Genetic Map Construction, QTL Mapping, and Candidate Genes Screening of Grain Size Traits in Common Buckwheat (Fagopyrum esculentum M.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Mapping Population and Grain Size Traits Evaluation
2.2. Primer Development and Marker Validation
2.3. Genetic Map Construction and QTL Analysis
2.4. Validation of Candidate Genes by RNA-Seq
3. Results
3.1. The Phenotypic Data Analysis of Parents and F1 Population
3.2. Development of Primer
3.3. Construction of Genetic Linkage Map
3.4. Preliminary Mapping of QTL Related to Grain Size Traits
3.5. Candidate Gene Screening
4. Discussion
4.1. Development of Molecular Markers in Common Buckwheat
4.2. Construction of Genetic Map in Common Buckwheat
4.3. QTL Mapping and Candidate Genes Screening for Grain Size Related Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Li, Y.; Zhu, J.-K. Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 2018, 4, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Mir, N.A.; Riar, C.S.; Singh, S. Nutritional constituents of pseudo cereals and their potential use in food systems: A review. Trends Food Sci. Technol. 2018, 75, 170–180. [Google Scholar] [CrossRef]
- Sytar, O.; Brestic, M.; Zivcak, M.; Tran, L.-S.P. The Contribution of Buckwheat Genetic Resources to Health and Dietary Diversity. Curr. Genom. 2016, 17, 193–206. [Google Scholar] [CrossRef]
- Giménez-Bastida, J.A.; Zieliñski, H. Buckwheat as a functional food and its effects on health. J. Agric. Food Chem. 2015, 63, 7896–7913. [Google Scholar] [CrossRef]
- Fabjan, N.; Rode, J.; Kosir, I.J.; Wang, Z.; Zhang, Z.; Kreft, I. Tartary buckwheat (Fagopyrum tataricum Gaertn.) as a source of dietary rutin and quercitrin. J. Agric. Food Chem. 2003, 51, 6452–6455. [Google Scholar] [CrossRef]
- Bonafaccia, G.; Marocchini, M.; Kreft, I. Composition and technological properties of the flour and bran from common and Tartary buckwheat. Food Chem. 2003, 80, 9–15. [Google Scholar] [CrossRef]
- Lewis, D.; Jones, D.A. The Genetics of Heterostyly. In Evolution and Function of Heterostyly; Barrett, S.C.H., Ed.; Springer: Berlin, Germany, 1992; pp. 129–150. [Google Scholar]
- Campbell, C.G. Buckwheat, Fagopyrum esculentum Moench; Bioversity International: Rome, Italy, 1997; Volume 19. [Google Scholar]
- Acquaah, G. Principles of Plant Genetics and Breeding; John Wiley & Sons: West Sussex, UK, 2009. [Google Scholar]
- Yano, K.; Ohsawa, R.; Yonezawa, K. Cost Efficiency of Spatial Error Control in Single Plant Selection. Breed. Sci. 2002, 52, 177–184. [Google Scholar] [CrossRef]
- Kump, B.; Javornik, B. Evaluation of genetic variability among common buckwheat (Fagopyrum esculentum Moench) populations by RAPD markers. Plant Sci. 1996, 114, 149–158. [Google Scholar] [CrossRef]
- Aii, J.; Nagano, M.; Penner, A.G.; Campbell, G.C.; Adachi, T. Identification of RAPD markers linked to the homostylar (Ho) gene in buckwheat. Breed. Sci. 1998, 48, 59–62. [Google Scholar] [CrossRef][Green Version]
- Suvorova, G.N.; Funatsuki, H.; Terami, F. Phylogenetic relationships among cultivars, species, and hybrids in the genus Fagopyrum Mill. assessed by RAPD analysis. Russ. J. Genet. 1999, 35, 1428–1432. [Google Scholar]
- Sharma, T.; Jana, S. Species relationships in Fagopyrum revealed by PCR-based DNA fingerprinting. Theor. Appl. Genet. 2002, 105, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Yasui, Y.; Wang, Y.; Ohnishi, O.; Campbell, C.G. Amplified fragment length polymorphism linkage analysis of common buckwheat (Fagopyrum esculentum) and its wild self-pollinated relative Fagopyrum homotropicum. Genome 2004, 47, 345–351. [Google Scholar] [CrossRef]
- Matsui, K.; Kiryu, Y.; Komatsuda, T.; Kurauchi, N.; Ohtani, T.; Tetsuka, T. Identification of AFLP makers linked to non-seed shattering locus (sht1) in buckwheat and conversion to STS markers for marker-assisted selection. Genome 2004, 47, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Konishi, T.; Yasui, Y.; Ohnishi, O. Original birthplace of cultivated common buckwheat inferred from genetic relationships among cultivated populations and natural populations of wild common buckwheat revealed by AFLP analysis. Genes Genet. Syst. 2005, 80, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Imon, K.; Tsumura, Y.; Ohsawa, R. Genetic diversity among Japanese indigenous common buckwheat (Fagopyrum esculentum) cultivars as determined from amplified fragment length polymorphism and simple sequence repeat markers and quantitative agronomic traits. Genome 2005, 48, 367–377. [Google Scholar] [CrossRef]
- Konishi, T.; Ohnishi, O. A linkage map for common buckwheat based on microsatellite and AFLP markers. Fagopyrum 2006, 23, 1–6. [Google Scholar]
- Ma, K.H.; Kim, N.S.; Lee, G.A.; Lee, S.Y.; Lee, J.K.; Yi, J.Y.; Park, Y.J.; Kim, T.S.; Gwag, J.G.; Kwon, S.J. Development of SSR markers for studies of diversity in the genus Fagopyrum. Theor. Appl. Genet. 2009, 119, 1247–1254. [Google Scholar] [CrossRef]
- Pan, S.-J.; Chen, Q.-F. Genetic mapping of common buckwheat using DNA, protein and morphological markers. Hereditas 2010, 147, 27–33. [Google Scholar] [CrossRef]
- Shi, T.X.; Li, R.Y.; Chen, Q.J.; Li, Y.; Pan, F.; Chen, Q.F. De novo sequencing of seed transcriptome and development of genic-SSR markers in common buckwheat (Fagopyrum esculentum). Mol Breed. 2017, 37, 147. [Google Scholar] [CrossRef]
- Hara, T.; Iwata, H.; Okuno, K.; Matsui, K.; Ohsawa, R. QTL analysis of photoperiod sensitivity in common buckwheat by using markers for expressed sequence tags and photoperiod-sensitivity candidate genes. Breed. Sci. 2011, 61, 394–404. [Google Scholar] [CrossRef]
- Yabe, S.; Hara, T.; Ueno, M.; Enoki, H.; Kimura, T.; Nishimura, S.; Yasui, Y.; Ohsawa, R.; Iwata, H. Rapid genotyping with DNA micro-arrays for high-density linkage mapping and QTL mapping in common buckwheat (Fagopyrum esculentum Moench). Breed. Sci. 2014, 64, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Mizuno, N.; Ueno, M.; Takeshima, R.; Yasui, Y. Development of co-dominant markers linked to a hemizygous region that is related to the self-compatibility locus (S) in buckwheat (Fagopyrum esculentum). Breed. Sci. 2020, 70, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Grattapaglia, D.; Sederoff, R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudotestcross: Mapping strategy and RAPD markers. Genetics 1994, 137, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Serba, D.D.; Sykes, R.W.; Gjersing, E.L.; Decker, S.R.; Daverdlin, G.; Devos, K.M.; Brummer, E.C.; Saha, M.C. Cell Wall Composition and Underlying QTL in an F 1 Pseudo-Testcross Population of Switchgrass. Bioenerg. Res. 2016, 9, 836–850. [Google Scholar] [CrossRef]
- Xu, J.M.; Fan, W.; Jin, J.F.; Lou, H.Q.; Chen, W.W.; Yang, J.L.; Zheng, S.J. Transcriptome analysis of Al-induced genes in buckwheat (Fagopyrum esculentum Moench) root apex: New insight into Al toxicity and resistance mechanisms in an Al accumulating species. Front. Plant Sci. 2017, 8, 1141. [Google Scholar] [CrossRef]
- Koech, R.K.; Malebe, P.M.; Nyarukowa, C.; Mose, R.; Kamunya, S.M.; Apostolides, Z. Identification of novel QTL for black tea quality traits and drought tolerance in tea plants (Camellia sinensis). Tree Genet. Genomes 2018, 14, 9. [Google Scholar] [CrossRef]
- Yang, X.P.; Sood, S.; Glynn, N.; Islam, M.S.; Comstock, J.; Wang, J.P. Constructing high-density genetic maps for polyploid sugarcane (Saccharum spp.) and identifying quantitative trait loci controlling brown rust resistance. Mol. Breed. 2017, 37, 116. [Google Scholar] [CrossRef]
- McClure, K.A.; Gardner, K.M.; Toivonen, P.M.A.; Hampson, C.R.; Song, J.; Forney, C.F.; DeLong, J.; Rajcan, I.; Myles, S. QTL analysis of soft scald in two apple populations. Hortic. Res. 2016, 3, 16043. [Google Scholar] [CrossRef][Green Version]
- Liu, T.; Guo, L.L.; Pan, Y.L.; Zhao, Q.; Wang, J.H.; Song, Z.Q. Construction of the first high-density genetic linkage map of Salvia miltiorrhiza using specific length amplified fragment (SLAF) sequencing. Sci. Rep. 2016, 6, 24070. [Google Scholar] [CrossRef]
- Yabe, S.; Hara, T.; Ueno, M.; Enoki, H.; Kimura, T.; Nishimura, S.; Yasui, Y.; Ohsawa, R.; Iwata, H. Potential of genomic selection in mass selection breeding of an allogamous crop: An empirical study to increase yield of common buckwheat. Front. Plant Sci. 2018, 9, 276. [Google Scholar] [CrossRef]
- Hara, T.; Takeshima, R.; Matsui, K. Genes with different modes of inheritance regulate seed germination in preharvest-sprouting-tolerant lines of buckwheat (Fagopyrum esculentum). Jpn. Agric. Res. Q 2020, 54, 137–143. [Google Scholar] [CrossRef]
- Takeshima, R.; Ogiso, T.E.; Yasui, Y.; Matsui, K. Targeted amplicon sequencing + next-generation sequencing–based bulked segregant analysis identified genetic loci associated with preharvest sprouting tolerance in common buckwheat (Fagopyrum esculentum). BMC Plant Biol. 2021, 21, 18. [Google Scholar] [CrossRef]
- Fang, X.M.; Zhang, Y.L.; Zhang, Y.K.; Huang, K.H.; Yang, W.J.; Li, X.Y.; Zhang, Z.Y.; Wu, K.H.; Xu, X.; Ruan, R.W.; et al. De novo transcriptome assembly and identification of genes related to seed size in common buckwheat (Fagopyrum esculentum M.). Breed. Sci. 2019, 69, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fang, X.M.; Tang, T.; Wang, Y.D.; Wu, Y.H.; Luo, J.Y.; Wu, H.T.; Wang, Y.Q.; Zhang, J.; Ruan, R.W.; et al. Inflorescence Transcriptome Sequencing and Development of New EST-SSR Markers in Common Buckwheat (Fagopyrum esculentum). Plants 2022, 11, 742. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup (Subgroup GPDP). The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Zhang, Z.S.; Xiao, Y.H.; Luo, M.; Li, X.B.; Luo, X.Y.; Hou, L.; Li, D.M.; Pei, Y. Construction of a genetic linkage map and QTL analysis of fiber-related traits in upland cotton (Gossypium hirsutum L.). Euphytica 2005, 144, 91–99. [Google Scholar] [CrossRef]
- Van Ooijen, J.W.; Voorrips, R.E. JoinMap 4.0., Software for the Calculation of Genetic Linkage Maps; Plant Research International: Wageningen, The Netherlands, 2006. [Google Scholar]
- Van Ooijen, J.W. MapQTL 6.0., Software for the Mapping of Quantitative Trait Loci in Experimental Populations; Plant Research International: Wageningen, The Netherlands, 2009. [Google Scholar]
- Voorrips, R.E. MapChart 2.2: Software for the Graphical Presentation of Linkage Maps and QTLs; Plant Research International: Wageningen, The Netherlands, 2006. [Google Scholar]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef]
- Yasui, Y.; Hirakawa, H.; Ueno, M.; Matsui, K.; Katsube-Tanaka, T.; Yang, S.J.; Aii, J.; Sato, S.; Mori, M. Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 2016, 23, 215–224. [Google Scholar] [CrossRef]
- Hou, S.Y.; Sun, Z.X.; Bin, L.H.; Xu, D.M.; Wu, B.; Zhang, B.; Wang, X.C.; Han, Y.H.; Zhang, L.J.; Qiao, Z.J.; et al. Genetic diversity of buckwheat cultivars (Fagopyrum tartaricum Gaertn.) assessed with SSR markers developed from genome survey sequences. Plant Mollecular Biol. Report. 2016, 34, 233–241. [Google Scholar] [CrossRef]
- Liu, J.; Qu, J.T.; Yang, C.; Tang, D.G.; Li, J.W.; Lan, H.; Rong, T.Z. Development of genome-wide insertion and deletion markers for maize, based on next-generation sequencing data. BMC Genom. 2015, 16, 601. [Google Scholar] [CrossRef]
- Ren, Y.; Zhao, H.; Kou, Q.H.; Jiang, J.; Guo, S.G.; Zhang, H.Y.; Hou, W.J.; Zou, X.H.; Sun, H.H.; Gong, G.Y.; et al. A High Resolution Genetic Map Anchoring Scaffolds of the Sequenced Watermelon Genome. PLoS ONE 2012, 7, e29453. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.H.; Wu, H.P.; Wang, C.S.; Tseng, H.Y.; Hwu, K.K. Genome-wide InDel marker system for application in rice breeding and mapping studies. Euphytica 2013, 192, 131–143. [Google Scholar] [CrossRef]
- Lu, S.J.; Li, Y.; Wang, J.L.; Srinives, R.; Nan, H.Y.; Cao, D.; Wang, Y.P.; Li, J.L.; Li, X.M.; Fang, C.; et al. QTL mapping for flowering time in different latitude in soybean. Euphytica 2015, 206, 725–736. [Google Scholar] [CrossRef]
- Tan, S.; Cheng, J.W.; Zhang, L.; Cheng, Q.; Nong, D.G.; Li, W.P.; Tang, X.; Wu, Z.M.; Hu, K.L. Construction of an Interspecific Genetic Map Based on InDel and SSR for Mapping the QTLs Affecting the Initiation of Flower Primordia in Pepper (Capsicum spp.). PLoS ONE 2015, 10, e0119389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.J.; Li, X.X.; Ma, B.; Gao, Q.; Du, H.L.; Han, Y.H.; Li, Y.; Cao, Y.H.; Qi, M.; Zhu, Y.X.; et al. The Tartary Buckwheat Genome Provides Insights into Rutin Biosynthesis and Abiotic Stress Tolerance. Mol. Plant 2017, 10, 1224–1237. [Google Scholar] [CrossRef]
- Gao, Q.; Zhang, N.; Wang, W.Q.; Shen, S.Y.; Bai, C.; Song, X.J. The ubiquitin-interacting motif-type ubiquitin receptor HDR3 interacts with and stabilizes the histone acetyltransferase GW6a to control the grain size in rice. Plant Cell 2021, 33, 3331–3347. [Google Scholar] [CrossRef]
- Huangfu, L.X.; Chen, R.J.; Lu, Y.; Zhang, E.Y.; Miao, J.; Zuo, Z.H.; Zhao, Y.; Zhu, M.Y.; Zhang, Z.H.; Li, P.C.; et al. OsCOMT, encoding a caffeic acid O-methyltransferase in melatonin biosynthesis, increases rice grain yield through dual regulation of leaf senescence and vascular development. Plant Biotechnol. J. 2022, 20, 1122–1139. [Google Scholar] [CrossRef]
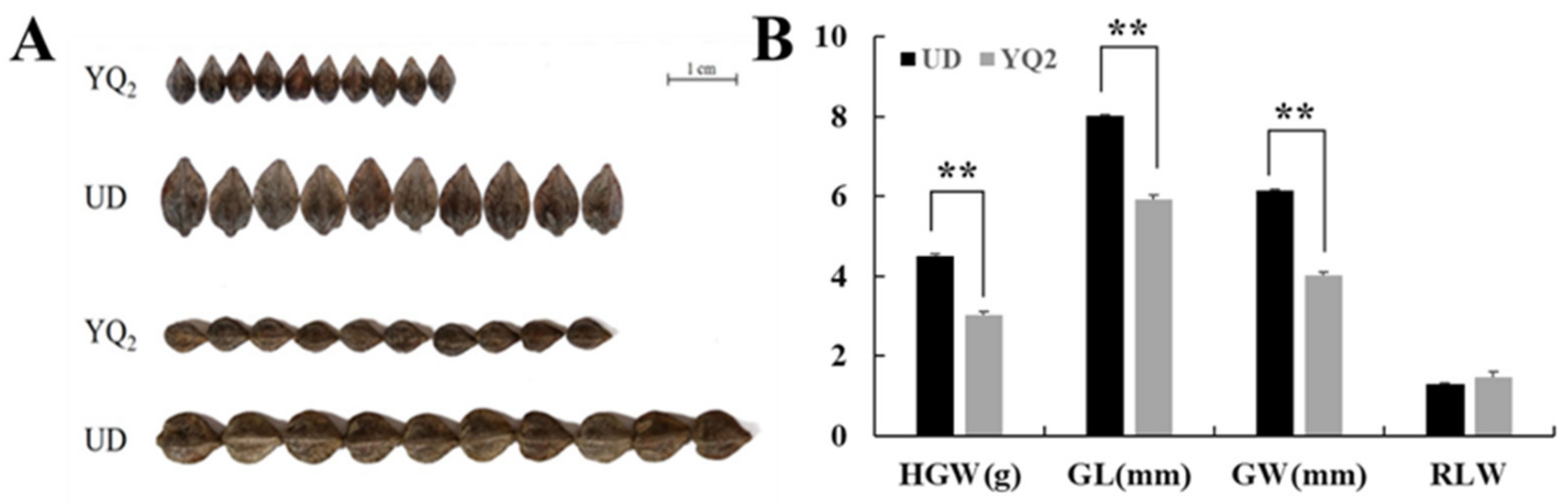



| Hundred-Grain-Weight (g) | Grain Length (mm) | Grain Width (mm) | ||
|---|---|---|---|---|
| Parent | UD | 4.500 ± 0.06 ** | 8.02 ± 0.03 ** | 6.15 ± 0.01 ** |
| YQ2 | 3.033 ± 0.09 | 5.92 ± 0.11 | 4.02 ± 0.08 | |
| F1 | Mean | 3.060 | 6.81 | 4.95 |
| Max | 4.767 | 9.43 | 7.30 | |
| Min | 1.200 | 5.00 | 3.23 | |
| Skewness | −0.119 | 0.23 | 0.20 | |
| Kurtosis | 0.052 | 0.94 | 0.97 | |
| Segregation Types | EST-SSR Loci | InDel Loci | Total | ||
|---|---|---|---|---|---|
| Number | Proportion (%) | Number | Proportion (%) | ||
| hk × hk | 33 | 29.20 | 80 | 70.80 | 113 |
| nn × np | 12 | 46.15 | 14 | 53.90 | 26 |
| lm × ll | 8 | 44.44 | 10 | 55.56 | 18 |
| ef × eg | 1 | 11.11 | 8 | 88.89 | 9 |
| ab × cd | 0 | 100.00 | 0 | 100.00 | 0 |
| Group | Length/cM | Total of Loci | No of EST-SSR | No of InDel | Average Intervals (cM) |
|---|---|---|---|---|---|
| LG1 | 243.75 | 16 | 9 | 7 | 15.23 |
| LG 2 | 208.24 | 25 | 7 | 18 | 8.33 |
| LG 3 | 173.37 | 15 | 3 | 12 | 11.56 |
| LG 4 | 181.19 | 18 | 3 | 15 | 10.07 |
| LG 5 | 185.76 | 18 | 2 | 16 | 10.32 |
| LG 6 | 157.67 | 18 | 4 | 14 | 8.76 |
| LG 7 | 133.27 | 13 | 8 | 5 | 10.25 |
| LG 8 | 115.09 | 9 | 3 | 6 | 12.79 |
| Total | 1398.33 | 132 | 39 | 93 | 10.59 |
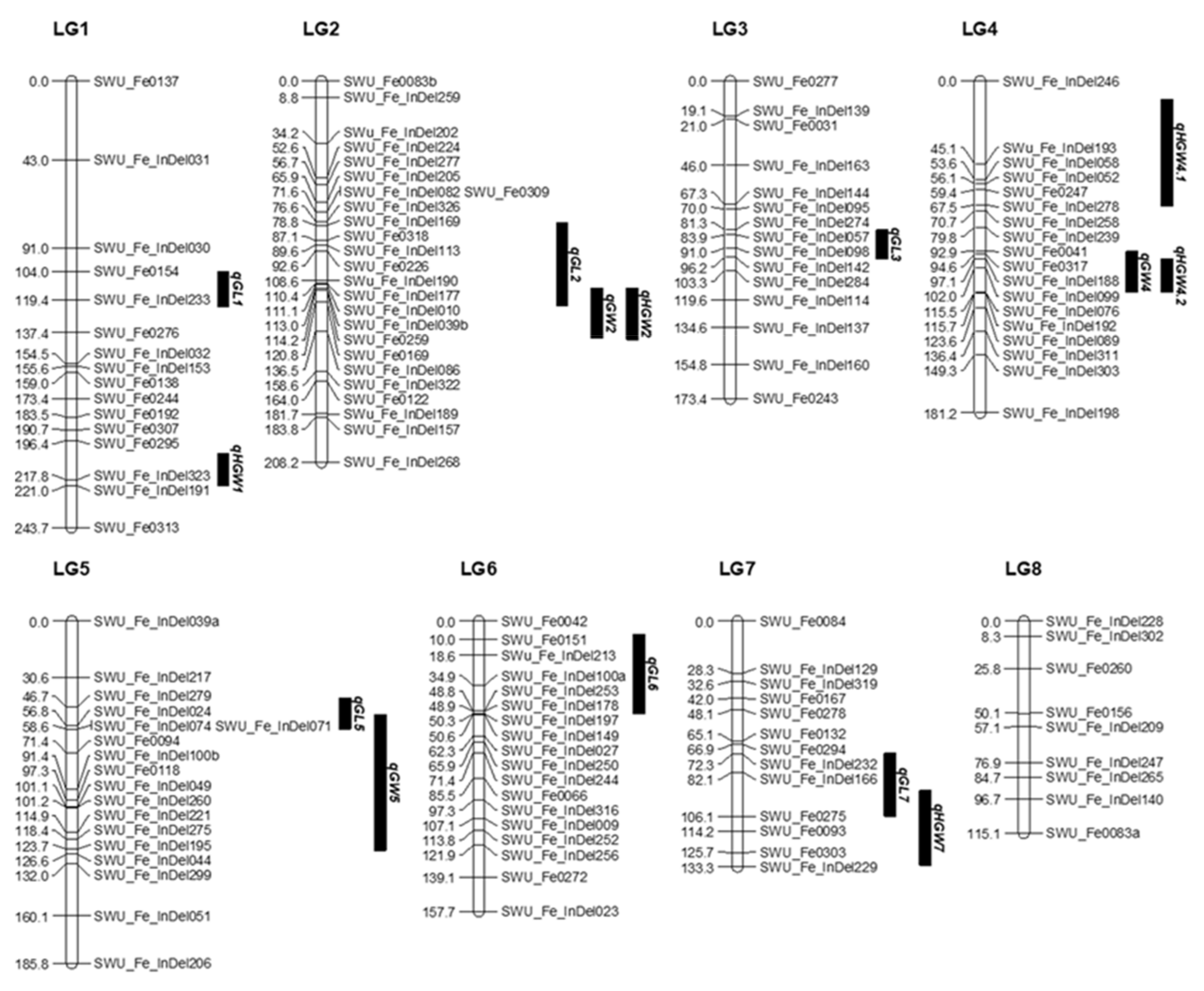
| Traits | QTL | Group | Nearest Marker | LOD | Var% | A |
|---|---|---|---|---|---|---|
| Grain length | qGL1 | LG1 | SWU_Fe_InDel233 | 3.13 | 6.8 | 0.622 |
| qGL2 | LG2 | SWU_Fe_InDel113 | 2.91 | 6.3 | 0.927 | |
| qGL3 | LG3 | SWU_Fe_InDel098 | 2.12 | 4.7 | 0.735 | |
| qGL5 | LG5 | SWU_Fe_InDel279 | 2.85 | 6.2 | 0.637 | |
| qGL6 | LG6 | SWU_Fe_InDel213 | 3.39 | 8 | 0.849 | |
| qGL7 | LG7 | SWU_Fe_InDel166 | 2.74 | 6 | 0.628 | |
| Grain width | qGW2 | LG2 | SWU_Fe_ InDel086 | 3.04 | 6.6 | 0.74 |
| qGW4 | LG4 | SWU_Fe_InDel076 | 2.61 | 5.7 | 0.618 | |
| qGW5 | LG5 | SWU_Fe_InDel195 | 3.23 | 7 | 0.665 | |
| Hundred-grain-weight | qHGW1 | LG1 | SWU_Fe_InDel323 | 3.43 | 7.4 | 0.705 |
| qHGW2 | LG2 | SWU_Fe_InDel086 | 5.04 | 10.7 | 0.795 | |
| qHGW4.1 | LG4 | SWU_Fe_InDel278 | 2.9 | 6.3 | 0.782 | |
| qHGW4.2 | LG4 | SWU_Fe_InDel076 | 2.09 | 4.6 | 0.618 | |
| qHGW7 | LG7 | SWU_Fe0303 | 5.66 | 11.9 | 0.847 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, X.; Zhang, Y.; Cui, J.; Yue, L.; Tao, J.; Wang, Y.; Zhang, R.; Liu, J.; Jiang, A.; Zhang, J.; et al. Genetic Map Construction, QTL Mapping, and Candidate Genes Screening of Grain Size Traits in Common Buckwheat (Fagopyrum esculentum M.). Agronomy 2022, 12, 2062. https://doi.org/10.3390/agronomy12092062
Fang X, Zhang Y, Cui J, Yue L, Tao J, Wang Y, Zhang R, Liu J, Jiang A, Zhang J, et al. Genetic Map Construction, QTL Mapping, and Candidate Genes Screening of Grain Size Traits in Common Buckwheat (Fagopyrum esculentum M.). Agronomy. 2022; 12(9):2062. https://doi.org/10.3390/agronomy12092062
Chicago/Turabian StyleFang, Xiaomei, Yuanli Zhang, Jingbin Cui, Lingqing Yue, Jianbo Tao, Yigang Wang, Ruifeng Zhang, Jiaqi Liu, Aohua Jiang, Jian Zhang, and et al. 2022. "Genetic Map Construction, QTL Mapping, and Candidate Genes Screening of Grain Size Traits in Common Buckwheat (Fagopyrum esculentum M.)" Agronomy 12, no. 9: 2062. https://doi.org/10.3390/agronomy12092062
APA StyleFang, X., Zhang, Y., Cui, J., Yue, L., Tao, J., Wang, Y., Zhang, R., Liu, J., Jiang, A., Zhang, J., Ruan, R., & Yi, Z. (2022). Genetic Map Construction, QTL Mapping, and Candidate Genes Screening of Grain Size Traits in Common Buckwheat (Fagopyrum esculentum M.). Agronomy, 12(9), 2062. https://doi.org/10.3390/agronomy12092062





