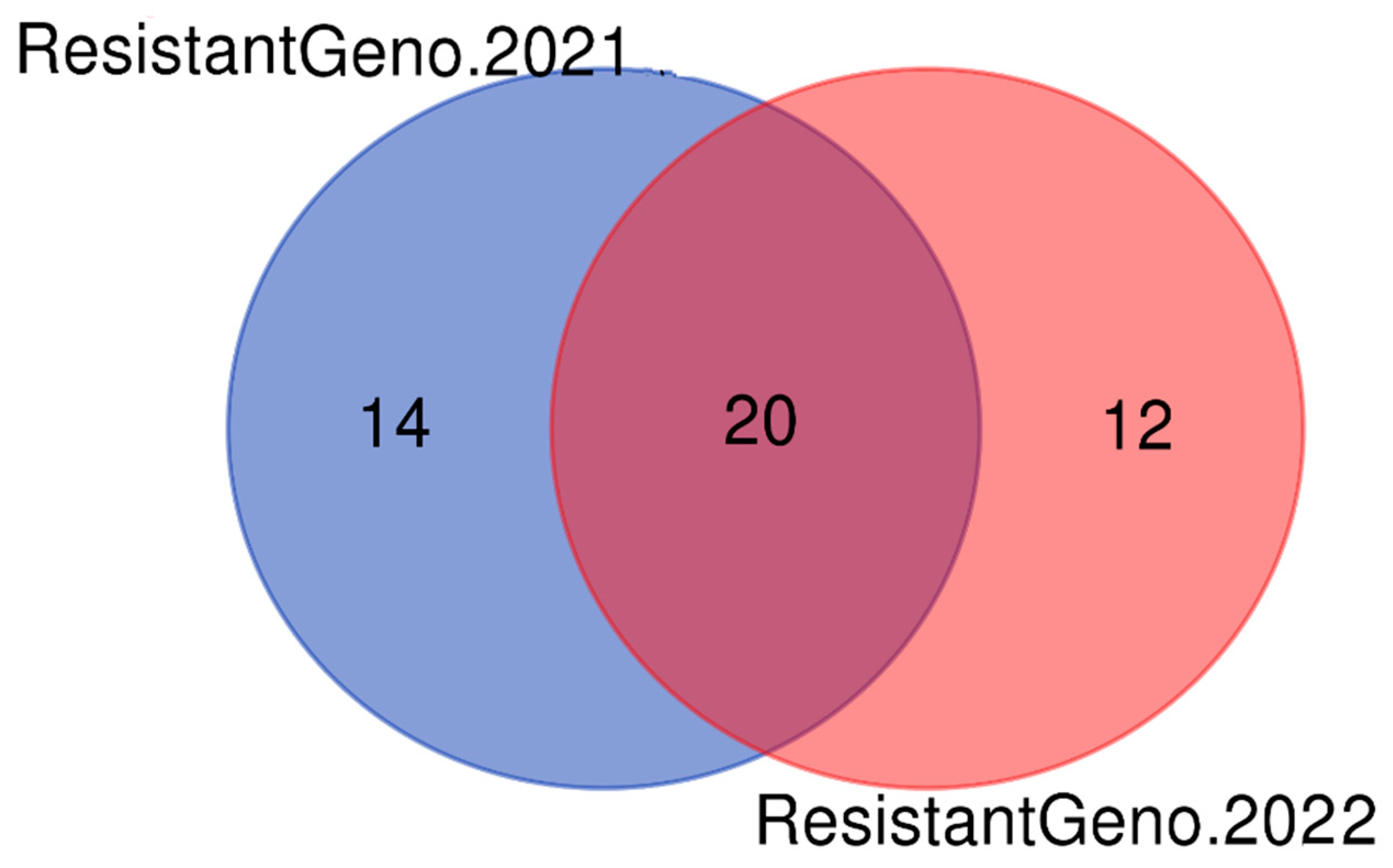
3.2.3. Identification of the Most Resistant and Susceptible Common Rice Genotypes over Two Years
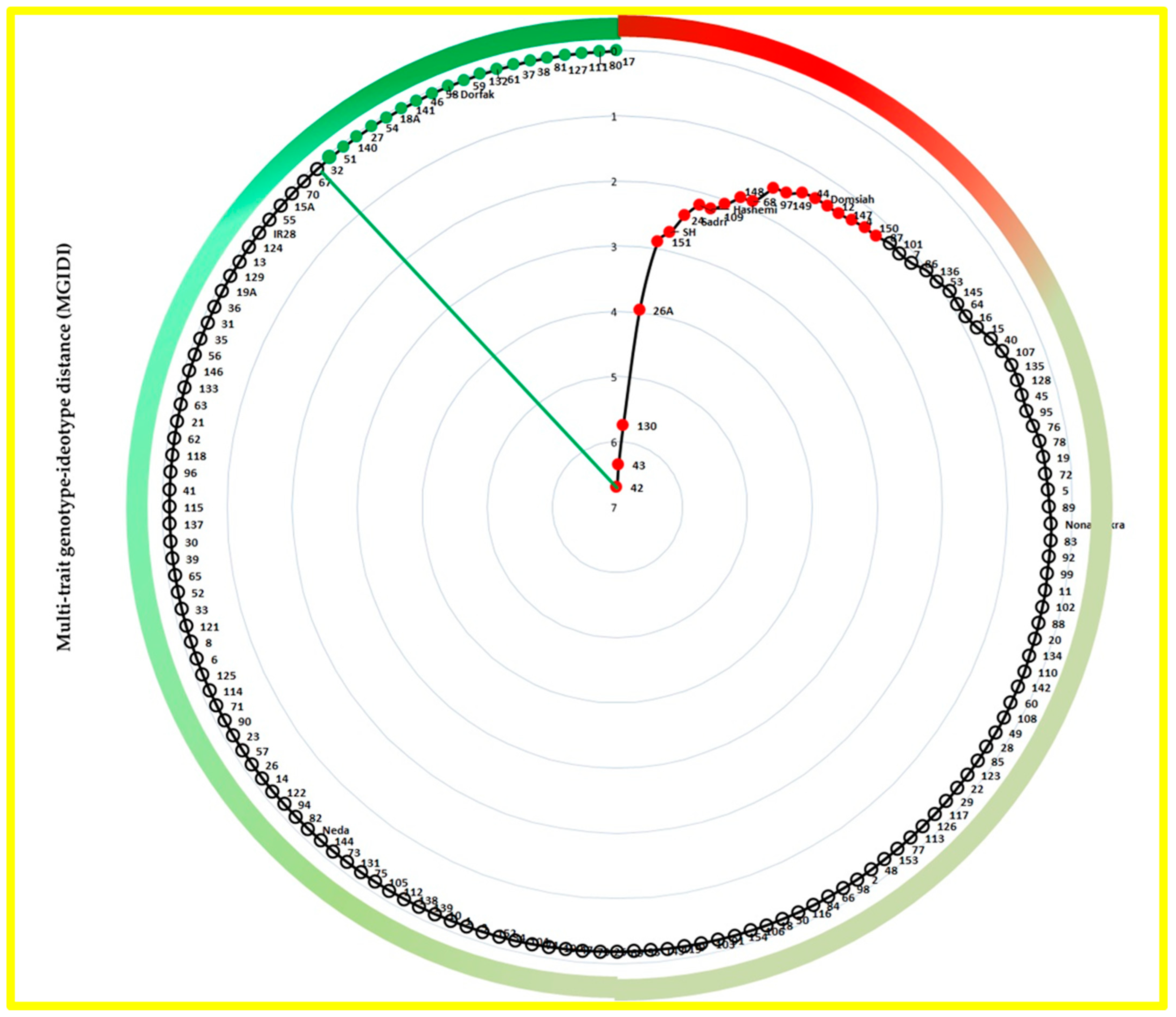
3.3. MGIDI Index Selections
3.4. Comparison of RILs with Control Varieties and Population Parents
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaiswal, S.; Gautam, R.K.; Singh, R.K.; Krishnamurthy, S.L.; Ali, S.; Sakthivel, K.; Iquebal, M.A.; Rai, A.; Kumar, D. Harmonizing technological advances in phenomics and genomics for enhanced salt tolerance in rice from a practical perspective. Rice 2019, 12, 89. [Google Scholar] [CrossRef] [PubMed]
- Mohamadi, S.F.; Bagheri, N.; Kiani, G.; Babaeian Jelodar, N. Evaluation of Reaction of some Rice Genotypes to Salinity Stress at Germination Stage. J. Crop Breed. 2018, 10, 20–30. [Google Scholar] [CrossRef]
- Ahmadi, K.; Ebadzadeh, H.R.; Hatami, F.; Hosseinpour, R.; Abdeshah, H. Volume I: Agricultural Crop Years 2018–2019. Deputy Director of Planning and Economic Affairs Center for Information and Communication Technology, Ministry of Jihad-e Agriculture. In Agricultural Statistics; Ministry of Jihad-e Agriculture: Tehran, Iran, 2022; Available online: https://www.maj.ir/Dorsapax/userfiles/Sub65/Amarnamehj1-97-98-site.pdf (accessed on 26 June 2023).
- Usama Younas, M.; Wang, G.; Du, H.; Zhang, Y.; Ahmad, I.; Rajput, N.; Li, M.; Feng, Z.H.; Hu, K.; Ullah Khan, N.; et al. Approaches to reduce rice blast disease using knowledge from host resistance and pathogen pathogenicity. Int. J. Mol. Sci. 2023, 24, 4985. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Suk Chung, Y.; Lee, E.; Tripathi, P.; Heo, S. Root response to drought stress in rice (Oryza sativa L.). Int. J. Mol. Sci. 2020, 21, 1513. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, M.G.; Alhasan, U.; Mary, Y.; Munsur, Y.; Olufunmilayo, A. Screening of rice germplasm for blast resistance in Nigeria. Asian J. Agric. 2022, 6, 1–6. [Google Scholar] [CrossRef]
- Roustaee, A. Plant Disease Management, 1st ed.; Jahad Daneshgahi Publication: Tehran, Iran, 2002; 400p. [Google Scholar]
- Kumar, I.S.; Nadarajah, K. A meta-analysis of quantitative trait loci associated with multiple disease resistance in rice (Oryza sativa L.). Plants 2020, 9, 1491. [Google Scholar] [CrossRef] [PubMed]
- Agbowuro, G.O.; Afolabi, M.S.; Olamiriki, E.F.; Awoyemi, S.O. Rice blast disease (Magnaporthe oryzae): A menace to rice production and humanity. Int. J. Pathog. Res. 2020, 4, 32–39. [Google Scholar] [CrossRef]
- Rijala, S.; Devkota, Y. A review on various management method of rice blast disease. Malays. J. Sustain. Agric. 2020, 4, 29–33. [Google Scholar] [CrossRef]
- Sahu, P.K.; Sao, R.; Choudhary, D.K.; Thada, A.; Kumar, V.; Mondal, S.; Das, B.K.; Jankuloski, L.; Sharma, D. Advancement in the breeding, biotechnological and genomic tools towards development of durable genetic resistance against the rice blast disease. Plants 2022, 11, 2386. [Google Scholar] [CrossRef]
- Mahdian, S.A. Transferring of resistance genes Pi-1 and Pi-2 to blast in Tarom-dylamani rice cultivar. J. Crop Breed. 2009, 1, 67–77. [Google Scholar]
- Bastiaans, L. Effects of leaf blast on growth and production of a rice crop. 1. Determining the mechanism of yield reduction. Neth. J. Plant Pathol. 1993, 99, 323–334. [Google Scholar] [CrossRef]
- Korinsak, S.; Sriprakhon, S.; Sirithunya, K.; Sriwongchai, T.; Wongsaprom, C.; Pabpla, A.; Vanavichit, A.; Toonjinda, T. Resistance QTLs controlling leaf and neck blast disease identified in a doubled haploid rice population. Euphytica 2023, 219, 1–15. [Google Scholar] [CrossRef]
- Amanzadeh, M.; Moumeni, A.; Okhovat, M.; Javan Nikkhah, M.; Khosravi, V. Evaluation of Resistance of Rice to Leaf and Panicle Blast in Mazandaran Province. Agric. Sci. Tech. Nat. Res. 2008, 11, 209–2019. [Google Scholar]
- Abedi, F.; Babaeiyan, N.; Moumeni, A.; Nemat Zadeh, G.A. Evaluation of partial resistance to Magnaporthe grisea Sacc. in rice cultivars at the seedling stage under upland nursery and greenhouse conditions. Agric. Knowl. 2011, 4, 31–42. [Google Scholar]
- Ghimire, P.; Giri, B.; Gautam, P.; Shrestha, P.; Shrestha, S.H. Screening of different rice genotypes against rice blast (Pyricularia oryzae) at Gokuleshwor Baitadi. Int. J. Sci. Res. Publ. 2019, 9, 809–812. [Google Scholar] [CrossRef]
- Alinezhad, F.; Sabouri, A.; Mousanejad, S. Evaluation of genetic diversity of resistance to blast disease in some Iranian and aerobic rice genotypes. Iran. J. Field Crops Res. 2020, 51, 63–74. [Google Scholar]
- Arooj, S.; Ahmad, S.; Ejaz Ashraf, E.; Ehetisham Ul Haq, M.; Abdul Rehman, M.; Ali, Y.; Atiq, M.; Said, F.; Haq, I.; Raza, W. Field evaluation of rice germplasm for resistance against Pyricularia Oryzae, the cause of rice blast. Ann. Romanian Soc. Cell Biol. 2022, 26, 690–704. [Google Scholar]
- McCouch, S. Diversifying selection in plant breeding. PLoS Biol. 2004, 2, 1507–1512. [Google Scholar] [CrossRef]
- Bertan, I.; Carvalho, F.I.F.; Oliveira, A.C.D. Parental selection strategies in plant breeding programs. J. Crop Sci. Biotechnol. 2007, 10, 211–222. [Google Scholar]
- IRRI. Standard Evaluation System for Rice, 5th ed.; International Rice Research Institute: Los Baños, Philippines, 2013; 55p. [Google Scholar]
- Mukherjee, A.K.; Mohapatra, N.K.; Nayak, P. Assessment of partial resistance to rice blast disease. Oryza 2018, 55, 363–382. [Google Scholar] [CrossRef]
- Van der plank, J.E. Plant Diseases: Epidemics and Control; Academic Press: New York, NY, USA, 1963; 349p. [Google Scholar]
- SAS Institute Inc. SAS/STAT. User’s Guide, Version 9; SAS Institute Inc.: Cary, NC, USA, 2003. [Google Scholar]
- IBM Corp. IBM SPSS Statistics for Windows, Version 24.0; IBM Corp: Armonk, NY, USA, 2016. [Google Scholar]
- Rubin, A. Statistics for Evidence Based Practice and Evaluation; Cengage Learning: Boston, MA, USA, 2012; 368p. [Google Scholar]
- Olivoto, T.; Nardino, M. MGIDI: Toward an effective multivariate selection in biological experiments. Bioinformatics 2021, 37, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Moumeni, A.; Mousanejad, S. Genetic analysis of resistance to races of Magnaporthe grisea the causal agent of blast disease in some Iranian rice cultivars. Seedl. Seed Breed. J. 2013, 1, 423–441. [Google Scholar]
- Sabouri, A.; Alinezhad, F.; Mousanejad, S. Association analysis using SSR markers and identification of resistant aerobic and Iranian rice cultivars to blast disease. Eur. J. Plant Pathol. 2020, 2, 561–570. [Google Scholar] [CrossRef]
- DeLacy, I.; Basford, K.; Cooper, M.; Bull, J.; McLaren, C. Analysis of multi-environment trials—An historical perspective. Plant Adapt. Crop Improv. 1996, 39124, 39–124. [Google Scholar]
- Triki, T.; Bennani, L.; Boussora, F.; Tlahig, S.; Ben Ali, S.; Gasmi, A.; Yahia, H.; Belhouchette, K.; Loumerem, M.; Guasmi, F. Characterization and Trait Association Analysis of 27 Pearl Millet Landraces in Southern Tunisia. Agronomy 2023, 13, 2128. [Google Scholar] [CrossRef]
- Mamun, A.A.; Islam, M.M.; Adhikary, S.K.; Sultana, M.S. Resolution of Genetic Variability and Selection of Novel Genotypes in EMS Induced Rice Mutants Based on Quantitative Traits through MGIDI. Int. J. Agric. Biol. 2022, 28, 100–112. [Google Scholar]
- Rani, R.; Raza, G.; Ashfaq, H.; Rizwan, M.; Shimelis, H.; Tung, M.H.; Arif, M. Analysis of genotype× environment interactions for agronomic traits of soybean (Glycine max [L.] Merr.) using association mapping. Front. Genet. 2023, 13, 1090994. [Google Scholar] [CrossRef]
- Patel, R.; Parmar, D.J.; Kumar, S.; Patel, D.A.; Memon, J.; Patel, M.B.; Patel, J.K. Dissection of genotype× environment interaction for green cob yield using AMMI and GGE biplot with MTSI for selection of elite genotype of sweet corn (Zea mays conva. Saccharata var. rugosa). Indian J. Genet. Plant Breed. 2023, 83, 59–68. [Google Scholar]
- Dias, A.H.; Spanholi, L.F.; Souza, A.L.K.D.; Brighenti, A.F.; Welter, L.J.; Nodari, R.O. Phenology and viticultural performance of different fungus-resistant grapevine advanced selections at three different altitudes in southern Brazil. Rev. Bras. Frutic. 2023, 45, e988. [Google Scholar] [CrossRef]
- Amrate, P.K.; Shrivastava, M.K.; Bhale, M.S.; Agrawal, N.; Kumawat, G.; Shivakumar, M.; Nataraj, V. Identification and genetic diversity analysis of high-yielding charcoal rot resistant soybean genotypes. Sci. Rep. 2023, 13, 8905. [Google Scholar] [CrossRef]
- Ahmed, S.R.; Ali, Z.; Ijaz, I.; Khan, Z.; Gul, N.; Ahmed, S.R.; Ali, Z.; Ijaz, I.; Khan, Z.; Gul, N.; et al. Multi-Trait Selection of Quinoa Ideotypes at Different Levels of Cutting and Spacing. Sustainability 2023, 15, 11446. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Widmer, A.; Arntz, A.M.; Burke, J.M. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003, 358, 1141–1147. [Google Scholar] [CrossRef]
- Ilyas, M.; Rafique, K.; Ahmed, S.; Zulfiqar, S.; Afzal, F.; Khalid, M.; Kazi, A.G.; Mujeeb-Kazi, A. Preventing potential diseases of crop plants under the impact of a changing environment. In Emerging Technologies and Management of Crop Stress Tolerance; Elsevier: Amsterdam, The Netherlands, 2014; pp. 193–214. [Google Scholar]
- Bolek, Y.; El-Zik, K.M.; Pepper, A.E.; Bell, A.A.; Magill, C.W.; Thaxton, P.M.; Reddy, O.U.K. Mapping of verticillium wilt resistance genes in cotton. Plant Sci. 2005, 168, 1581–1590. [Google Scholar] [CrossRef]
- Sabouri, A.; Dadras, A.R.; Azari, M.; Saberi Kouchesfahani, A.; Taslimi, M.; Jalalifar, R. Screening of rice drought-tolerant lines by introducing a new composite selection index and competitive with multivariate methods. Sci. Rep. 2022, 12, 2163. [Google Scholar] [CrossRef]

| Code or Name | Designation | Parentage or Origin |
|---|---|---|
| 1-154 1 | F11 Recombinant inbred lines (RILs) | Iran |
| 15A | IR 82635-B-B-82-2 | IR 78875-176-B-2/IR 78875-207-B-3 |
| 18A | IR 82639-B-B-140-1 | IR 78875-176-B-2/IR 78908-143-B-4 |
| 19A | IR 83749-B-B-46-1 | IR 71524-44-1-1/2*IR 74371-54-1-1 |
| 26A | IR 82635-B-B-32-4 | IR 78875-176-B-2/IR 78875-207-B-3 |
| 31A | Nona Bokra | India |
| Domsiah | Domsiah | Iran (native) |
| Dorfak | Dorfak | Iran (improved) |
| Neda | Neda | Iran (improved) |
| Sadri | Sadri | Iran (native) |
| Hashemi | Hashemi | Iran (native) |
| SH | Shaahpasand | Iran (native) |
| IR28 | IR28 | IRRI |
| Year | Month | Rainfall (mm) | Absolute Temperature (°C) | Mean Temperature (°C) | Relative Humidity (%) | Evaporation (mm) | |
|---|---|---|---|---|---|---|---|
| Minimum | Maximum | ||||||
| 2021 | April | 11.9 | 11.94 | 24.07 | 18 | 73.17 | 92.1 |
| May | 38 | 16.42 | 26.12 | 21.27 | 79.73 | 104.6 | |
| June | 5.1 | 20.96 | 31.13 | 26.05 | 75.77 | 162.1 | |
| July | 108.1 | 22.35 | 30.93 | 26.64 | 78.87 | 117.2 | |
| August | 1.6 | 21.55 | 34.74 | 28.14 | 68.31 | 161.6 | |
| September | 80.9 | 18.86 | 27.9 | 23.38 | 79.32 | 79.1 | |
| 2022 | April | 18.9 | 10.82 | 23 | 16.92 | 71.65 | 91 |
| May | 46.7 | 13.47 | 22.54 | 18 | 80.66 | 83 | |
| June | 1.2 | 20.2 | 30.75 | 25.47 | 71.42 | 166.5 | |
| July | 35.7 | 20.67 | 30.82 | 25.74 | 74.4 | 135.7 | |
| August | 15.7 | 20.56 | 33.06 | 26.81 | 71.53 | 152.8 | |
| September | 129.3 | 18.45 | 29.67 | 24.05 | 78.98 | 90.7 | |
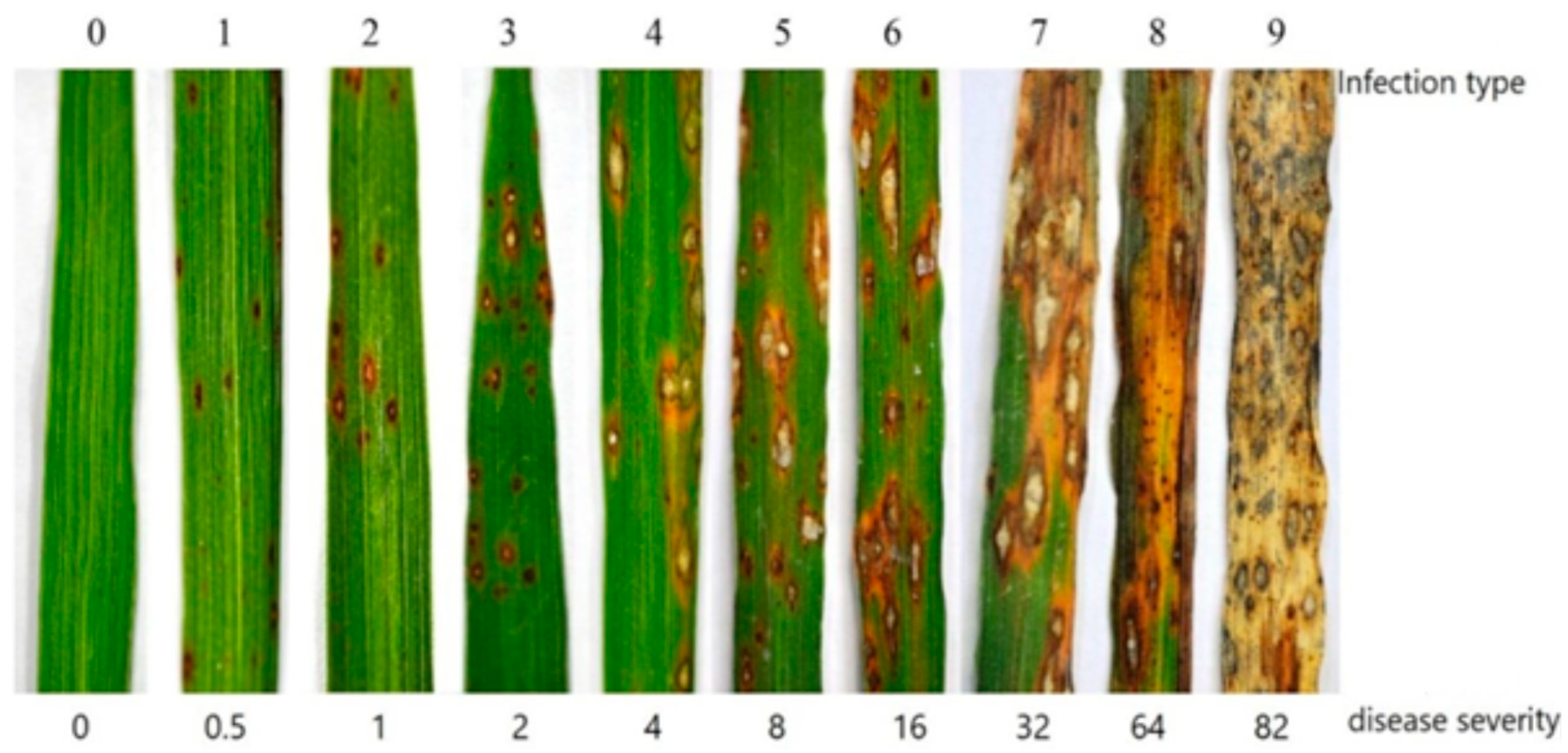
| 0 | No lesions were observed |
| 1 | Small brown specks of pin-point size or larger brown specks without a sporulating center |
| 2 | Small, roundish to slightly elongated, necrotic gray spots, about 1–2 mm in diameter, with a distinct brown margin |
| 3 | The lesion type is the same as in scale 2, but a significant number of lesions are on the upper leaves |
| 4 | Typical susceptible blast lesions are 3 mm or longer, infecting less than 4% of the leaf area |
| 5 | Typical blast lesions infect 4–10% of the leaf area |
| 6 | Typical blast lesions infect 11–25% of the leaf area |
| 7 | Typical blast lesions infect 26–50% of the leaf area |
| 8 | Typical blast lesions infect 51–75% of the leaf area, and many leaves are dead |
| 9 | More than 75% of the leaf area affected |
| Mean Squares | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| S.O.V | df | Severity 1 2 | Severity 2 | Severity 3 | Severity 4 | Severity 5 | AUDPC | r | IT |
| Year | 1 | 5.90 × 10−6 ns 1 | 7.67 × 10−4 ns | 2.27 × 10−3 ns | 3.38 × 10−3 ns | 1.70 × 10−1 ** | 6.14 × 10−1 ns | 4.70 × 10−4 ** | 2.40 × 10−1 ** |
| Block(Year) | 4 | 3.58 × 10−6 | 1.81 × 10−4 | 1.37 × 10−3 | 3.56 × 10−3 | 1.28 × 10−3 | 8.26 × 10−1 | 1.72 × 10−5 | 1.97 × 10−3 |
| Genotype | 164 | 1.55 × 10−6 ** | 1.75 × 10−4 ** | 1.39 × 10−3 ** | 4.37 × 10−3 ** | 1.33 × 10−2 ** | 1.22 ** | 8.1 × 10−5 ** | 1.46 × 10−2 ** |
| Genotype×Year | 164 | 8.30 × 10−7 ns | 7.86 × 10−5 ** | 1.68 × 10−4 ns | 6.91 × 10−4 * | 4.74 × 10−3 ** | 1.08 × 10−1 ** | 2.35 × 10−5 ** | 1.87 × 10−3 * |
| Erorr | 656 | 7.30 × 10−7 | 4.92 × 10−5 | 2.45 × 10−4 | 5.46 × 10−4 | 8.47 × 10−4 | 4.57 × 10−2 | 6.41 × 10−6 | 1.49 × 10−3 |
| CV% | 0.26 | 2.65 | 4.74 | 13.64 | 5.047 | 14.27 | 5.77 | 17.65 | |
| h2mg | 0.46 | 0.54 | 0.81 | 0.81 | 0.59 | 0.84 | 0.71 | 0.84 | |
| Accuracy | 0.68 | 0.73 | 0.90 | 0.90 | 0.77 | 0.92 | 0.84 | 0.92 | |
| Group Information | Trait | Group Mean | Z-Score |
|---|---|---|---|
| Group name: 1.1 Memberships: 34 | Severity 1 1 | 0.0000 | −0.1793 |
| Severity 2 | 0.0000 | −0.2144 | |
| Severity 3 | 0.0705 | −0.3105 | |
| Severity 4 | 0.1121 | −0.3400 | |
| Severity 5 | 0.3324 | −0.3962 | |
| AUDPC | 2.4419 | −0.3704 | |
| r | 0.0016 | −0.4856 | |
| IT | 0.5882 | −0.7183 | |
| Group name: 1.2 Memberships: 30 | Severity 1 | 0.0013 | −0.1333 |
| Severity 2 | 0.0114 | −0.1815 | |
| Severity 3 | 0.1877 | −0.2092 | |
| Severity 4 | 0.2592 | −0.2870 | |
| Severity 5 | 0.5702 | −0.3569 | |
| AUDPC | 5.2087 | −0.3136 | |
| r | 0.0026 | −0.4121 | |
| IT | 1.4444 | −0.2004 | |
| Group name: 1.3 Memberships 60 | Severity 1 | 0.0005 | −0.1615 |
| Severity 2 | 0.0024 | −0.2076 | |
| Severity 3 | 0.1316 | −0.2577 | |
| Severity 4 | 0.1790 | −0.3159 | |
| Severity 5 | 0.5321 | −0.3632 | |
| AUDPC | 4.0550 | −0.3373 | |
| r | 0.0026 | −0.4131 | |
| IT | 0.9944 | −0.4726 | |
| Group name: 2 Memberships: 15 | Severity 1 | 0.0012 | −0.1381 |
| Severity 2 | 0.0070 | −0.1944 | |
| Severity 3 | 0.3195 | −0.0954 | |
| Severity 4 | 1.0673 | 0.0042 | |
| Severity 5 | 2.9782 | 0.0416 | |
| AUDPC | 20.1839 | −0.0062 | |
| r | 0.0129 | 0.3477 | |
| IT | 2.5111 | 0.4448 | |
| Group name: 3 Memberships: 3 | Severity 1 | 0.1385 | 4.7052 |
| Severity 2 | 1.7693 | 4.8747 | |
| Severity 3 | 7.6764 | 6.2596 | |
| Severity 4 | 18.6184 | 6.3278 | |
| Severity 5 | 36.0052 | 5.5067 | |
| AUDPC | 322.9518 | 6.2097 | |
| r | 0.0428 | 2.5525 | |
| IT | 7.5556 | 3.4960 | |
| Group name: 4 Memberships: 23 | Severity 1 | 0.0146 | 0.3367 |
| Severity 2 | 0.2784 | 0.5864 | |
| Severity 3 | 1.1824 | 0.6499 | |
| Severity 4 | 3.4782 | 0.8728 | |
| Severity 5 | 10.3013 | 1.2533 | |
| AUDPC | 70.6786 | 1.0305 | |
| r | 0.0322 | 1.7733 | |
| IT | 4.7681 | 1.8100 |
| Group Information | Trait | Group Mean | Z-Score |
|---|---|---|---|
| Group name: 1 Memberships: 32 | Severity 1 1 | 0.0009 | −0.2941 |
| Severity 2 | 0.0113 | −0.3505 | |
| Severity 3 | 0.1534 | −0.4072 | |
| Severity 4 | 0.1934 | −0.3963 | |
| Severity 5 | 0.1595 | −0.3528 | |
| AUDPC | 3.0683 | −0.3944 | |
| r | 0.0014 | −0.4591 | |
| IT | 0.2188 | −0.9526 | |
| Group name: 2 Memberships: 20 | Severity 1 | 0.0314 | 0.3594 |
| Severity 2 | 0.2408 | 0.1190 | |
| Severity 3 | 0.6237 | 0.0106 | |
| Severity 4 | 0.7702 | 0.0187 | |
| Severity 5 | 0.5891 | −0.0854 | |
| AUDPC | 13.6149 | 0.0096 | |
| r | 0.0052 | 0.0766 | |
| IT | 1.3500 | 0.3214 | |
| Group name: 3 Memberships: 41 | Severity 1 | 0.0022 | −0.2673 |
| Severity 2 | 0.0248 | −0.3229 | |
| Severity 3 | 0.2391 | −0.3310 | |
| Severity 4 | 0.2934 | −0.3243 | |
| Severity 5 | 0.2080 | −0.3226 | |
| AUDPC | 4.6365 | −0.3343 | |
| r | 0.0021 | −0.3585 | |
| IT | 0.6667 | −0.4482 | |
| Group name: 4 Memberships: 51 | Severity 1 | 0.0033 | −0.2426 |
| Severity 2 | 0.0454 | −0.2806 | |
| Severity 3 | 0.2968 | −0.2798 | |
| Severity 4 | 0.3557 | −0.2795 | |
| Severity 5 | 0.2551 | −0.2933 | |
| AUDPC | 5.7907 | −0.2901 | |
| r | 0.0026 | −0.2959 | |
| IT | 1.0000 | −0.0728 | |
| Group name: 5 Memberships: 3 | Severity 1 | 0.2359 | 4.7472 |
| Severity 2 | 3.1416 | 6.0511 | |
| Severity 3 | 7.1521 | 5.8101 | |
| Severity 4 | 9.0822 | 5.9984 | |
| Severity 5 | 10.3343 | 5.9812 | |
| AUDPC | 172.6270 | 6.1013 | |
| r | 0.0446 | 5.6402 | |
| IT | 4.5556 | 3.9314 | |
| Group name: 6 Memberships: 8 | Severity 1 | 0.0651 | 1.0821 |
| Severity 2 | 0.7228 | 1.1045 | |
| Severity 3 | 2.7194 | 1.8723 | |
| Severity 4 | 3.4860 | 1.9725 | |
| Severity 5 | 4.0725 | 2.0831 | |
| AUDPC | 62.9790 | 1.9008 | |
| r | 0.0203 | 2.2066 | |
| IT | 3.2500 | 2.4611 | |
| Group name: 7 Memberships: 10 | Severity 1 | 0.0270 | 0.2657 |
| Severity 2 | 0.6422 | 0.9398 | |
| Severity 3 | 1.5403 | 0.8248 | |
| Severity 4 | 1.5898 | 0.6083 | |
| Severity 5 | 1.7816 | 0.6570 | |
| AUDPC | 32.7360 | 0.7422 | |
| r | 0.0106 | 0.8377 | |
| IT | 2.3667 | 1.4663 |
| Traits Related to Resistance to Leaf Blast Disease | ||||||||
|---|---|---|---|---|---|---|---|---|
| Genotypes | Severity 1 1 | Severity 2 | Severity 3 | Severity 4 | Severity 5 | AUDPC | r | IT |
| Most resistants | 0.001 ± 0.001 | 0.006 ± 0.002 | 0.097 ± 0.008 | 0.149 ± 0.009 | 0.241 ± 0.011 | 2.611 ± 0.144 | 0.001 ± 0.000 | 0.383 ± 0.024 |
| Most susceptibles | 0.050 ± 0.016 | 0.789 ± 0.164 | 2.541 ± 0.506 | 4.863 ± 0.912 | 9.458 ± 1.446 | 90.633 ± 15.686 | 0.028 ± 0.002 | 4.272 ± 0.217 |
| Genotypes | Xo | Xs | SD | SD% | h2 | SG | SG% | Sense |
|---|---|---|---|---|---|---|---|---|
| Severity 1 1 | 0.010 | 0.005 | −0.005 | −46.113 | 0.461 | −0.002 | −21.264 | decrease |
| Severity 2 | 0.129 | 0.062 | −0.067 | −51.948 | 0.539 | −0.036 | −28.005 | decrease |
| Severity 3 | 0.521 | 0.169 | −0.352 | −67.580 | 0.810 | −0.285 | −54.767 | decrease |
| Severity 4 | 0.900 | 0.283 | −0.617 | −68.605 | 0.808 | −0.499 | −55.456 | decrease |
| Severity 5 | 1.727 | 0.846 | −0.880 | −50.995 | 0.588 | −0.518 | −30.007 | decrease |
| AUDPC | 16.924 | 4.665 | −12.259 | −72.437 | 0.844 | −10.346 | −61.131 | decrease |
| r | 0.006 | 0.003 | −0.004 | −54.688 | 0.706 | −0.002 | −38.629 | decrease |
| IT | 1.420 | 0.560 | −0.861 | −60.591 | 0.843 | −0.726 | −51.106 | decrease |
| mean | −59.120 | −1.552 | −42.546 |
| Resistant Control Varieties | Susceptible Control Varieties | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | Trait | RILsMean | 15A | 18A | 19A | Nona Bokra | Dorfak | Neda | IR28 2 | SH 3 | Hashemi | Domsiah | 26A | Sadri |
| Severity 1 1 | 0.005 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 2021 | Severity 2 | 0.067 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.519 | 1.499 | 0.000 | 0.000 | 0.000 |
| Severity 3 | 0.417 | 0.154 | 0.035 | 0.033 | 0.120 | 0.000 | 0.011 | 0.018 | 1.106 | 2.776 | 0.297 | 1.720 | 0.867 | |
| Severity 4 | 0.982 | 0.043 | 0.069 | 0.154 | 0.180 | 0.058 | 0.342 | 0.020 | 8.002 | 6.834 | 1.175 | 4.436 | 2.604 | |
| Severity 5 | 2.461 | 0.280 | 0.192 | 0.491 | 1.021 | 0.296 | 0.640 | 0.245 | 17.043 | 9.098 | 4.384 | 16.445 | 23.262 | |
| AUDPC | 18.898 | 2.363 | 1.399 | 3.029 | 5.678 | 1.444 | 4.710 | 1.116 | 127.037 | 109.603 | 25.646 | 100.653 | 105.709 | |
| r | 0.007 | 0.002 | 0.001 | 0.002 | 0.006 | 0.002 | 0.002 | 0.002 | 0.058 | 0.014 | 0.019 | 0.046 | 0.076 | |
| IT | 1.697 | 0.333 | 0.667 | 1.000 | 1.333 | 0.667 | 1.333 | 1.000 | 5.000 | 5.333 | 4.667 | 6.333 | 5.667 | |
| 2022 | Severity 1 | 0.012 | 0.000 | 0.000 | 0.000 | 0.014 | 0.000 | 0.000 | 0.000 | 0.083 | 0.000 | 0.167 | 0.200 | 0.177 |
| Severity 2 | 0.166 | 0.000 | 0.028 | 0.008 | 0.110 | 0.033 | 0.000 | 0.000 | 0.855 | 0.484 | 0.697 | 2.149 | 0.369 | |
| Severity 3 | 0.557 | 0.189 | 0.144 | 0.155 | 0.525 | 0.100 | 0.233 | 0.175 | 2.855 | 1.789 | 1.389 | 7.236 | 0.933 | |
| Severity 4 | 0.665 | 0.239 | 0.236 | 0.205 | 0.968 | 0.147 | 0.276 | 0.265 | 3.897 | 3.110 | 1.366 | 9.423 | 0.887 | |
| Severity 5 | 0.634 | 0.139 | 0.182 | 0.135 | 0.429 | 0.118 | 0.273 | 0.180 | 3.874 | 4.761 | 2.140 | 10.064 | 0.544 | |
| AUDPC | 11.977 | 3.483 | 3.494 | 3.057 | 12.775 | 2.370 | 4.517 | 3.709 | 67.101 | 54.342 | 32.237 | 167.581 | 17.850 | |
| r | 0.004 | 0.002 | 0.002 | 0.002 | 0.006 | 0.001 | 0.002 | 0.002 | 0.022 | 0.020 | 0.008 | 0.046 | 0.005 | |
| IT | 1.031 | 0.667 | 0.333 | 0.333 | 0.667 | 0.333 | 0.667 | 0.333 | 2.667 | 3.333 | 2.333 | 4.333 | 2.000 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jalalifar, R.; Sabouri, A.; Mousanejad, S.; Dadras, A.R. Estimation of Genetic Parameters and Identification of Leaf Blast-Resistant Rice RILs Using Cluster Analysis and MGIDI. Agronomy 2023, 13, 2730. https://doi.org/10.3390/agronomy13112730
Jalalifar R, Sabouri A, Mousanejad S, Dadras AR. Estimation of Genetic Parameters and Identification of Leaf Blast-Resistant Rice RILs Using Cluster Analysis and MGIDI. Agronomy. 2023; 13(11):2730. https://doi.org/10.3390/agronomy13112730
Chicago/Turabian StyleJalalifar, Reza, Atefeh Sabouri, Sedigheh Mousanejad, and Ahmad Reza Dadras. 2023. "Estimation of Genetic Parameters and Identification of Leaf Blast-Resistant Rice RILs Using Cluster Analysis and MGIDI" Agronomy 13, no. 11: 2730. https://doi.org/10.3390/agronomy13112730
APA StyleJalalifar, R., Sabouri, A., Mousanejad, S., & Dadras, A. R. (2023). Estimation of Genetic Parameters and Identification of Leaf Blast-Resistant Rice RILs Using Cluster Analysis and MGIDI. Agronomy, 13(11), 2730. https://doi.org/10.3390/agronomy13112730





