Intra- and Inter-Cultivar Variability of Lavandin (Lavandula × intermedia Emeric ex Loisel.) Landraces from the Island of Hvar, Croatia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. AFLP Analysis
2.3. Data Analysis
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Upson, T.; Andrews, S. The Genus Lavandula; Timber Press Inc.: Portland, OR, USA, 2004; ISBN 0881926426. [Google Scholar]
- Adal, A.M.; Demissie, Z.A.; Mahmoud, S.S. Identification, Validation and Cross-Species Transferability of Novel Lavandula EST-SSRs. Planta 2015, 241, 987–1004. [Google Scholar] [CrossRef] [PubMed]
- Moja, S.; Guitton, Y.; Nicolè, F.; Legendre, L.; Pasquier, B.; Upson, T.; Jullien, F. Genome Size and Plastid TrnK-MatK Markers Give New Insights into the Evolutionary History of the Genus Lavandula L. Plant Biosyst. 2015, 150, 1216–1224. [Google Scholar] [CrossRef]
- Meneghetti, S.; Costacurta, A.; Morreale, G.; Calò, A. Study of Intra-Varietal Genetic Variability in Grapevine Cultivars by PCR-Derived Molecular Markers and Correlations with the Geographic Origins. Mol. Biotechnol. 2012, 50, 72–85. [Google Scholar] [CrossRef]
- Hind, K.R.; Adal, A.M.; Upson, T.M.; Mahmoud, S.S. An Assessment of Plant DNA Barcodes for the Identification of Cultivated Lavandula (Lamiaceae) Taxa. Biocatal. Agric. Biotechnol. 2018, 16, 459–466. [Google Scholar] [CrossRef]
- Lesage-Meessen, L.; Bou, M.; Sigoillot, J.C.; Faulds, C.B.; Lomascolo, A. Essential Oils and Distilled Straws of Lavender and Lavandin: A Review of Current Use and Potential Application in White Biotechnology. Appl. Microbiol. Biotechnol. 2015, 99, 3375–3385. [Google Scholar] [CrossRef]
- Kıvrak, Ş. Essential Oil Composition and Antioxidant Activities of Eight Cultivars of Lavender and Lavandin from Western Anatolia. Ind. Crop. Prod. 2018, 117, 88–96. [Google Scholar] [CrossRef]
- Devetak, Z.; Cenci, A. Lavandini Di Produzione Jugoslava. Riv. Ital. Essenze, Profum. Piante Off. Aromi, Sapon. Cosmet. 1965, 47, 588–592. (In Italian) [Google Scholar]
- Devetak, Z. Agrobiological Research of Lavender and Lavandin and Biochemical of Their Essential Oils. Ph.D. Thesis, University of Sarajevo, Sarajevo, Bosnia and Herzegovina, 1965. (In Croatian). [Google Scholar]
- Petrić, K. The Island of Hvar—Velo Grablje: People and Events in the 20th Century; Udruga “Pjover”: Hvar, Croatia, 2016; ISBN 978-953-57406-12. (In Croatian) [Google Scholar]
- Devetak, Z. Lavanda (Lavandula Vera D.C., Lavandula Latifolia Vill.). Farm. Glas. 1949, 5, 1–5. (In Croatian) [Google Scholar]
- Vernazza, N. Lavender Hybrid Oil from the Island of Hvar. Arh. Za Poljopr. Nauk. 1957, 10, 1–6. [Google Scholar]
- Walters, S. Essential Role of Crop Landraces for World Food Security. Mod. Concepts Dev. Agron. 2018, 1, 1–14. [Google Scholar] [CrossRef]
- Zdunić, G.; Šimon, S.; Malenica, N.; Budić-Leto, I.; Maletić, E.; Karoglan Kontić, J.; Pejić, I. Intravarietal Variability of “Crljenak Kastelanski” and Its Relationship with “Zinfandel” and “Primitivo” Selections. Acta Hortic. 2014, 1046, 573–580. [Google Scholar] [CrossRef]
- Schüller, E.; Fernández, F.F.; Antanaviciute, L.; Anhalt-brüderl, U.; Spornberger, A.; Forneck, A. Autochthonous Austrian Varieties of Prunus Avium L. Represent a Regional Gene Pool, Assessed Using SSR and AFLP Markers. Genes 2021, 12, 322. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Vandelee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP—A New Technique for DNA-Fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grdiša, M.; Liber, Z.; Radosavljevic, I.; Carovic-Stanko, K.; Kolak, I.; Satovic, Z. Genetic Diversity and Structure of Dalmatian Pyrethrum (Tanacetum Cinerariifolium Trevir./Sch./Bip., Asteraceae) within the Balkan Refugium. PLoS ONE 2014, 9, e105265. [Google Scholar] [CrossRef] [Green Version]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Hammer, D.A.T.; Ryan, P.D.; Hammer, Ø.; Harper, D.A.T. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 178. [Google Scholar]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Champaign, IL, USA, 1949. [Google Scholar]
- Lewontin, R.C. The Apportionment of Human Diversity. Evol. Biol. 1972, 6, 381–398. [Google Scholar]
- SAS Institute. SAS/STAT® 9.1 User’s Guide; SAS Institute: Cary, NC, USA, 2004. [Google Scholar]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Corander, J.; Waldmann, P.; Sillanpaa, M.J. Bayesian Analysis of Genetic Differentiation between Populations. Genetics 2003, 163, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Corander, J.; Waldmann, P.; Marttinen, P.; Sillanpaa, M.J. BAPS 2: Enhanced Possibilities for the Analysis of Genetic Population Structure. Bioinformatics 2004, 20, 2363–2369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Puechmaille, S.J. The Program Structure Does Not Reliably Recover the Correct Population Structure When Sampling Is Uneven: Subsampling and New Estimators Alleviate the Problem. Mol. Ecol. Resour. 2016, 16, 608–627. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. StructureSelector: A Web-Based Software to Select and Visualize the Optimal Number of Clusters Using Multiple Methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A Program for Identifying Clustering Modes and Packaging Population Structure Inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [Green Version]
- Corander, J.; Marttinen, P. Bayesian Identification of Admixture Events Using Multilocus Molecular Markers. Mol. Ecol. 2006, 15, 2833–2843. [Google Scholar] [CrossRef]
- Zdunić, G.; Preece, J.E.; Dangl, G.S.; Koehmstedt, A.; Mucalo, A.; Maletić, E.; Pejić, I. Genetic Characterization of Grapevine Cultivars Collected throughout the Dalmatian Region. Am. J. Enol. Vitic. 2013, 64, 285–290. [Google Scholar] [CrossRef]
- Giakoni, J. The Letter to Sinčić, May 15th 1956, Wienna. Ph.D. Thesis, Faculty of Agriculture, University of Sarajevo, Sarajevo, Bosnia and Herzegovina, 1965. [Google Scholar]
- Lazović, B.; Klepo, T.; Adakalić, M.; Šatović, Z.; Arbeiter, A.B.; Hladnik, M.; Strikić, F.; Liber, Z.; Bandelj, D. Intra-Varietal Variability and Genetic Relationships among the Homonymic East Adriatic Olive (Olea Europaea L.) Varieties. Sci. Hortic. 2018, 236, 175–185. [Google Scholar] [CrossRef]
- Sales, H.; Šatović, Z.; Alves, M.L.; Fevereiro, P.; Nunes, J.; Vaz Patto, M.C. Accessing Ancestral Origin and Diversity Evolution by Net Divergence of an Ongoing Domestication Mediterranean Olive Tree Variety. Front. Plant Sci. 2021, 12, 1098. [Google Scholar] [CrossRef] [PubMed]
- McKey, D.; Elias, M.; Pujol, M.E.; Duputié, A. The Evolutionary Ecology of Clonally Propagated Domesticated Plants. New Phytol. 2010, 186, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Strikic, F.; Bandelj Mavsar, D.; Perica, S.; Cmelik, Z.; Satovic, Z.; Javornik, B. Genetic Variation within the Olive (Olea Europaea L.) Cultivar Oblica Detected Using Amplified Fragment Length Polymorphism (AFLP) Markers. African J. Biotechnol. 2010, 9, 2880–2883. [Google Scholar] [CrossRef]
- Strikić, F.; Liber, Z.; Bandelj Mavsar, D.; Čmelik, Z.; Perica, S.; Radunić, M.; Javornik, B.; Šatović, Z. Intra-Cultivar Diversity in the Croatian Olive Cultivar, ‘Lastovka’. J. Hortic. Sci. Biotechnol. 2011, 86, 305–311. [Google Scholar] [CrossRef]
- Mihaljević, M.Ž.; Anhalt, U.C.M.; Rühl, E.; Mugoša, M.T.; Maraš, V.; Forneck, A.; Zdunić, G.; Preiner, D.; Pejić, I. Cultivar Identity, Intravarietal Variation, and Health Status of Native Grapevine Varieties in Croatia and Montenegro. Am. J. Enol. Vitic. 2015, 66, 531–541. [Google Scholar] [CrossRef]
- Gil-Vega, K.; Díaz, C.; Nava-Cedillo, A.; Simpson, J. AFLP Analysis of Agave Tequilana Varieties. Plant Sci. 2006, 170, 904–909. [Google Scholar] [CrossRef]
- Díaz-Martínez, M.; Nava-Cedillo, A.; Guzmán-López, J.A.; Escobar-Guzmán, R.; Simpson, J. Polymorphism and Methylation Patterns in Agave Tequilana Weber Var. “Azul” Plants Propagated Asexually by Three Different Methods. Plant Sci. 2012, 185–186, 321–330. [Google Scholar] [CrossRef]
- Zhou, L.; Matsumoto, T.; Tan, H.W.; Meinhardt, L.W.; Mischke, S.; Wang, B.; Zhang, D. Developing Single Nucleotide Polymorphism Markers for the Identification of Pineapple (Ananas Comosus) Germplasm. Hortic. Res. 2015, 2, 15056. [Google Scholar] [CrossRef] [Green Version]
- Matsuoka, Y.; Vigouroux, Y.; Goodman, M.M.; Sanchez, J.G.; Buckler, E.; Doebley, J. A Single Domestication for Maize Shown by Multilocus Microsatellite Genotyping. Proc. Natl. Acad. Sci. USA 2002, 99, 6080–6084. [Google Scholar] [CrossRef] [Green Version]
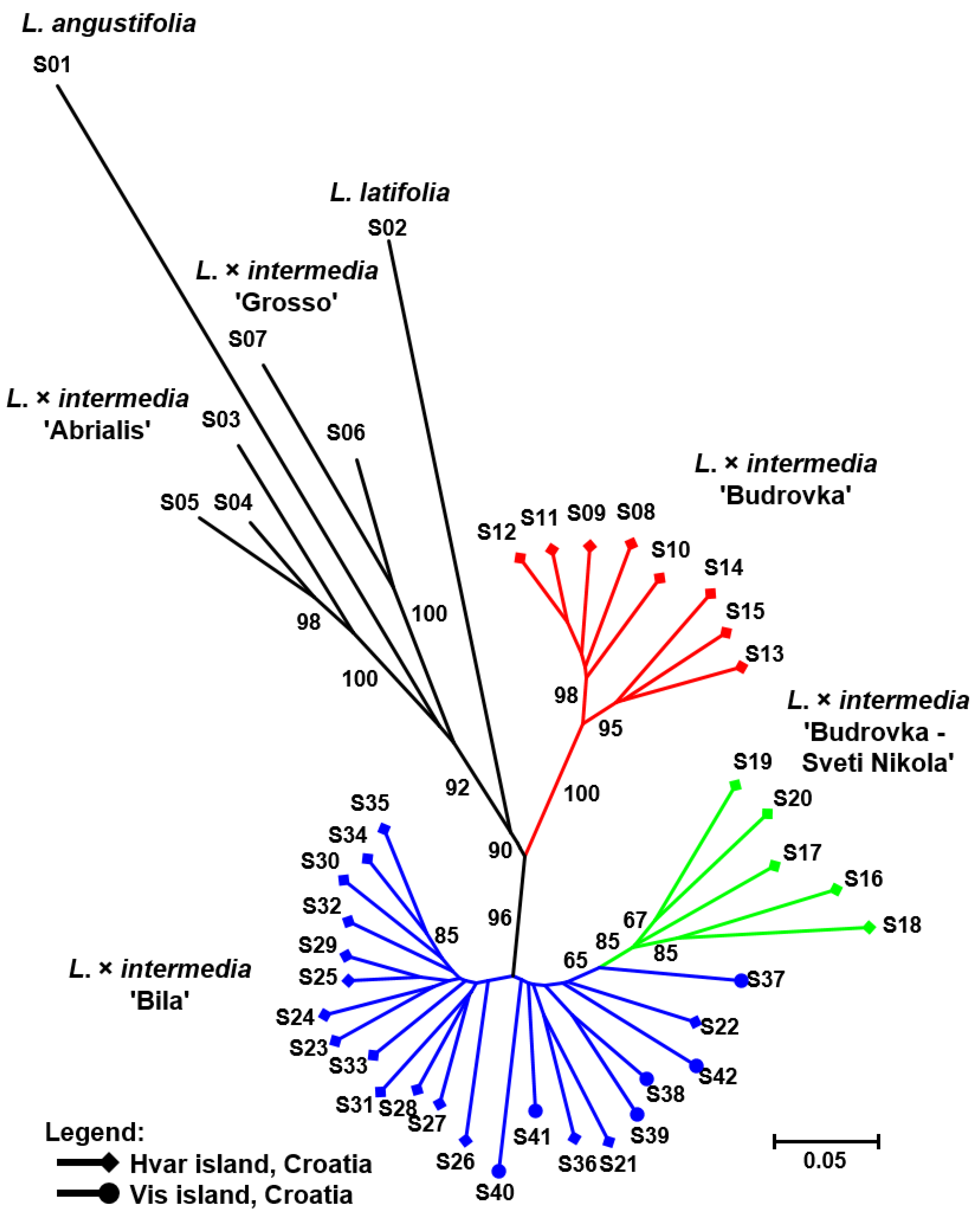
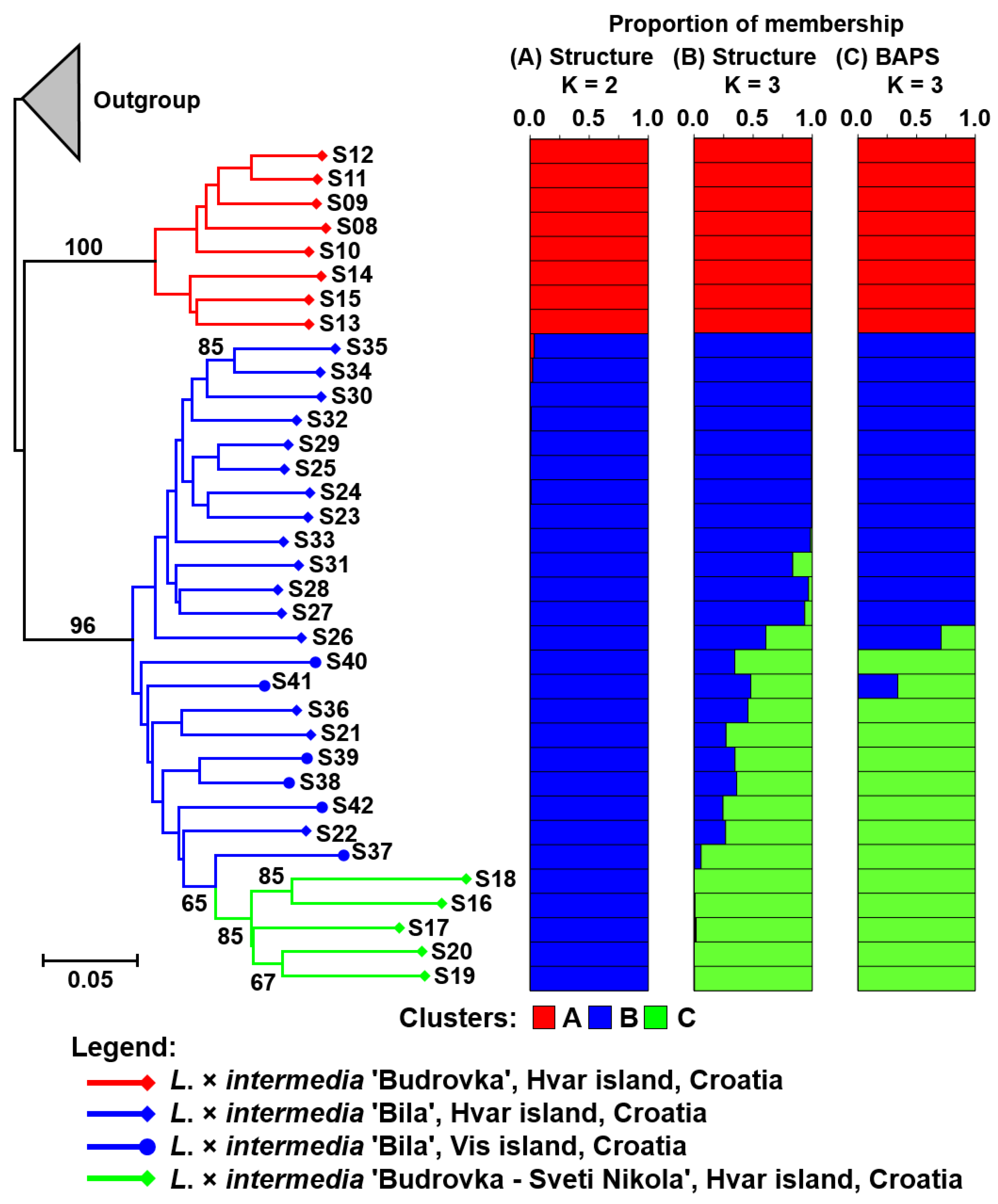
| No. | Landrace | n | %P | Npr | Ia | SDice |
|---|---|---|---|---|---|---|
| 1 | ‘Budrovka’ | 8 | 56.87 | 21 | 0.445 b | 0.855 a |
| 2 | ‘Budrovka Sveti Nikola’ | 5 | 48.47 | 1 | 0.408 b | 0.818 c |
| 3 | ‘Bila’ | 22 | 76.34 | 11 | 0.547 a | 0.838 b |
| P(KW) | *** | ** | ||||
| 3.1 | - ‘Bila’, Island of Hvar | 16 | 73.28 | 7 | 0.513 a | 0.851 a |
| 3.2 | - ‘Bila’, Island of Vis | 6 | 51.15 | 0 | 0.419 b | 0.843 a |
| P(KW) | * | ns |
| Analysis | Source of Variation | df | Variance Components | % Total Variation | φST | P(φST) |
|---|---|---|---|---|---|---|
| (A) | Among landraces | 2 | 25.44 | 43.37 | 0.434 | <0.0001 |
| Within landraces | 32 | 33.27 | 56.63 | |||
| (B) | Between islands | 1 | 5.44 | 14.50 | 0.145 | 0.0004 |
| Within islands | 20 | 32.08 | 85.50 |
| No. | Landrace | Landrace | |||
|---|---|---|---|---|---|
| 1 | 2 | 3.1 | 3.2 | ||
| 1 | ‘Budrovka’ | *** | *** | *** | |
| 2 | ‘Budrovka Sveti Nikola’ | 0.577 | *** | ** | |
| 3.1 | ‘Bila’, Island of Hvar | 0.480 | 0.337 | *** | |
| 3.2 | ‘Bila’, Island of Vis | 0.531 | 0.176 | 0.145 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jug-Dujaković, M.; Ninčević Runjić, T.; Grdiša, M.; Liber, Z.; Šatović, Z. Intra- and Inter-Cultivar Variability of Lavandin (Lavandula × intermedia Emeric ex Loisel.) Landraces from the Island of Hvar, Croatia. Agronomy 2022, 12, 1864. https://doi.org/10.3390/agronomy12081864
Jug-Dujaković M, Ninčević Runjić T, Grdiša M, Liber Z, Šatović Z. Intra- and Inter-Cultivar Variability of Lavandin (Lavandula × intermedia Emeric ex Loisel.) Landraces from the Island of Hvar, Croatia. Agronomy. 2022; 12(8):1864. https://doi.org/10.3390/agronomy12081864
Chicago/Turabian StyleJug-Dujaković, Marija, Tonka Ninčević Runjić, Martina Grdiša, Zlatko Liber, and Zlatko Šatović. 2022. "Intra- and Inter-Cultivar Variability of Lavandin (Lavandula × intermedia Emeric ex Loisel.) Landraces from the Island of Hvar, Croatia" Agronomy 12, no. 8: 1864. https://doi.org/10.3390/agronomy12081864
APA StyleJug-Dujaković, M., Ninčević Runjić, T., Grdiša, M., Liber, Z., & Šatović, Z. (2022). Intra- and Inter-Cultivar Variability of Lavandin (Lavandula × intermedia Emeric ex Loisel.) Landraces from the Island of Hvar, Croatia. Agronomy, 12(8), 1864. https://doi.org/10.3390/agronomy12081864







