Bilinear Attention Network for Image-Based Fine-Grained Recognition of Oil Tea (Camellia oleifera Abel.) Cultivars
Abstract
:1. Introduction
2. Materials and Methods
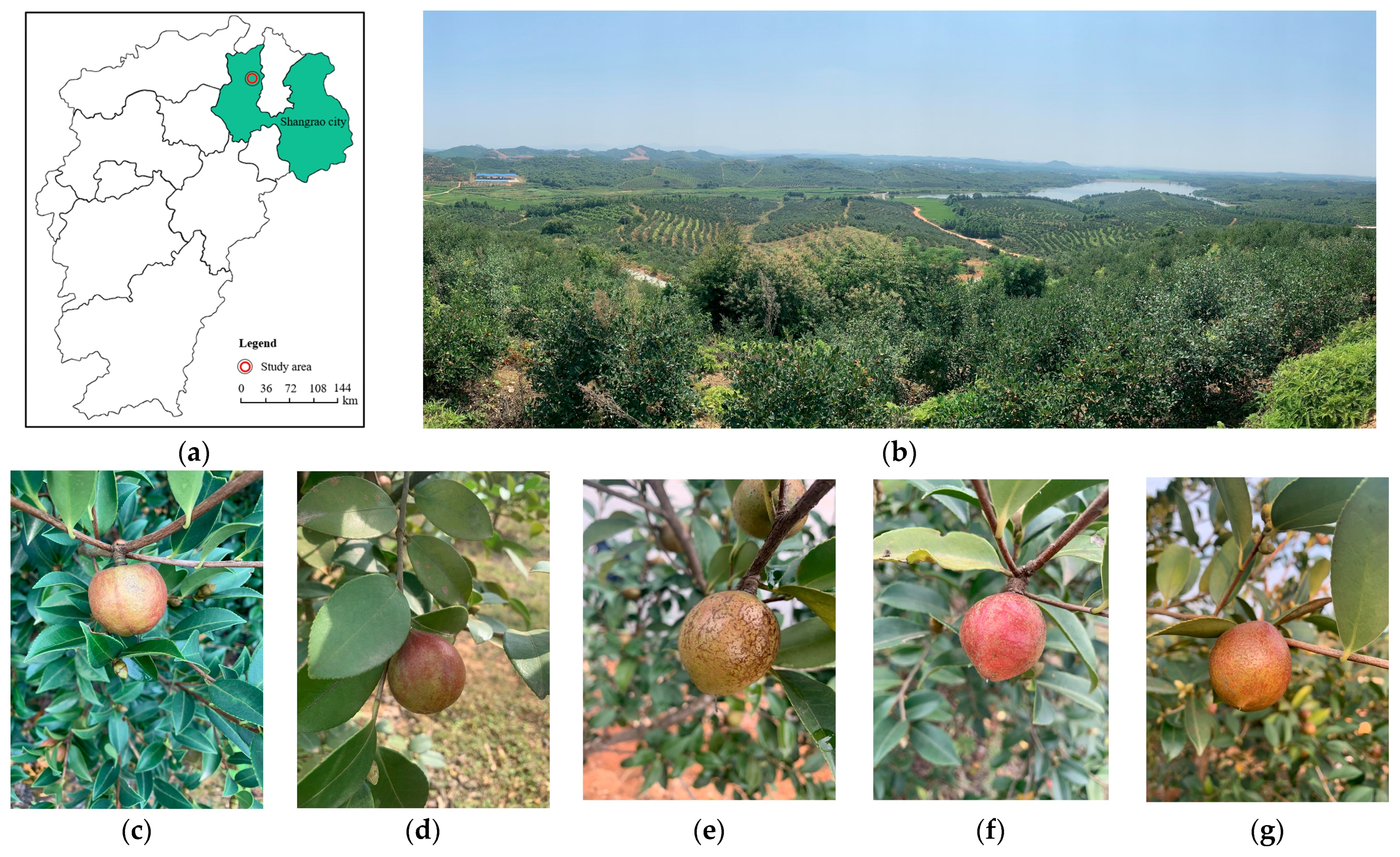
2.1. Study Area
2.2. Data Collection and Dataset Construction
2.3. Fine-Grained Oil Tea Cultivar Recognition Model
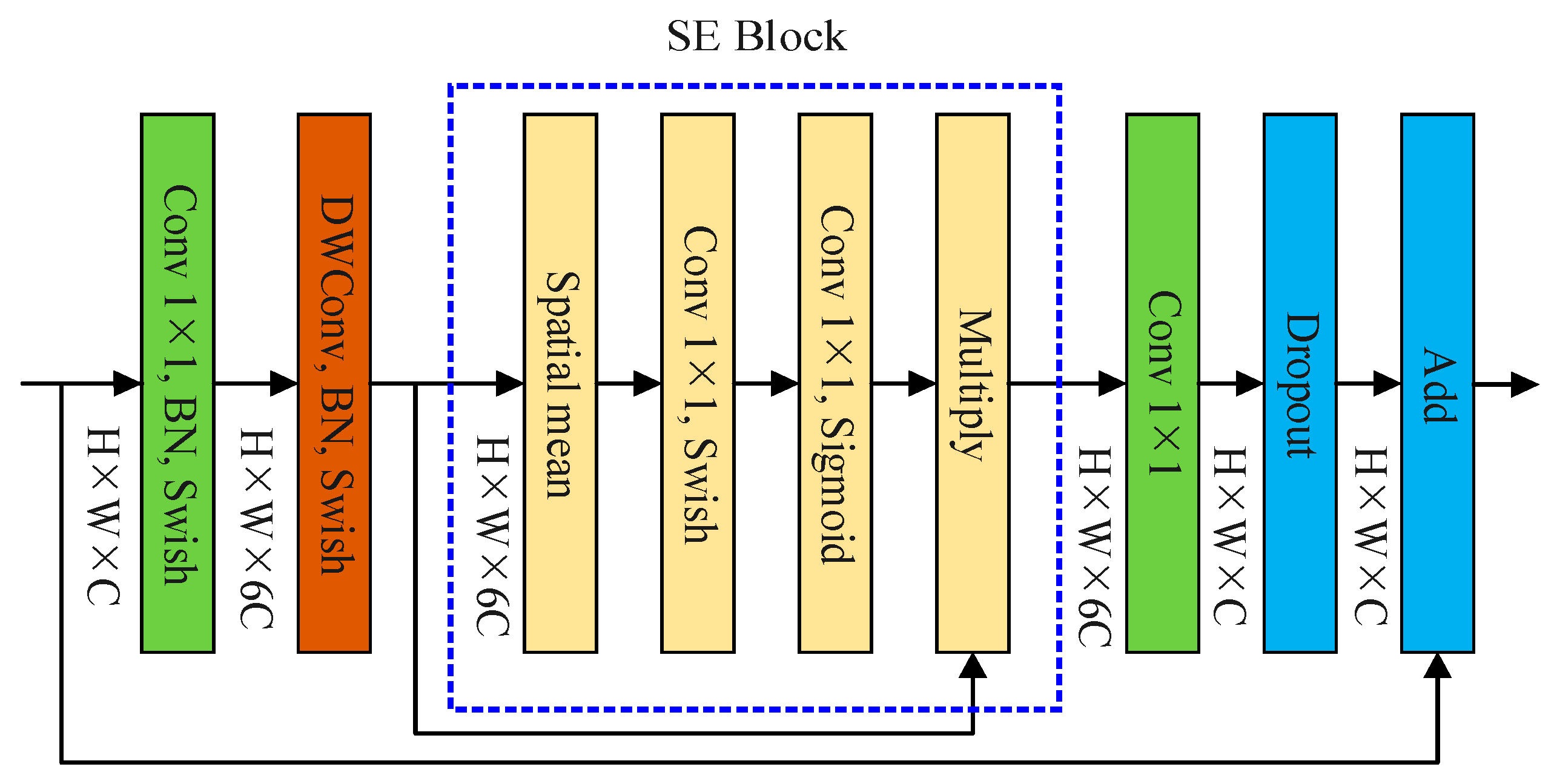
2.3.1. EfficientNet-B0 Network
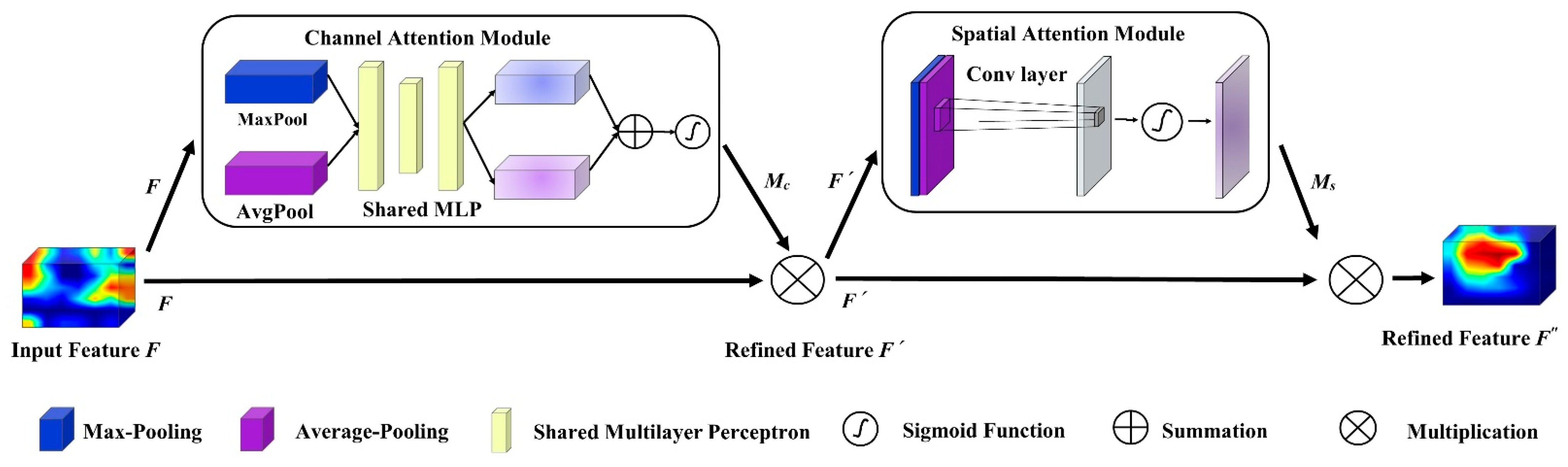
2.3.2. CBAM Module
2.3.3. Bilinear Pooling
3. Results
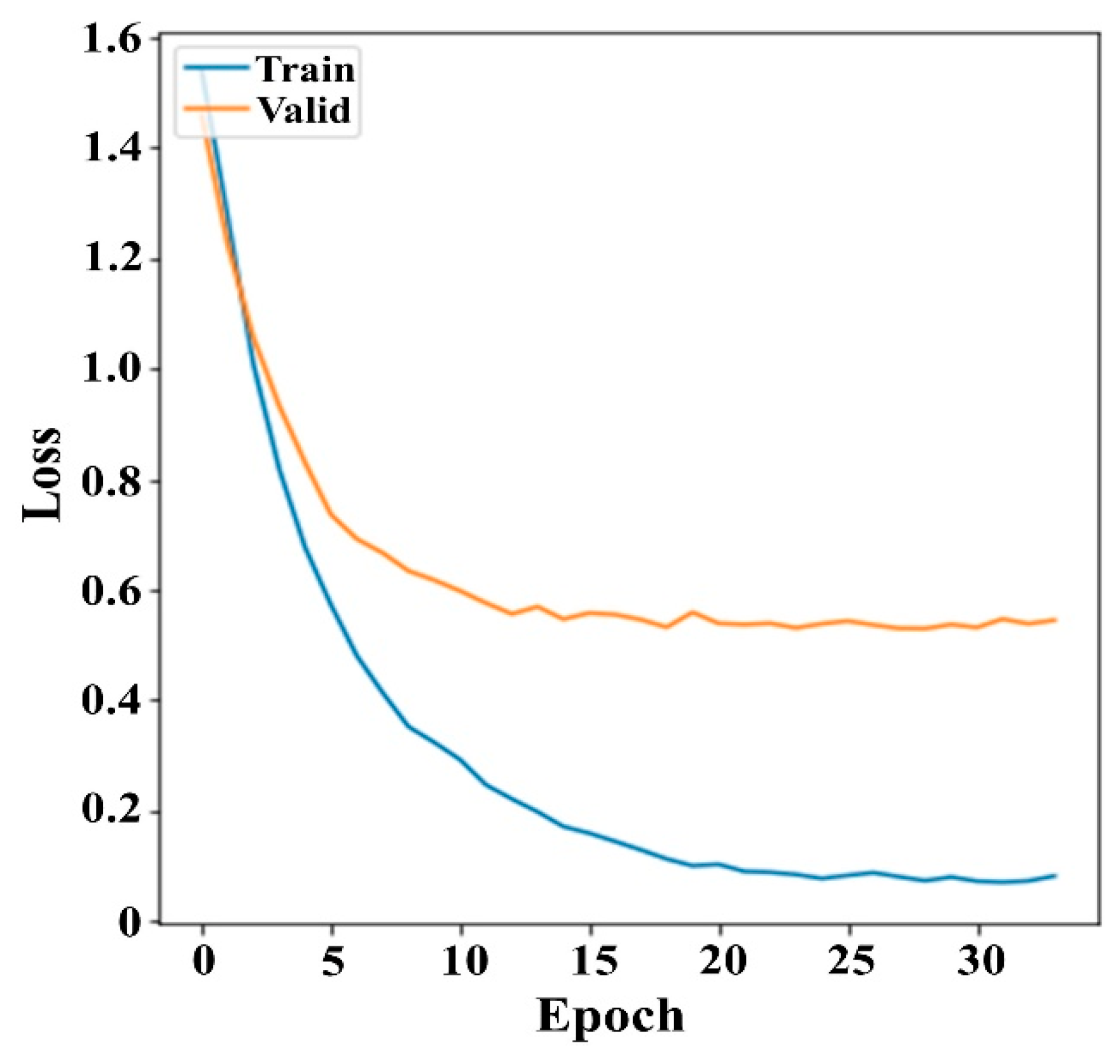
3.1. Experiment Setup
3.2. Cultivar Recognition Results and Analysis
4. Discussion
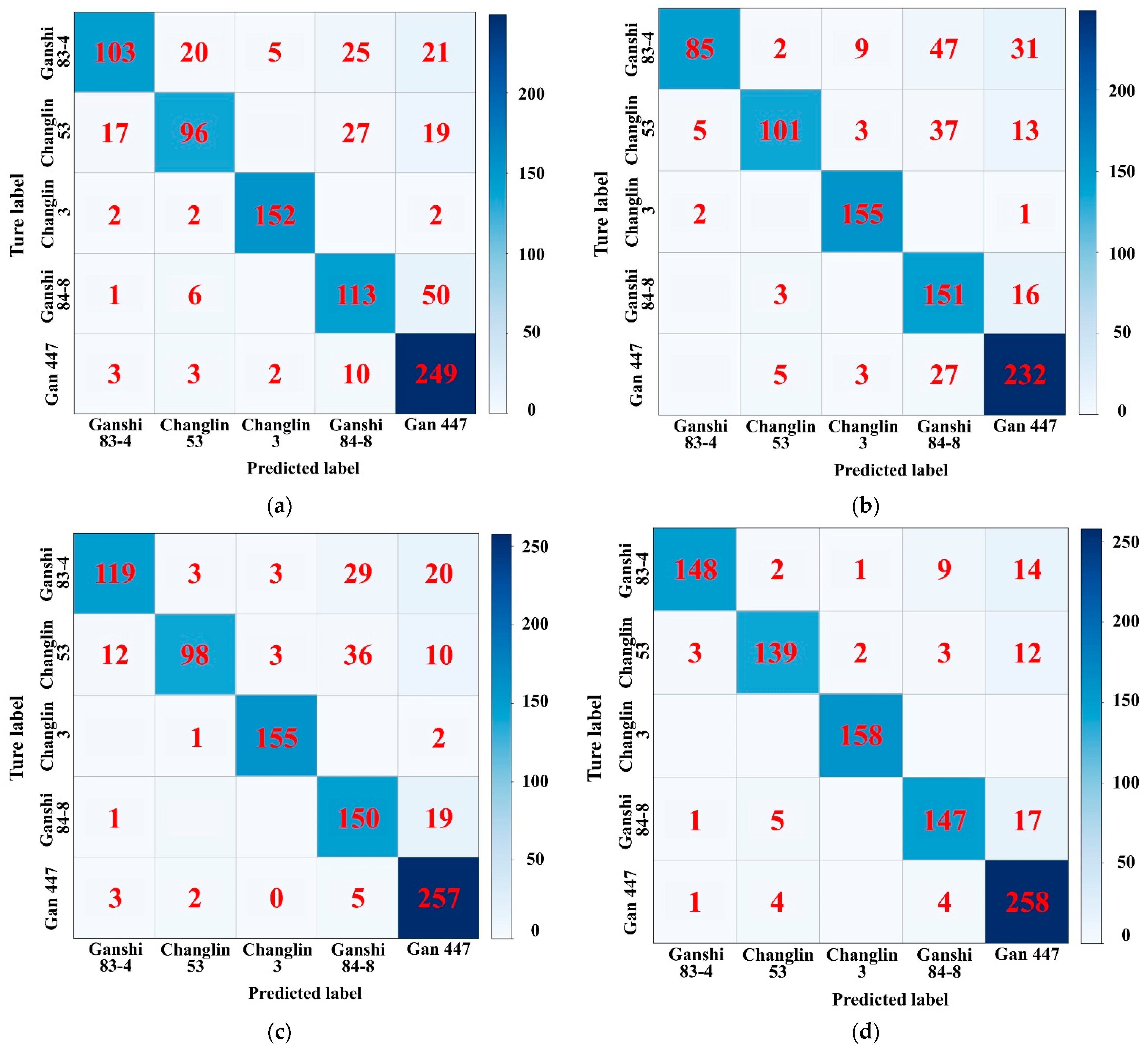
4.1. Comparison and Analysis of Cultivar Recognition Results
4.2. Ablation Experiments
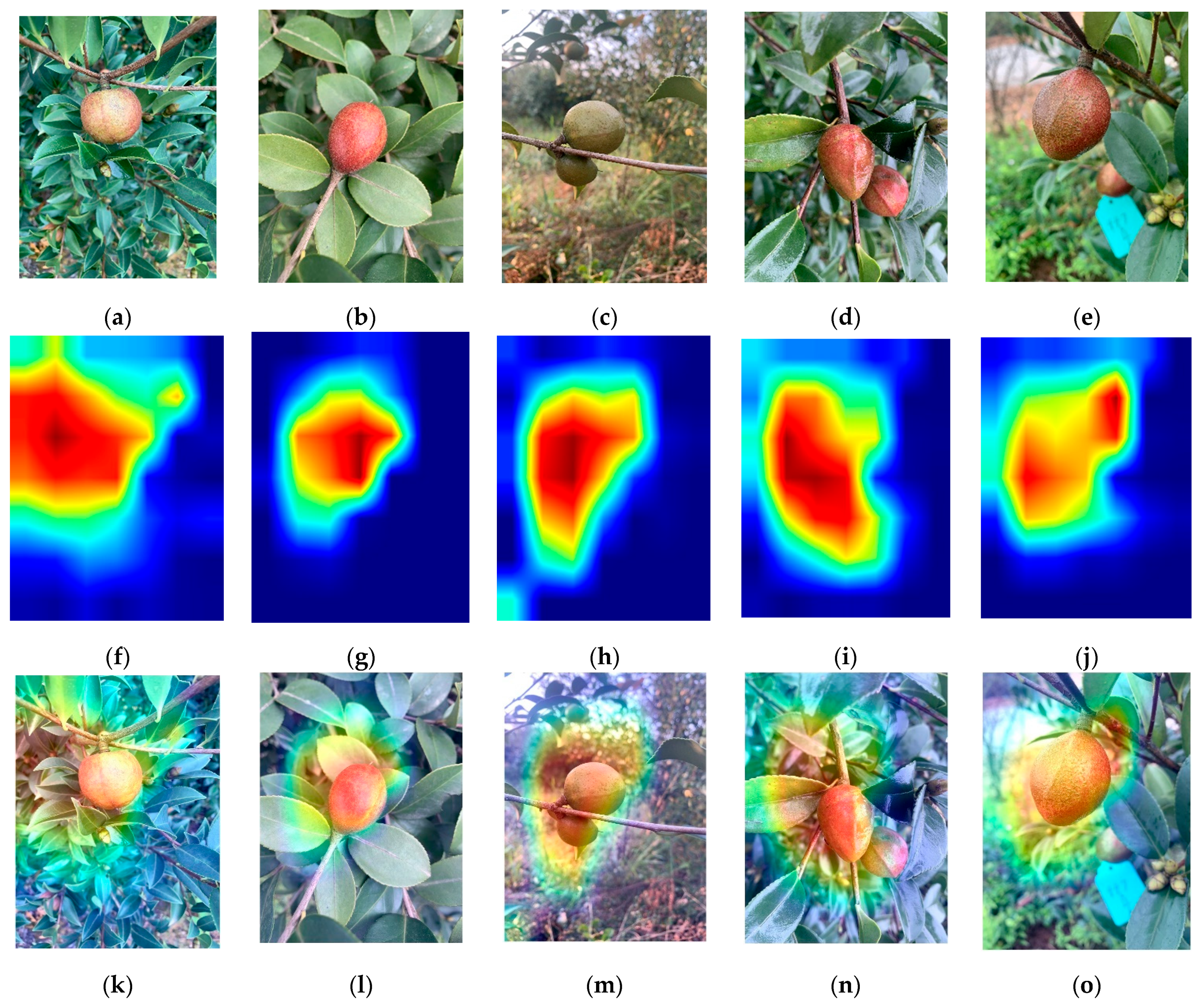
4.3. Visual Analysis of Oil Tea Cultivar Recognition
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Deng, Q.; Li, J.; Gao, C.; Cheng, J.; Deng, X.; Jiang, D.; Li, L.; Yan, P. New perspective for evaluating the main Camellia oleifera cultivars in China. Sci. Rep. 2020, 10, 20676. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, L.; Tang, W.; Peng, S.; Li, M.; Deng, N.; Chen, Y. Predicting potential distribution and evaluating suitable soil condition of oil tea Camellia in China. Forests 2018, 9, 487. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Li, Z.; Zhou, J.; Gu, Y.; Tan, X. Comparative study on fruit development and oil synthesis in two cultivars of Camellia oleifera. BMC Plant Biol. 2021, 21, 348. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Su, S.; Ma, L.; Yang, S.; Wang, Y.; Wang, X. Effects of canopy microclimate on fruit yield and quality of Camellia oleifera. Sci. Hortic. 2018, 235, 132–141. [Google Scholar] [CrossRef]
- Zeng, W.; Endo, Y. Effects of cultivars and geography in China on the lipid characteristics of Camellia oleifera seeds. J. Oleo Sci. 2019, 68, 1051–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Jiang, D.; Cheng, H.; Zhou, X.; Fang, Y.; Zhang, X.; Xiao, X.; Deng, X.; Li, L. Determination of Camellia oleifera Abel. germplasm resources of genetic diversity in China using ISSR markers. Not. Bot. Horti Agrobot. Cluj-Napoca. 2018, 46, 501–508. [Google Scholar] [CrossRef] [Green Version]
- Chao, W.; Tang, C.; Zhang, J.; Yu, L.; Yoichi, H. Development of a stable SCAR marker for rapid identification of Ganoderma lucidum Hunong 5 cultivar using DNA pooling method and inter-simple sequence repeat markers. J. Integr. Agric. 2018, 17, 130–138. [Google Scholar] [CrossRef]
- Ding, P.; Zhou, H.; Shang, J.; Zou, X.; Wang, M. Object detection via flexible anchor generation. Int. J. Pattern Recogn. 2021, 35, 2155012. [Google Scholar] [CrossRef]
- Tang, Y.; Cheng, Z.; Miao, A.; Zhuang, J.; Hou, C.; He, Y.; Chu, X.; Luo, S. Evaluation of cultivar identification performance using feature expressions and classification algorithms on optical images of sweet corn seeds. Agronomy 2020, 10, 1268. [Google Scholar] [CrossRef]
- Kim, M.; Jung, J.; Shim, E.; Chung, S.; Park, Y.; Lee, G.; Sim, S. Genome-wide SNP discovery and core marker sets for DNA barcoding and variety identification in commercial tomato cultivars. Sci. Hortic. 2021, 276, 109734. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, B.; Chen, J.; Wang, X.; Wang, R.; Peng, S.; Chen, L.; Ma, L.; Luo, J. Identification of rubisco rbcL and rbcS in Camellia oleifera and their potential as molecular markers for selection of high tea oil cultivars. Front Plant Sci. 2015, 6, 189. [Google Scholar] [CrossRef] [Green Version]
- Calzone, A.; Cotrozzi, L.; Lorenzini, G.; Nali, C.; Pellegrini, E. Hyperspectral detection and monitoring of salt stress in pomegranate cultivars. Agronomy 2021, 11, 1038. [Google Scholar] [CrossRef]
- Naeem, S.; Ali, A.; Chesneau, C.; Tahir, M.H.; Jamal, F.; Sherwani, R.A.K.; Ul Hassan, M. The classification of medicinal plant leaves based on multispectral and texture feature using machine learning approach. Agronomy 2021, 11, 263. [Google Scholar] [CrossRef]
- Zhu, S.; Zhang, J.; Chao, M.; Xu, X.; Song, P.; Zhang, J.; Huang, Z. A rapid and highly efficient method for the identification of soybean seed varieties: Hyperspectral images combined with transfer learning. Molecules. 2020, 25, 152. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Wang, L.; Sun, D.; Zeng, X.; Qu, J.; Ma, J. Lychee variety discrimination by hyperspectral imaging coupled with multivariate classification. Food Anal. Methods 2014, 7, 1848–1857. [Google Scholar] [CrossRef]
- Li, H.; Zhang, L.; Sun, H.; Rao, Z.; Ji, H. Identification of soybean varieties based on hyperspectral imaging technology and one-dimensional convolutional neural network. J. Food Process Eng. 2021, 44, e13767. [Google Scholar] [CrossRef]
- Zhang, J.; Dai, L.; Cheng, F. Corn seed variety classification based on hyperspectral reflectance imaging and deep convolutional neural network. Food Meas. 2021, 15, 484–494. [Google Scholar] [CrossRef]
- Liu, Y.; Su, J.; Shen, L.; Lu, N.; Fang, Y.; Liu, F.; Song, Y.; Su, B. Development of a mobile application for identification of grapevine (Vitis vinifera L.) cultivars via deep learning. Int. J. Agric. Biol. Eng. 2021, 14, 172–179. [Google Scholar] [CrossRef]
- Kaya, A.; Keceli, A.; Catal, C.; Yalic, H.; Temucin, H.; Tekinerdogan, B. Analysis of transfer learning for deep neural network based plant classification models. Comput. Electron Agric. 2019, 158, 20–29. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, Y.; Yan, T.; Bai, X.; Xiao, Q.; Gao, P.; Li, M.; Huang, W.; Bao, Y.; He, Y.; et al. Application of near-infrared hyperspectral imaging for variety identification of coated maize kernels with deep learning. Infrared Phys. Technol. 2020, 111, 103550. [Google Scholar] [CrossRef]
- Abbaspour-Gilandeh, Y.; Molaee, A.; Sabzi, S.; Nabipur, N.; Shamshirband, S.; Mosavi, A. A combined method of image processing and artificial neural network for the identification of 13 Iranian rice cultivars. Agronomy 2020, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Osako, Y.; Yamane, H.; Lin, S.; Chen, P.; Tao, R. Cultivar discrimination of litchi fruit images using deep learning. Sci. Hortic. 2020, 269, 109360. [Google Scholar] [CrossRef]
- Barré, P.; Stöver, B.; Müller, K.; Steinhage, V. LeafNet: A computer vision system for automatic plant species identification. Ecol. Inform. 2017, 40, 50–56. [Google Scholar] [CrossRef]
- Taner, A.; Öztekin, Y.B.; Duran, H. Performance analysis of deep learning CNN models for variety classification in hazelnut. Sustainability 2021, 13, 6527. [Google Scholar] [CrossRef]
- Franczyk, B.; Hernes, M.; Kozierkiewicz, A.; Kozina, A.; Pietranik, M.; Roemer, I.; Schieck, M. Deep learning for grape variety recognition. Procedia Comput. Sci. 2020, 176, 1211–1220. [Google Scholar] [CrossRef]
- Nasiri, A.; Taheri-Garavand, A.; Fanourakis, D.; Zhang, Y.; Nikoloudakis, N. Automated grapevine cultivar identification via leaf imaging and deep convolutional neural networks: A proof-of-concept study employing primary iranian varieties. Plants 2021, 10, 1628. [Google Scholar] [CrossRef]
- Yang, G.; He, Y.; Yang, Y.; Xu, B. Fine-grained image classification for crop disease based on attention mechanism. Front. Plant Sci. 2020, 11, 600854. [Google Scholar] [CrossRef]
- Zhang, C.; Li, T.; Zhang, W. The detection of impurity content in machine-picked seed cotton based on image processing and improved YOLO V4. Agronomy 2022, 12, 66. [Google Scholar] [CrossRef]
- Su, W.-H.; Zhang, J.; Yang, C.; Page, R.; Szinyei, T.; Hirsch, C.D.; Steffenson, B.J. Automatic evaluation of wheat resistance to fusarium head blight using dual Mask-RCNN deep learning frameworks in computer vision. Remote Sens. 2021, 13, 26. [Google Scholar] [CrossRef]
- Kumar, M.; Gupta, S.; Gao, X.; Singh, A. Plant species recognition using morphological features and adaptive boosting methodology. IEEE Access 2019, 7, 163912–163918. [Google Scholar] [CrossRef]
- Mi, Z.; Zhang, X.; Su, J.; Han, D.; Su, B. Wheat stripe rust grading by deep learning with attention mechanism and images from mobile devices. Front. Plant Sci. 2020, 11, 558126. [Google Scholar] [CrossRef]
- Pang, C.; Wang, W.; Lan, R.; Shi, Z.; Luo, X. Bilinear pyramid network for flower species categorization. Multimed. Tools Appl. 2021, 80, 215–225. [Google Scholar] [CrossRef]
- Rzanny, M.; Mäder, P.; Deggelmann, A.; Chen, M.; Widchen, J. Flowers, leaves or both? How to obtain suitable images for automated plant identification. Plant Methods 2019, 15, 77. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L. MobileNetV2: Inverted residuals and linear bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference of Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.; Park, J.; Lee, J.; Kweon, I. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Zhong, X.; Shih, F. Joint learning for pneumonia classification and segmentation on medical images. Int. J. Pattern Recogn. 2021, 35, 2157003. [Google Scholar] [CrossRef]
- Men, H.; Yuan, H.; Shi, Y.; Liu, M.; Wang, Q.; Liu, J. A residual network with attention module for hyperspectral information of recognition to trace the origin of rice. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 263, 120155. [Google Scholar] [CrossRef]
- Duong, L.; Nguyen, P.; Sipio, C.; Ruscio, D. Automated fruit recognition using EfficientNet and MixNet. Comput. Electron Agric. 2020, 171, 105326. [Google Scholar] [CrossRef]
- Liu, J.; Wang, M.; Bao, L.; Li, X. EfficientNet based recognition of maize diseases by leaf image classification. J. Phys. Conf. Ser. 2020, 1693, 012148. [Google Scholar] [CrossRef]
- Zhang, P.; Yang, L.; Li, D. EfficientNet-B4-Ranger: A novel method for greenhouse cucumber disease recognition under natural complex environment. Comput. Electron Agric. 2020, 176, 105652. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the European Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference of Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wang, J.; Tian, Y.; Dai, S. Deep learning for image-based large-flowered chrysanthemum cultivar recognition. Plant Methods 2019, 4, 146. [Google Scholar] [CrossRef]
- Mirzazadeh, A.; Azizi, A.; Abbaspour-Gilandeh, Y.; Hernández-Hernández, J.L.; Hernández-Hernández, M.; Gallardo-Bernal, I. A novel technique for classifying bird damage to tapeseed plants based on a deep learning algorithm. Agronomy 2021, 11, 2364. [Google Scholar] [CrossRef]
- Azizi, A.; Gilandeh, Y.A.; Mesri-Gundoshmian, T.; Saleh-Bigdeli, A.A.; Moghaddam, H.A. Classification of soil aggregates: A novel approach based on deep learning. Soil Tillage Res. 2020, 199, 104586. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, X.; Sun, Z.; Zheng, Y.; Su, S.; Chen, F. Identification of oil tea (Camellia oleifera C.Abel) cultivars using EfficientNet-B4 CNN model with attention mechanism. Forests 2022, 13, 1. [Google Scholar] [CrossRef]
- Prakash, A.; Prakasam, P. An intelligent fruits classification in precision agriculture using bilinear pooling convolutional neural networks. Vis. Comput. 2022. [Google Scholar] [CrossRef]
- Wang, P.; Liu, J.; Xu, L.; Huang, P.; Luo, X.; Hu, Y.; Kang, Z. Classification of Amanita species based on bilinear networks with attention mechanism. Agriculture 2021, 11, 393. [Google Scholar] [CrossRef]
- Selvaraju, R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef] [Green Version]

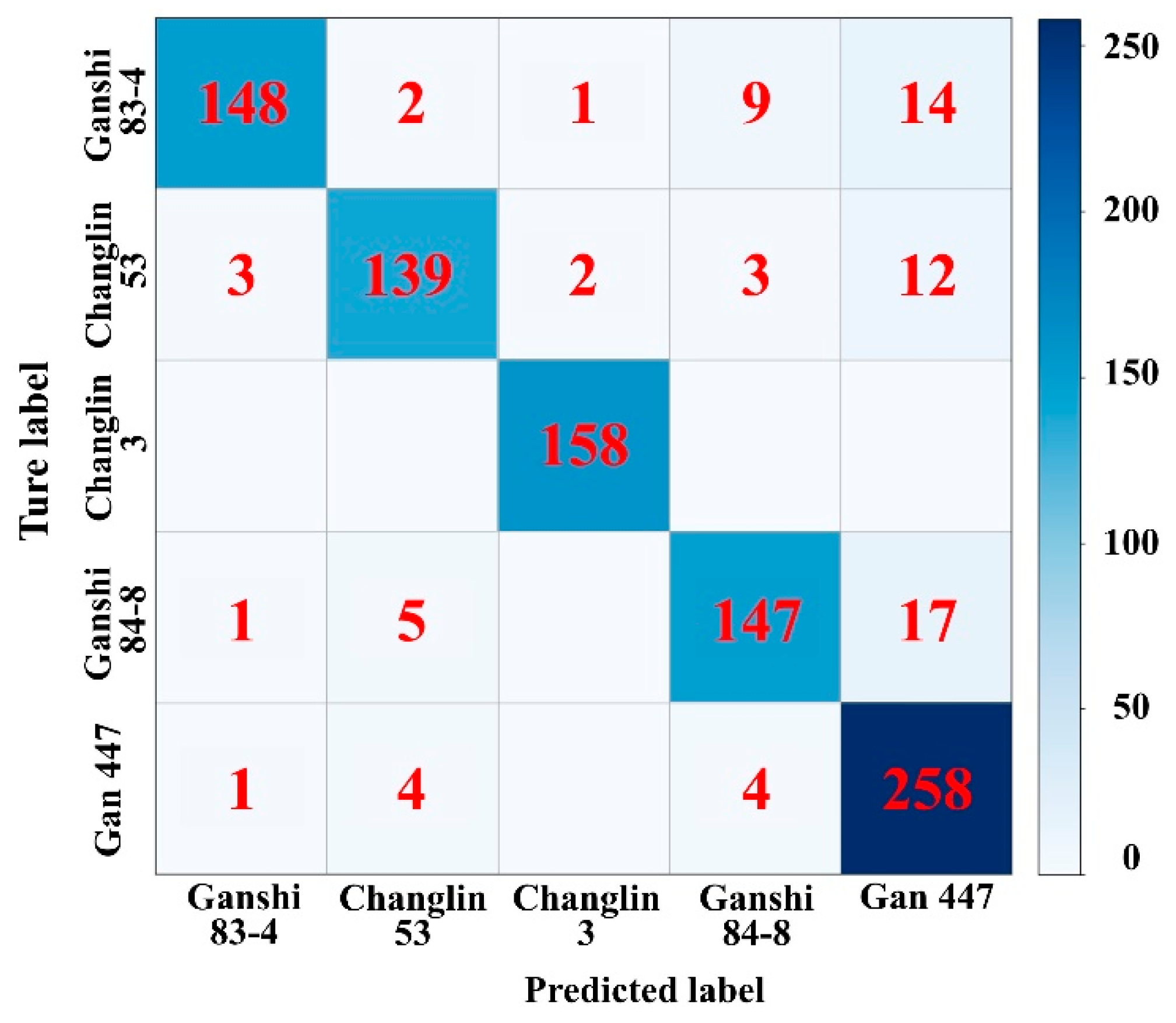
| Cultivar | TP | TN | FP | FN | Acc (%) | P (%) | R (%) | F1-score (%) | OA (%) | Kc |
|---|---|---|---|---|---|---|---|---|---|---|
| Ganshi 83-4 | 148 | 749 | 5 | 26 | 96.66 | 96.73 | 85.06 | 90.52 | 91.59 | 0.89 |
| Changlin 53 | 139 | 758 | 11 | 20 | 96.66 | 92.67 | 87.42 | 89.97 | ||
| Changlin 3 | 158 | 767 | 3 | 0 | 99.68 | 98.14 | 100 | 99.06 | ||
| Ganshi 84-8 | 147 | 742 | 16 | 23 | 95.80 | 90.18 | 86.47 | 88.29 | ||
| Gan 447 | 258 | 618 | 43 | 9 | 94.40 | 85.71 | 96.63 | 90.84 |
| Model | Cultivars | TP | TN | FP | FN | ACC (%) | P (%) | R (%) | F1-score (%) | OA (%) | Kc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| InceptionV3 | Ganshi 83-4 | 103 | 731 | 23 | 71 | 89.87 | 81.75 | 59.20 | 68.67 | 76.83 | 0.70 |
| Changlin 53 | 96 | 738 | 31 | 63 | 89.87 | 75.59 | 60.38 | 67.13 | |||
| Changlin 3 | 152 | 763 | 7 | 6 | 98.60 | 95.60 | 96.20 | 95.90 | |||
| Ganshi 84-8 | 113 | 696 | 62 | 57 | 87.18 | 64.57 | 66.47 | 65.51 | |||
| Gan 447 | 249 | 569 | 92 | 18 | 88.15 | 73.02 | 93.26 | 81.91 | |||
| VGG16 | Ganshi 83-4 | 85 | 747 | 7 | 89 | 89.66 | 92.39 | 48.85 | 63.91 | 78.02 | 0.72 |
| Changlin 53 | 101 | 759 | 10 | 58 | 92.67 | 90.99 | 63.52 | 74.81 | |||
| Changlin 3 | 155 | 755 | 15 | 3 | 98.06 | 91.18 | 98.10 | 94.51 | |||
| Ganshi 84-8 | 151 | 647 | 111 | 19 | 85.99 | 57.63 | 88.82 | 69.90 | |||
| Gan 447 | 232 | 600 | 61 | 35 | 89.66 | 79.18 | 86.89 | 82.86 | |||
| ResNet50 | Ganshi 83-4 | 119 | 738 | 16 | 55 | 92.35 | 88.15 | 68.39 | 77.02 | 83.94 | 0.80 |
| Changlin 53 | 98 | 763 | 6 | 61 | 92.78 | 94.23 | 61.64 | 74.53 | |||
| Changlin 3 | 155 | 764 | 6 | 3 | 99.03 | 96.27 | 98.10 | 97.18 | |||
| Ganshi 84-8 | 150 | 688 | 70 | 20 | 90.30 | 68.18 | 88.24 | 76.92 | |||
| Gan 447 | 257 | 610 | 51 | 10 | 93.43 | 83.44 | 96.25 | 89.39 | |||
| BA-EfficientNet | Ganshi 83-4 | 148 | 749 | 5 | 26 | 96.66 | 96.73 | 85.06 | 90.52 | 91.59 | 0.89 |
| Changlin 53 | 139 | 758 | 11 | 20 | 96.66 | 92.67 | 87.42 | 89.97 | |||
| Changlin 3 | 158 | 767 | 3 | 0 | 99.68 | 98.14 | 100 | 99.06 | |||
| Ganshi 84-8 | 147 | 742 | 16 | 23 | 95.80 | 90.18 | 86.47 | 88.29 | |||
| Gan 447 | 258 | 618 | 43 | 9 | 94.40 | 85.71 | 96.63 | 90.84 |
| Model | Cultivars | TP | TN | FP | FN | ACC (%) | P (%) | R (%) | F1-score (%) | OA (%) | Kc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EfficientNet-B0 | Ganshi 83-4 | 133 | 743 | 11 | 41 | 94.40 | 92.36 | 76.44 | 83.65 | 86.75 | 0.83 |
| Changlin 53 | 132 | 753 | 16 | 27 | 95.37 | 89.19 | 83.02 | 85.99 | |||
| Changlin 3 | 158 | 759 | 11 | 0 | 98.81 | 93.49 | 100 | 96.64 | |||
| Ganshi 84-8 | 151 | 707 | 51 | 19 | 92.46 | 74.75 | 88.82 | 81.18 | |||
| Gan 447 | 231 | 627 | 34 | 36 | 92.46 | 87.17 | 86.52 | 86.84 | |||
| EfficientNet-CBAM | Ganshi 83-4 | 136 | 750 | 4 | 38 | 95.47 | 97.14 | 78.16 | 86.62 | 88.36 | 0.85 |
| Changlin 53 | 126 | 765 | 4 | 33 | 96.01 | 96.92 | 79.25 | 87.20 | |||
| Changlin 3 | 158 | 762 | 8 | 0 | 99.14 | 95.18 | 100 | 97.53 | |||
| Ganshi 84-8 | 144 | 732 | 26 | 26 | 94.40 | 84.71 | 84.71 | 84.71 | |||
| Gan 447 | 256 | 595 | 66 | 11 | 91.70 | 79.50 | 95.88 | 86.93 | |||
| Bilinear EfficientNet | Ganshi 83-4 | 138 | 745 | 9 | 36 | 95.15 | 93.88 | 79.31 | 85.98 | 88.90 | 0.86 |
| Changlin 53 | 136 | 751 | 18 | 23 | 95.58 | 88.31 | 85.53 | 86.90 | |||
| Changlin 3 | 158 | 765 | 5 | 0 | 99.46 | 96.93 | 100 | 98.44 | |||
| Ganshi 84-8 | 153 | 728 | 30 | 17 | 94.94 | 83.61 | 90 | 86.69 | |||
| Gan 447 | 240 | 620 | 41 | 27 | 92.67 | 85.41 | 89.89 | 87.59 | |||
| BA-EfficientNet | Ganshi 83-4 | 148 | 749 | 5 | 26 | 96.66 | 96.73 | 85.06 | 90.52 | 91.59 | 0.89 |
| Changlin 53 | 139 | 758 | 11 | 20 | 96.66 | 92.67 | 87.42 | 89.97 | |||
| Changlin 3 | 158 | 767 | 3 | 0 | 99.68 | 98.14 | 100 | 99.06 | |||
| Ganshi 84-8 | 147 | 742 | 16 | 23 | 95.80 | 90.18 | 86.47 | 88.29 | |||
| Gan 447 | 258 | 618 | 43 | 9 | 94.40 | 85.71 | 96.63 | 90.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Yu, Y.; Zheng, Y.; Su, S.; Chen, F. Bilinear Attention Network for Image-Based Fine-Grained Recognition of Oil Tea (Camellia oleifera Abel.) Cultivars. Agronomy 2022, 12, 1846. https://doi.org/10.3390/agronomy12081846
Zhu X, Yu Y, Zheng Y, Su S, Chen F. Bilinear Attention Network for Image-Based Fine-Grained Recognition of Oil Tea (Camellia oleifera Abel.) Cultivars. Agronomy. 2022; 12(8):1846. https://doi.org/10.3390/agronomy12081846
Chicago/Turabian StyleZhu, Xueyan, Yue Yu, Yili Zheng, Shuchai Su, and Fengjun Chen. 2022. "Bilinear Attention Network for Image-Based Fine-Grained Recognition of Oil Tea (Camellia oleifera Abel.) Cultivars" Agronomy 12, no. 8: 1846. https://doi.org/10.3390/agronomy12081846
APA StyleZhu, X., Yu, Y., Zheng, Y., Su, S., & Chen, F. (2022). Bilinear Attention Network for Image-Based Fine-Grained Recognition of Oil Tea (Camellia oleifera Abel.) Cultivars. Agronomy, 12(8), 1846. https://doi.org/10.3390/agronomy12081846







