Abstract
β-amylase activity is related to the polymorphisms of Bmy1 intron III; however, no attention has been given to such relationships under environmental stresses such as drought. In this study, 73 cultivated barley genotypes and 52 Tibetan wild barley accessions were used to test the association between Bmy1 gene intron III polymorphisms and β-amylase activity under drought stress. Our results showed that three alleles, Bmy1.a, Bmy1.b, and Bmy1.c, existed in the examined barley genotypes. Tibetan wild barley had a higher proportion of Bmy1.b, whereas cultivated barley showed a higher proportion of Bmy1.a. Impressively, barley genotypes with Bmy1.b showed a significant increase in β-amylase activity under drought stress, compared with those with Bmy1.a or Bmy1.c, indicating that the Bmy1.b allele might provide more chances for developing barley cultivars with higher β-amylase activity under water stress than both the Bmy1.a and Bmy1.c alleles. Furthermore, the Tibetan wild barley XZ147, belonging to the Bmy1.b allele type, showed significantly higher β-amylase activity than Triumph under drought stress. This might be the result of the unique amino acid substitution M527 or the amino acid composition of R115, D165, A233, S347, and M527 of XZ147.
1. Introduction
Barley (Hordeum vulgare L.) is widely used for malting to produce beer and whisky. In the brewing industry, one of the most important malt quality parameters is diastatic power (DP), which refers to the starch-degrading ability of barley. DP represents a general ability of four major enzymes, i.e., α-amylase, β-amylase, limit dextrinase, and α-glucosidase. β-amylase (EC 3.2.1.2; 1, 4-β-glucan maltohydrolase), which catalyzes β-maltose released from the non-reducing end of poly-glucan chain, is the predominant enzyme affecting DP [1,2,3].
Genes controlling β-amylase synthesis differ among plant tissues; however, their functional domain is similar because all of them belong to the same gene family. In malting barley, β-amylase activity is an important quality trait, being closely associated with malt quality [2]. There are two forms of β-amylase in barley. The endosperm-specific form is the dominant one, which is encoded by the Bmy1 gene, located on the long arm telomere of the 4H chromosome [4,5]. It consists of 7 extrons and 6 introns, and encodes a polypeptide chain of 535 amino acids [6,7]. The Bmy1 gene is closely correlated with DP [8]. The polymorphisms in the intron III of Bmy1 are extremely abundant. There are four different types according to the presence and absence of four insertion/deletions (INDELs) (126-, 38-, 11-, and 21-bp), namely Bmy1.a, Bmy1.b, Bmy1.c, and Bmy1.d. Erkkilä et al. [9,10] identified two different indels (126-bp and 38-bp) through Southern blot analysis using the first 320 bp located in the 5′ region of Bmy1 intron III. Currently, all these four types derived from Bmy1 intron III INDELs have been identified, including the insertion of 126-bp and 38-bp in the cultivated barley Adorra, and the deletion of 126-bp and 38-bp INDELs in the wild barley PI 296,897 [9]. Sjakste and Zhuk [11] also observed abundant polymorphisms of Bmy1 intron III and found a potential binding site for a transcript factor. Numerous studies have confirmed that the polymorphisms of Bmy1 intron III are correlated with the enzyme activity, thermal stability, and enzymatic kinetics of β-amylase [9,12,13,14,15]. Erkkilä and Ahokas [13] and Gunkel et al. [16] reported that the presence or deletion of a 126-bp INDEL in the 5′ end of Bmy1 intron III was associated with low activity and high thermal stability of β-amylase, respectively. Coventry et al. [17] determined the activity and thermal stability of β-amylase and the DP value and identified a primer pair, which could discriminate between the presence or absence of the 126-bp INDEL. Meanwhile, the single-nucleotide polymorphisms (SNPs) in the coding region of Bmy1 also affected the morphology, activity, and thermal stability of β-amylase [18]. All these studies indicate that it is practicable to identify the enzyme activity and thermal stability of β-amylase only by detecting the polymorphisms of either the coding or noncoding regions of Bmy1 under normal environmental conditions. However, little attention has been given to this issue under environmental stresses such as drought.
Tibetan wild barley is considered as one of ancestors of modern cultivated barley and is rich in genetic diversity [19]. However, most studies about wild barley have mainly focused on salinity tolerance [20], aluminum tolerance [21], and grain protein content [22], with no research investigating the genetic variations in the malt quality under drought stress. In this study, we used 73 cultivated and 52 Tibetan wild barley genotypes to investigate the correlation between Bmy1 intron III polymorphisms and grain β-amylase activity under drought stress and compare the differences between wild and cultivated barley.
2. Materials and Methods
2.1. Plant Cultivation and Drought Treatment
In the present study, 73 cultivated barley genotypes and 52 Tibetan wild barley accessions were used (Supplementary Materials Table S1). All genotypes or accessions were sown in mid-November 2018 and grew in two rain shelters (60 m × 20 m) in Changxing experimental station of Zhejiang University (Huzhou, Zhejiang Province, China) for the control and drought stress treatments, respectively. The experiment was arranged in a random block design, with three replicates for each treatment. In each replicate, every barley genotype was sown in 3 rows with a row length of 2 m and row space of 0.33 m, and only the grains from the middle row line were harvested for the further investigations.
After germination, all barley plants in the two rain shelters were well-irrigated with a sprinkling irrigation system to keep the soil water content around 35% (equaling to a water potential of −0.15 MPa, monitored by an HH2 Moisture Meter, Delta-T Devices, Cambridge, UK). For drought treatment, when approximately 85% of the barley genotypes reached the heading stage, drought stress was started and water supply was stopped until the soil water content dropped to 14% (equaling a water potential of −0.75 MPa), and thereafter, this water level was maintained until the maturity stage. The control treatment maintained a normal water supply to keep the soil water content around 35%. During drought treatment, the soil water contents in the two rain shelters were monitored every 3 days by randomly measuring the soil water content at 30 positions over the whole shelter.
To analyze the polymorphisms of the Bmy1 gene, seeds of each barley genotype were surface sterilized with 12.5% NaClO solution, thoroughly rinsed with tap water (for at least 30 min), and then grown using paper roll with 1/5-strength Hoagland solution in the well-controlled growth chamber, with a day length of 14 h, light/dark temperatures of 23/16 °C, and relative humidity of 65% at Zhejiang University, China. The first fully expanded leaf was collected to extract DNA for further investigations.
2.2. Measurement of β-Amylase Activity
Grains of each barley genotype were harvested at maturity, dried at 40 °C, milled to pass through a 0.5-mm sieve, and then stored at −20 °C for further use.
β-amylase activity was measured using a Betamyl assay kit (Megazyme International Ireland Ltd., Bray, Ireland) according to McCleary and Codd [23].
2.3. DNA Extraction
DNA extraction was conducted according to the CTAB (hexadecyltrimethyl ammonium bromide) protocol with the following procedures: approximately 0.5 g leaf tissues was finely ground in liquid nitrogen and transferred to a 2-mL centrifuge tube containing 1 mL CTAB buffer (2% w/v CTAB, 1.42 M NaCl, 20 mM EDTA, 100 mM Tris-HCl, and 0.2% β-mercaptoethanol, preheated to 60 °C). After incubation in the water bath (65 °C) for 30 min, the leaf extract was thoroughly mixed with 750 μL chloroform:isoamylol (24:1) solution by vortex shaking and then centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatant was transferred to a new tube and treated with chloroform:isoamylol solution to purify the DNA extraction. The supernatant was then well mixed with 0.7 v/v isopropanol, placed at room temperature for 10 min, and centrifuged again at 12,000 rpm for 10 min at 4 °C. The supernatant was discarded, and the pellet was washed with 1000 µl 70% ethanol and then centrifuged at 12,000 rpm for 15 min at room temperature. The clean pellet was later dried in the air, suspended with 15–20 µL of 0.1 M TE, and then stored at −20 °C for the following detection.
2.4. Polymorphism Analysis of the Bmy1 Gene, Cloning, and Sequencing of Bmy1 Intron III
According to Hayden et al. [24], 12 simple sequence repeat (SSR) marker primers (Table 1) were used to screen Bmy1 diversity. PCR products were analyzed by scoring the presence of a band as 1 and the absence as 0. All data was imported into SPSS using hierarchical cluster analysis with the default settings to conduct cluster analysis (Figure 1).

Table 1.
Primer sequences of the SSR assays.

Figure 1.
Genetic cluster of 125 barley accessions by SSR markers.
Intron III was cloned by PCR using the primers of forward 5′-GTTATCGTCGACATTGAAGTAGGACT-3′ and reverse 5′-GCTTTGAAGTCTGCTTGTAGGTATTT-3′ [10]. PCR was conducted in 50-ul reactions in a T100 Thermal Cycler (Bio-Rad Laboratories, Inc., Shanghai, China) using Ex TaqTM DNA polymerase (Takara Bio, Tokyo, Japan) with dNTPs, Ex TaqTM, and buffer (with Mg2+) concentrations according to the manufacturer’s protocol. Primers were used at a final concentration of 0.5 μM. In total, 5 uL template was added to each reaction (~100 ng/reaction). Amplification was conducted using the following cycling conditions: 95 °C for 5 min, 95 °C for 45 s, 50 °C for 30 s, 72 °C for 1 min (35 cycles), 72 °C for 10 min, and 12 °C forever. PCR products were sequenced using the Sanger method. All the fragments were aligned in DNAStar using Clustal W software with the sequence of Huruna Nijo (D49999) as the reference. All the genotypes or accessions in this study could be divided into three allele types: Bmy1.a, Bmy1.b, and Bmy1.c (Table 2) [10].

Table 2.
Bmy1 intron III polymorphisms for 125 barley genotypes. Haruna Nijo was used as the reference.
In a previous study, we found that the Tibetan wild barley XZ147 was a drought-tolerant genotype with the largest increase in β-amylase activity and smallest grain weight loss under water stress compared with other genotypes, including Triumph, a drought-sensitive malting barley [25]. In the present study, therefore, we sequenced the full Bmy1 gene of XZ147 and Triumph to analyze the molecular differences between the wild and cultivated barley [26], and identify the mRNA differences in SNP and amino acid substitution based on the cDNA of Huruna Nijo using Clustal W (Table 3).

Table 3.
mRNA and amino acid sequence alignment of the Bmy1 gene from a cultivated barley Triumph and a wild barley XZ147. Huruna Nijo was used as the reference.
2.5. Statistical Analysis
Statistical analysis was performed using SPSS Statistics 20 (IBM, New York, NY, USA). Hierarchical cluster analysis of 125 genotypes based on SSR markers was conducted with the method of average-linkage-between-groups. Two-way variance analysis (ANOVA) was carried out to evaluate the significance among barley genotypes (G), drought treatments (E), and the interaction between genotype and drought treatment (G × E).
3. Results
3.1. Polymorphisms of Bmy1 DNA
SSR markers based on the Bmy1 intron III DNA sequence divided the cultivated barley into numerous small groups (Figure 1), and the wild accessions were scattered over these groups. Unfortunately, there was no outstanding cluster found in the present study. Thus, based on our previous findings, XZ147 and Triumph were selected from the 125 barley genotypes for further analysis [25].
3.2. Bmy1 Gene Intron III Alleles
Four INDELs of Bmy1 intron III alleles, 126-bp, 38-bp, 11-bp, and 21-bp, were identified in this study (Table 2). According to the different combinations of the INDELs, the 125 barley accessions used in this study were classified into three allele types: Bmy1.a, Bmy1.b, and Bmy1.c, with no Bmy1.d allele being detected. Among these three allele types, the Bmy1.c group occupied the largest proportion of the barley accessions (consisting of 36 cultivated and 19 wild barley accessions), followed by Bmy1.b (consisting of 10 cultivated and 26 wild barley accessions) and Bmy1.a (consisting of 27 cultivated and 7 wild barley accessions). Half of the wild accessions were grouped into group Bmy1.b, but most of the cultivated barley were classified into group Bmy1.a and c.
3.3. Polymorphisms of Bmy1 cDNA and Amino Acid Composition
In this study, six SNPs and the corresponding amino acid substitutions were identified based on the alignment of cDNA between wild barley XZ147 and cultivated barley Triumph (Table 3). Wild barley XZ147 showed a great difference from the Triumph and Huruna Nijo mRNA and amino acid composition, especially in D165E (495C → D) and V430A (1289T → C).
3.4. The Effect of Drought on β-Amylase Activity
Under drought stress, the changing trend in the β-amylase activities differed greatly between subspecies (cultivated vs. wild), allele types, and among genotypes (Figure 2, Table 4, Table 5 and Table 6). Impressively, the β-amylase activities in all wild barley accessions were dramatically increased by 6.4–81.2% during drought stress, whereas around 20% of the cultivated genotypes showed a significant decrease (Figure 2). Furthermore, the impact of drought stress on β-amylase activity also showed differences among the three allele types. For the Bmy1.b group, none of the cultivated barley genotypes showed a decrease in β-amylase activity under drought stress compared with the control (Figure 2B, Table 5) while the β-amylase activities decreased in 30% and 20% of the cultivated genotypes of the Bmy1.a and Bmy1.c group, respectively (Figure 2A,C, Table 4 and Table 6). XZ147 and Triumph, which belong to the Bmy1.b group, showed increases in β-amylase activity of 76.8% and 20.3% under drought stress in comparison with the control, respectively (Figure 3).
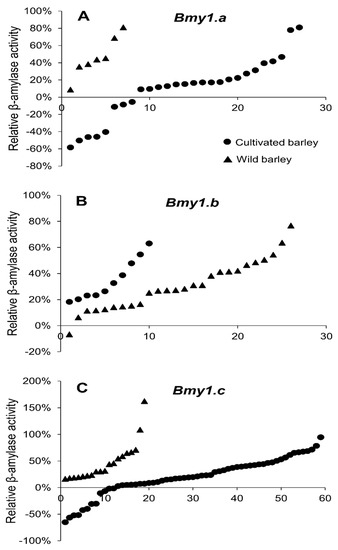
Figure 2.
The effect of drought stress on the grain β-amylase activity of 125 barley genotypes compared with control plants. The genotypes were arranged from low to high relative β-amylase activity along the X axis. (A) Bmy1.a alleles; (B) Bmy1.b alleles; (C) Bmy1.c alleles.

Table 4.
The distribution of the grain β-amylase activity of Bmy1.a alleles.

Table 5.
The distribution of the grain β-amylase activity of Bmy1.b alleles.

Table 6.
The distribution of the grain β-amylase activity of Bmy1.c alleles.
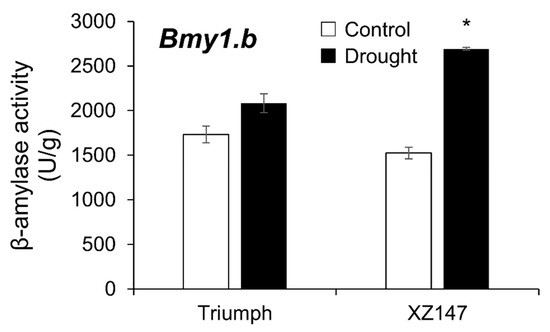
Figure 3.
The effect of drought stress on β-amylase activity in barley grains. Data show mean + SD. *, significant at 95% probability.
4. Discussion
Marker-assisted selection (MAS) has already been widely used in breeding, phyletic evolution, comparative genetics, and gene mapping [27,28,29]. Sufficient evidence has demonstrated the usefulness of MAS in accelerating breeding efficiency [30,31]. In this study, an SSR assay of Bmy1 DNA sequences showed abundant polymorphisms of the gene (Figure 1). The genetic cluster and the distribution of β-amylase activity (Table 2, 4–6) showed that the Tibetan wild barley is randomly scattered over various Bmy1 groups of cultivated barley rather than a specific cluster group. Moreover, more abundant polymorphisms of the wild barley (H. vulgare subsp. spontaneum) could be observed in comparison with the cultivated barley (Hordeum vulgare L.).
According to the presence and absence of the 126-, 38-, 11-, and 21-bp INDELs, the 125 barley accessions could be divided into 3 allele types: Bmy1.a, Bmy1.b, and Bmy1.c (Table 2). The allele type with the presence of the 126-bp, 38-bp, and 21-bp INDELs, and the absence of the 11-bp INDEL was defined as Bmy1.a; the type with the presence of the 38-bp and the absence of the 126-bp, 11-bp, and 21-bp INDELs was defined as Bmy1.b; and the type with the absence of the 126-bp and the presence of the 38-bp, 11-bp, and 21-bp INDELs was defined as Bmy1.c [10 ]. Bmy1.d, which was once identified in wild barley PI 2,976,897 [9], shows the presence of the 11- and 21-bp, and the absence of the 126- and 38-bp INDELs. In this study, no Bmy1.d was detected. The impact of drought on β-amylase activity in grains varied greatly among genotypes. All genotypes belonging to Bmy1.b and the wild barley accessions belonging to Bmy1.a and Bmy1.c showed an increase in β-amylase activity while some of the cultivated barley genotypes belonging to Bmy1.a and Bmy1.c showed a decrease in β-amylase activity. Bmy1 intron III has been reported to be a useful marker in barley breeding for the selection of high malt quality [12] and the 126-bp INDEL is closely correlated with β-amylase activity and thermo-stability [9,12,16,17,32]. The 126-bp fragment in the Bmy1.a allele type may be a site of a negatively regulated transcription factor, and could be linked with the low β-amylase activity [13]. The current result also confirmed that the Bmy1 intron III allele type could be a good indicator of β-amylase activity under drought stress. In this study, all genotypes of the Bmy1.b allele type showed an increase in grain β-amylase activity while those of the Bmy1.a and Bmy1.c allele types showed less changes, indicating that the genotypes belonging to the Bmy1.b allele type could be more useful in developing barley cultivars with drought tolerance and high malt quality.
It has been documented that some amino acid substitutions derived from specific SNPs of mRNA in the Bmy1 gene were highly correlated with the β-amylase activity in barley grains [14,33,34,35]. Zhang et al. [35] and Ma et al. [14] reported that 115R→C amino acid substitution was the main reason for the high β-amylase activity of W127 and Ashqeleon. Chiapparino et al. [33] found that the genotypes containing the amino acid composition of C115, E165, and V233 had higher β-amylase activity than the ones with the composition of R115, D165, and A233. Based on the alignment of the Bmy1 gene DNA, Filichkin et al. [34] identified two genotypes only differing in V233A, with the A233 genotype having higher β-amylase activity than the V233 genotype. R115 or C115 alone had no effect on β-amylase activity, but their co-existence increased the β-amylase activity and thermo-stability significantly [14,36]. In this study, β-amylase activity was higher in cultivated barley Triumph than wild barley XZ147 under normal conditions, which may be attributed to the amino acid composition of C115, D165, and V233. Surprisingly, β-amylase activity was higher in XZ147, which showed less loss in grain yield and grain weight than Triumph under drought stress [25], which might have resulted from its unique M527 or the composition of R115, D165, A233, S347, and M527. In addition, a drought-induced abundance increase of β-amylase might also cause such an increase in its activity. Therefore, more investigations need to be carried out to analyze the association of the amino acid M527 or the composition of the R115, A233, S347, and M527 allele and the increase in β-amylase activity under drought stress, and the A233 potentiality in breeding barley cultivars with both drought tolerance and high β-amylase activity in grains.
5. Conclusions
Novel genetic variation is essential for successful breeding. In this study, based on the comparison of the genetic variations of the Bmy1 gene from cultivated and wild barley, we evaluated the potential value of wild barley in breading cultivars with drought tolerance and high β-amylase activity. In this study, the β-amylase activity of the Bmy1.b allele-type accessions increased under drought treatment, indicating its higher implication value in malt barley breeding than the other two allele types. Meanwhile, wild barley belonging to the Bmy1.b allele type displayed higher β-amylase activity and minimum yield loses under drought stress [25]. This might result from its unique amino acid substitution M527 or the amino acid composition of R115, D165, A233, S347, and M527. We conclude that the gene pool of Tibetan wild barley germplasms provides a unique resource for improving the drought tolerance and grain β-amylase activity of malt barley.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy12081737/s1, Table S1: The accession number, Chinese name and geographic origin of barley used in this study.
Author Contributions
Conceptualization, X.W.; validation, X.W., H.W., W.Y. and K.C.; formal analysis, X.W., W.Y. and K.C.; investigation, X.W., H.W., W.Y. and K.C.; data curation, X.W., J.W. and F.Z.; writing—original draft preparation, X.W.; writing—review and editing, X.W., J.W. and F.Z.; supervision, J.W.; project administration, J.W.; funding acquisition, X.W and J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Zhejiang Provincial Natural Science Foundation [LY19C130004], National Natural Science Foundation of China [31671678], Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding [2021C02064-3] and China Agriculture Research System of MOF and MARA [CARS-05-01A-06].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict to interest.
References
- Delcour, J.A.; Versciiaeve, S. Malt Diastatic Activity. Part ii. A Modified Ebc Diastatic Power Assay for the Selective Estimation of Jie7vi-Amylase Activity. Time and Temperature Dependence Ofthe Release of Reducing Sugars. J. Inst. Brew. 1987, 33, 296–301. [Google Scholar] [CrossRef]
- Arends, A.M.; Fox, G.P.; Henry, R.J.; Marschke, R.J.; Symons, M.H. Genetic and Environmental Variation in the Diastatic Power of Australian Barley. J. Cereal Sci. 1995, 21, 63–70. [Google Scholar] [CrossRef]
- Solah, T.S. Diastatic power in malted barley-contributions of malt parameters to its development and the potential of barley grain β-amylase to predict malt diastatic power. J. Inst. Brew. 1995, 101, 277–280. [Google Scholar]
- Powling, A.; Islam, A.K.; Shepherd, K.W. Isozymes in wheat-barley hybrid derivative lines. Biochem. Genet. 1981, 19, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, G.; Johansen, H.; Jensen, J.; Hejgaard, J. Localization on barley chromosome 4 of genes coding for β-amylase (Bmy1) and protein Z (Paz1). Barley Genet. Newsl. 1983, 13, 55–57. [Google Scholar]
- Kreis, M.; Williamson, M.S.; Shewry, P.R.; Sharp, P.; Gale, M. Identification of a second locus encoding β-amylase on chromosome 2 of barley. Genet. Res. 1988, 51, 13–16. [Google Scholar] [CrossRef]
- Yoshigi, N.; Okada, Y.; Sahara, H.K.S. PCR cloning and sequencing of the β-amylase cDNA from barley. J. Biochem. 1994, 115, 47–51. [Google Scholar] [CrossRef]
- Hayes, P.M.; Liu, B.H.; Knapp, S.J.; Chen, F.; Jones, B.; Blake, T.; Franckowiak, J.; Rasmusson, D.; Sorrells, M.; Ullrich, S.E.; et al. Quantitative trait locus effects and environmental interaction in a sample of North American barley germ plasm. TAG. Theor. Appl. Genetics. Theor. Angew. Genet. 1993, 87, 392–401. [Google Scholar] [CrossRef]
- Erkkila, M.J.; Leah, R.; Ahokas, H.; Cameron-Mills, V. Allele-dependent barley grain beta-amylase activity. Plant Physiol. 1998, 117, 679–685. [Google Scholar] [CrossRef][Green Version]
- Vinje, M.A.; Duke, S.H.; Henson, C.A. Utilization of Different Bmy1 Intron III Alleles for Predicting β-Amylase Activity and Thermostability in Wild and Cultivated Barley. Plant Mol. Biol. Rep. 2010, 28, 491–501. [Google Scholar] [CrossRef]
- Sjakste, T.G.; Zhuk, A.F. Novel haplotype description and structural background of the eventual functional significance of the barley beta-amylase gene intron III rearrangements. TAG. Theor. Appl. Genetics. Theor. Und Angew. Genet. 2006, 113, 1063–1079. [Google Scholar] [CrossRef]
- Kaneko, T.; Kihara, M.; Ito, K. Genetic analysis of β-amylase thermostability to develop a DNA marker for malt fermentability improvement in barley, Hordeum vulgare. Plant Breed. 2000, 119, 197–201. [Google Scholar] [CrossRef]
- Erkkila, M.J.; Ahokas, H. Special barley P-amylase allele in a Finnish landrace line HA52 with high grain enzyme activity. Hereditas 2001, 134, 91–95. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, Y.F.; Evans, D.E.; Logue, S.J.; Langridge, P. Mutations of barley beta-amylase that improve substrate-binding affinity and thermostability. Mol. Genet. Genom. MGG 2001, 266, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Pairs, M.; Jones, M.G.; Eglinton, J.K. Genotyping Single Nucleotide Polymorphisms for Selection of Barley-amylase Alleles. Plant Mol. Biol. Rep. 2002, 20, 149–159. [Google Scholar] [CrossRef]
- Gunkel, J.; Voetz, M.; Rath, F. Effect of the Malting Barley Variety Hordeum vulgare L. on Fermentability. J. Inst. Brew. 2002, 108, 355–361. [Google Scholar] [CrossRef]
- Coventry, S.J.; Collins, H.M.; Barr, A.R.; Jefferies, S.P.; Chalmers, K.J.; Logue, S.J.; Langridge, P. Use of putative QTLs and structural genes in marker assisted selection for diastatic power in malting barley (Hordeum vulgare L.). Aust. J. Agric. Res. 2003, 54, 1241–1250. [Google Scholar] [CrossRef]
- Li, C.D.; Langridge, P.; Zhang, X.Q.; Eckstein, P.E.; Rossnagel, B.G.; Lance, R.C.M.; Lefol, E.B.; Lu, M.Y.; Harvey, B.L.; Scoles, G.J. Mapping of Barley (Hordeum vulgare L.) Beta-amylase Alleles in which an Amino Acid Substitution Determines Beta -amylase Isoenzyme Type and the Level of Free Beta-amylase. J. Cereal Sci. 2002, 35, 39–50. [Google Scholar] [CrossRef]
- Dai, F.; Nevo, E.; Wu, D.; Comadran, J.; Zhou, M.; Qiu, L.; Chen, Z.; Beiles, A.; Chen, G.; Zhang, G. Tibet is one of the centers of domestication of cultivated barley. Proc. Natl. Acad. Sci. USA 2012, 109, 16969–16973. [Google Scholar] [CrossRef]
- Qiu, L.; Wu, D.; Ali, S.; Cai, S.; Dai, F.; Jin, X.; Wu, F.; Zhang, G. Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. TAG. Theor. Appl. Genetics. Theor. Und Angew. Genet. 2011, 122, 695–703. [Google Scholar] [CrossRef]
- Dai, H.; Shan, W.; Zhao, J.; Zhang, G.; Li, C.; Wu, F. Difference in response to aluminum stress among Tibetan wild barley genotypes. J. Plant Nutr. Soil Sci. 2011, 174, 952–960. [Google Scholar] [CrossRef]
- Wei, K.; Jin, X.; Chen, X.; Wu, F.; Zhou, W.; Qiu, B.; Qiu, L.; Wang, X.; Li, C.; Zhang, G. The effect of H2O2 and abscisic acid (ABA) interaction on beta-amylase activity under osmotic stress during grain development in barley. Plant Physiol. Biochem. 2009, 47, 778–784. [Google Scholar] [CrossRef] [PubMed]
- McCleary, B.V.; Codd, R. Measurement of β-amylase in cereal flours and commercial enzyme preparations. J. Cereal Sci. 1989, 9, 17–33. [Google Scholar] [CrossRef]
- Hayden, M.J.; Nguyen, T.M.; Waterman, A.; McMichael, G.L.; Chalmers, K.J. Application of multiplex-ready PCR for fluorescence-based SSR genotyping in barley and wheat. Mol. Breed. 2008, 21, 271–281. [Google Scholar] [CrossRef]
- Wu, X.J.; Chen, X.; Zeng, F.R.; Zhang, G.P. The genotypic difference in the effect of water stress after anthesis on the malt quality parameters in barley. J. Cereal Sci. 2015, 65, 209–214. [Google Scholar] [CrossRef]
- Gong, X.; Westcott, S.; Zhang, X.Q.; Yan, G.; Lance, R.; Zhang, G.; Sun, D.; Li, C. Discovery of novel Bmy1 alleles increasing beta-amylase activity in Chinese landraces and Tibetan wild barley for improvement of malting quality via MAS. PLoS ONE 2013, 8, e7287. [Google Scholar] [CrossRef]
- Prasad, M.; Varshney, R.K.; Roy, J.K.; Balyan, H.S.; Gupta, P.K. The use of microsatellites for detecting DNA polymorphism, genotype identification and genetic diversity in wheat. TAG. Theor. Appl. Genet. Theor. Und Angew. Genet. 2000, 100, 584–592. [Google Scholar]
- Stein, N.; Graner, A. Map-Based Gene Isolation in Cereal Genomes. In Cereal Genomics; Gupta, P.K., Varshney, R.K., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 331–360. [Google Scholar]
- Varshney, R.K.; Sigmund, R.; Börner, A.; Korzun, V.; Stein, N.; Sorrells, M.E.; Langridge, P.; Graner, A. Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci. 2005, 168, 195–202. [Google Scholar] [CrossRef]
- Koebner, R.M.; Summers, R.W. 21st century wheat breeding: Plot selection or plate detection? Trends Biotechnol. 2003, 21, 59–63. [Google Scholar] [CrossRef]
- Crepieux, S.; Lebreton, C.; Flament, P.; Charmet, G. Application of a new IBD-based QTL mapping method to common wheat breeding population: Analysis of kernel hardness and dough strength. TAG. Theor. Appl. Genet. Theor. Und Angew. Genet. 2005, 111, 1409–1419. [Google Scholar] [CrossRef]
- Erkkilä, M.J. Intron III-Specific Markers for Screening of β-amylase Alleles in Barley Cultivars. Plant Mol. Biol. Rep. 1999, 17, 139–147. [Google Scholar] [CrossRef]
- Chiapparino, E.; Donini, P.; Reeves, J.; Tuberosa, R.; O’Sullivan, D.M. Distribution of β-amylase I haplotypes among European cultivated barleys. Mol. Breed. 2006, 18, 341–354. [Google Scholar] [CrossRef]
- Filichkin, T.P.; Vinje, M.A.; Budde, A.D.; Corey, A.E.; Duke, S.H.; Gallagher, L.; Helgesson, J.; Henson, C.A.; Obert, D.E.; Ohm, J.B.; et al. Phenotypic Variation for Diastatic Power, β-Amylase Activity, and β-Amylase Thermostability vs. Allelic Variation at the Bmy1 Locus in a Sample of North American Barley Germplasm. Crop Sci. 2010, 50, 826–834. [Google Scholar] [CrossRef]
- Zhang, W.S.; Li, X.; Liu, J.B. Genetic variation of Bmy1 alleles in barley (Hordeum vulgare L.) investigated by CAPS analysis. TAG. Theor. Appl. Genet. Theor. Und Angew. Genet. 2007, 114, 1039–1050. [Google Scholar] [CrossRef]
- Clark, S.E.; Hayes, P.M.; Henson, C.A. Effects of single nucleotide polymorphisms in β-amylase1 alleles from barley on functional properties of the enzymes. Plant Physiol. Biochem. 2003, 41, 798–804. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).