Functional Analysis and Precise Location of m-1a in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Genetic Transformation of Exogenous Genes
2.3. DNA Southern Blot Analysis
2.4. Insertion Site Analysis
2.5. Grain Trait Measurements
2.6. Analysis of Starch Properties
2.7. Observation of Endosperm Starch Granules
2.8. Gene Expression Analyses
2.9. Statistical Approach
3. Results
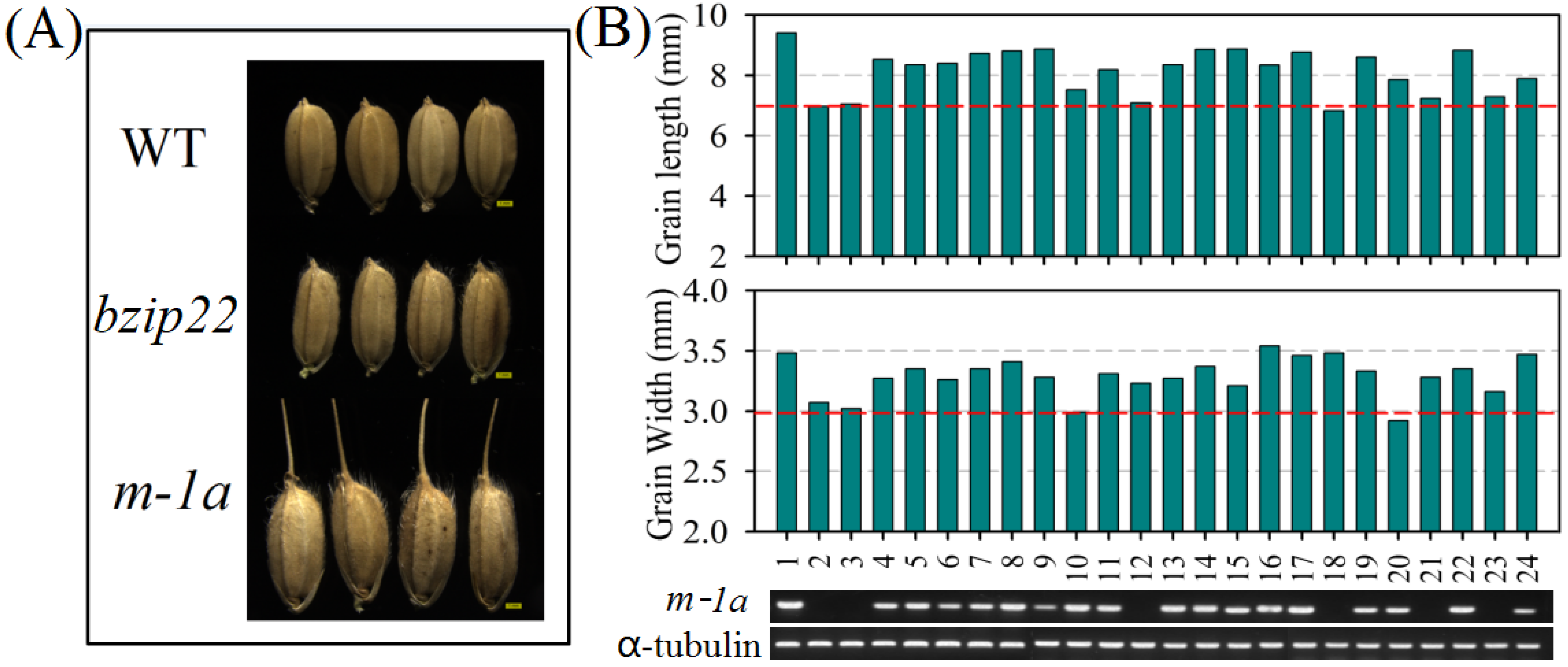
3.1. Analysing of m-1a Grain Characteristics
3.2. Analysis of m-1a Insertion Site
3.3. Insertion Position and Phenotypic Relationship Analysis
3.4. Expression Levels of Related Genes in Rice Starch Synthesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agrawal, G.K.; Yamazaki, M.; Kobayashi, M.; Hirochika, R.; Miyao, A.; Hirochika, H. Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol. 2001, 125, 1248–1257. [Google Scholar] [CrossRef] [Green Version]
- Marcon, C.; Altrogge, L.; Win, Y.N.; Stöcker, T.; Gardiner, J.M.; Portwood, J.L.; Opitz, N.; Kortz, A.; Baldauf, J.A.; Hunter, C.T.; et al. BonnMu: A Sequence-indexed resource of transposon-induced maize mutations for functional genomics studies. Plant Physiol. 2020, 184, 620–631. [Google Scholar] [CrossRef]
- Kim, E.Y.; Fan, W.; Cho, J. Determination of TE insertion positions using transposon display. Methods Mol. Biol. 2021, 2250, 115–121. [Google Scholar]
- Settles, A.M. EMS mutagenesis of maize pollen. Methods Mol. Biol. 2020, 2122, 25–33. [Google Scholar]
- Krysan, P.J.; Young, J.C.; Sussman, M.R. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 1999, 11, 2283–2290. [Google Scholar] [CrossRef] [Green Version]
- Pucker, B.; Kleinboelting, N.; Weisshaar, B. Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis. BMC Genom. 2021, 22, 599. [Google Scholar] [CrossRef]
- Nakano, A.; Suzuki, G.; Yamamoto, M.; Turnbull, K.; Rahman, S.; Mukai, Y. Rearrangements of large-insert T-DNAs in transgenic rice. Mol. Genet. Genom. 2005, 273, 123–129. [Google Scholar] [CrossRef]
- Gong, W.K.; Zhou, Y.; Wang, R.; Wei, X.L.; Zhang, L.; Dai, Y.; Zhu, Z. Analysis of T-DNA integration events in transgenic rice. J. Plant Physiol. 2021, 266, 153527. [Google Scholar] [CrossRef]
- Liu, Y.G.; Chen, Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 2007, 43, 649–650. [Google Scholar] [CrossRef]
- Cheng, F.M.; Zhong, L.J.; Wang, F.; Zhang, G.P. Differences in cooking and eating properties between chalky and translucent parts in rice grains. Food Chem. 2005, 90, 39–46. [Google Scholar] [CrossRef]
- Xi, M.; Wu, W.; Xu, Y.; Zhou, Y.J.; Chen, G.; Ji, Y.L.; Sun, X.Y. Grain chalkiness traits is affected by the synthesis and dynamic accumulation of the storage protein in rice. J. Sci. Food Agric. 2021, 101, 6125–6133. [Google Scholar] [CrossRef]
- Deng, F.; Li, Q.; Chen, H.; Zeng, Y.L.; Li, B.; Zhong, X.Y.; Wang, L. Relationship between chalkiness and the structural and thermal properties of rice starch after shading during grain-filling stage. Carbohydr. Polym. 2021, 252, 117212. [Google Scholar] [CrossRef]
- Yamakawa, H.; Hirose, T.; Kuroda, M.; Yamaguchi, T. Comprehensive expression profiling of rice grain filling related genes under high temperature using DNA microarray. Plant Physiol. 2007, 144, 258–277. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Liu, X.L.; Wan, X.Y.; Weng, J.F.; Liu, S.J.; Liu, X.; Chen, M.J.; Li, J.J.; Su, N.; Wu, F.Q.; et al. Identification of a stable quantitative trait locus for percentage grains with white chalkiness in rice (Oryza sativa). J. Integr. Plant Biol. 2011, 53, 598–607. [Google Scholar] [CrossRef]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Fujita, N.; Yoshida, M.; Kondo, T.; Saito, K.; Utsumi, Y.; Tokunaga, T.; Nishi, A.; Satoh, H.; Park, J.H.; Jane, J.L.; et al. Characterization of SSllla-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007, 144, 2009–2023. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.G.; Park, S.; Matsuoka, M.; An, G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 2005, 42, 901–911. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.P.; Xu, P.K.; Zhang, J.C.; Zhang, S.; Li, Z.W.; Yang, K.; Chang, X.Y.; Li, Y.B. OsbZIP60-mediated unfolded protein response regulates grain chalkiness in rice. J. Genet. Genom. 2022, 49, 414–426. [Google Scholar] [CrossRef]
- Southern, E. Southern blotting. Nat. Protoc. 2006, 1, 518–525. [Google Scholar] [CrossRef]
- Mao, H.L.; Sun, S.Y.; Yao, J.L.; Wang, C.G.; Yu, S.B.; Xu, C.G.; Li, X.H.; Zhang, Q.F. Linking differential domain functions of the gs3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.C.; Xu, H.; Zhu, Y.; Liu, Q.Q.; Cai, X.L. Osbzip58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 2013, 64, 3453–3466. [Google Scholar] [CrossRef]
- Fu, F.F.; Xue, H.W. Coexpression analysis identifies rice starch regulator1, a rice ap2/erebp family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef]
- Huang, X.Z.; Qian, Q.; Liu, Z.B.; Sun, H.Y.; He, S.Y.; Luo, D.; Xia, G.M.; Chu, C.C.; Li, J.Y.; Fu, X.D. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Zhang, X.J.; Wang, J.F.; Huang, J.; Lan, H.X.; Wang, C.L.; Yin, C.F.; Wu, Y.Y.; Tang, H.J.; Qian, Q.; Li, J.Y.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef] [Green Version]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Y.X.; Fang, Y.X.; Zeng, L.J.; Xu, J.; Yu, H.P.; Shi, Z.Y.; Pan, J.J.; Zhang, D.; Kang, S.J.; et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 2015, 8, 1455–1465. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.F.; Chen, J.; Zheng, X.M.; Wu, F.Q.; Lin, Q.B.; Heng, Y.Q.; Tian, P.; Cheng, Z.J.; Yu, X.W.; Zhou, K.N.; et al. GW5 acts in the brassinosteroid signaling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef]
- Yang, W.S.; Wu, K.; Wang, B.; Liu, S.Y.; Liu, H.H.; Guo, S.Y.; Guo, X.Y.; Luo, W.; Sun, S.Y.; Ouyang, Y.D.; et al. The RING E3 ligase CLG1 targets GS3 for degradation via the endosome pathway to determine grain size in rice. Mol. Plant 2021, 14, 1699–1713. [Google Scholar] [CrossRef]
- Wang, Y.H.; Ma, Y.; Lü, B.J.; Xu, E.P.; Huang, Q.; Lai, M. Differential expression of mimecan and thioredoxin domain–containing protein 5 in colorectal adenoma and cancer: A proteomic study. Exp. Biol. Med. 2007, 232, 1152–1159. [Google Scholar] [CrossRef]
- Ramírez-Torres, A.; Barceló-Batllori, S.; Martínez-Beamonte, R.; Navarro, M.A.; Surra, J.C.; Arnal, C.; Guillén, N.; Acín, S.; Osada, J. Proteomics and gene expression analyses of squalene-supplemented mice identify microsomal thioredoxindomain-containing protein 5 changes associated with hepatic steatosis. J. Proteom. 2012, 77, 27–39. [Google Scholar] [CrossRef]
- Mchale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biology 2006, 7, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.H.; Gu, T.T.; Pan, C.Y.; Feng, Z.M.; Ding, J.; Hang, Y.Y.; Chen, J.Q.; Tian, D.C. Genetic variation of NBS-LRR class resistance genes in rice lines. Theor. Appl. Genet. 2008, 116, 165–177. [Google Scholar] [CrossRef]
- Smidansky, E.D.; Clancy, M.; Meyer, F.D.; Lanning, S.P.; Blake, N.K.; Talbert, L.E.; Giroux, M.J. Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc. Natl. Acad. Sci. USA 2002, 99, 1724–1729. [Google Scholar] [CrossRef] [Green Version]
- Kawagoe, Y.; Kubo, A.; Satoh, H.; Takaiwa, F.; Nakamura, Y. Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J. 2010, 42, 164–174. [Google Scholar] [CrossRef]
- Saripalli, G.; Gupta, P.K. AGPase: Its role in crop productivity with emphasis on heat tolerance in cereals. Theor. Appl. Genet. 2015, 128, 1893–1916. [Google Scholar] [CrossRef]
- Lee, S.K.; Hwang, S.K.; Han, M.; Eom, J.S.; Kang, H.G.; Han, Y.; Choi, S.B.; Cho, M.H.; Bhoo, S.H.; An, G.; et al. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 531–546. [Google Scholar] [CrossRef]
- Li, Y.B.; Fan, C.C.; Xing, Y.Z.; Yun, P.; Luo, L.J.; Yan, B.; Peng, B.; Xie, W.B.; Wang, G.W.; Li, X.H.; et al. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 2014, 46, 398–404. [Google Scholar] [CrossRef]
- She, K.C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. Novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 2010, 22, 3280–3294. [Google Scholar] [CrossRef] [Green Version]
- Kuipers, G.J.; Vreem, J.T.M.; Meyer, H.; Jacobsen, E.; Feenstra, W.J.; Visser, R.G.F. Field evaluation of antisense RNA mediated inhibition of GBSS gene expression in potato. Euphytica 1991, 59, 83–91. [Google Scholar] [CrossRef]
- Edwards, A.; Vincken, J.P.; Suurs, L.C.J.M.; Visser, R.G.F.; Zeeman, S.; Smith, A.; Martin, C. Discrete forms of amylose are synthesized by isoforms of GBSSI in pea. Plant Cell 2002, 14, 1767–1785. [Google Scholar] [CrossRef] [Green Version]
- Burton, R.A.; Jenner, H.; Carrangis, L.; Fahy, B.; Fincher, G.B.; Hylton, C.; Laurie, D.A.; Parker, M.; Waite, D.; Wegen, S.V.; et al. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J. 2002, 31, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Kubo, A.; Myers, A.M. Functions of heteromeric and homomeric isoamylase-type starch-debranching enzymes in developing maize endosperm. Plant Physiol. 2010, 153, 956–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wattebled, F.; Dong, Y.; Dume, S.; Delvallé, D.; Planchot, V.; Berbezy, P.; Vyas, D.; Colonna, P.; Chatterjee, M.; Ball, S.; et al. Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 2005, 138, 184–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Q.; Shen, J.; Wang, F.; Qi, Y.; Jiang, C.; Zu, C.; Li, T. Functional Analysis and Precise Location of m-1a in Rice. Agronomy 2022, 12, 1706. https://doi.org/10.3390/agronomy12071706
Dong Q, Shen J, Wang F, Qi Y, Jiang C, Zu C, Li T. Functional Analysis and Precise Location of m-1a in Rice. Agronomy. 2022; 12(7):1706. https://doi.org/10.3390/agronomy12071706
Chicago/Turabian StyleDong, Qing, Jia Shen, Fang Wang, Yaocheng Qi, Chaoqiang Jiang, Chaolong Zu, and Tingchun Li. 2022. "Functional Analysis and Precise Location of m-1a in Rice" Agronomy 12, no. 7: 1706. https://doi.org/10.3390/agronomy12071706
APA StyleDong, Q., Shen, J., Wang, F., Qi, Y., Jiang, C., Zu, C., & Li, T. (2022). Functional Analysis and Precise Location of m-1a in Rice. Agronomy, 12(7), 1706. https://doi.org/10.3390/agronomy12071706




